Impact Factor
ISSN: 1449-1907
Int J Med Sci 2022; 19(5):893-900. doi:10.7150/ijms.73489 This issue Cite
Research Paper
Proteomic Profiling of Aqueous Humor Exosomes from Age-related Macular Degeneration Patients
1. Department of Ophthalmology, Taipei City Hospital Zhongxing Branch, Taipei, Taiwan
2. Institute of Public Health, National Yang Ming Chiao Tung University, Taipei, Taiwan
3. Department of Business Administration, Fu Jen Catholic University, New Taipei City, Taiwan
4. Institute of Traditional Medicine, National Yang Ming Chiao Tung University, Taipei, Taiwan
5. Cancer Progression Research Center, National Yang Ming Chiao Tung University, Taipei, Taiwan
6. Metabolomics-Proteomics Research Center, National Yang Ming Chiao Tung University, Taipei, Taiwan
7. Department of Chinese Medicine, Taipei City Hospital, Linsen, Chinese Medicine, and Kunming Branch, Taipei, Taiwan
Received 2022-3-31; Accepted 2022-5-1; Published 2022-5-13
Abstract
Purpose: The alteration of the exosomal proteins in the aqueous humor (AH) is linked to the development of eye diseases. The goal of this study was to examine the exosomal protein profile of patients with age-related macular degeneration (AMD) to better understand their role in the pathogenesis of AMD.
Methods: Exosomes were isolated from the AH of 28 AMD and 25 control eyes. The quality, concentration, and size distribution of exosomes were measured using a nanoparticle tracking analysis system (NTA). Total exosomal proteins from each sample were purified and digested with trypsin for liquid chromatography-tandem mass spectrometry (LC-MS/MS) analysis.
Results: Based on LC-MS/MS analysis, we got 105 exosomal peptides from AMD and control patients. Gene ontology (GO) analysis in the biology process revealed that exosomal proteins of AMD were enriched in the lipoprotein metabolic process. T-test analysis revealed six exosomal proteins in patients with AMD were significantly different from controls. Comparing the exosomal protein profile of AMD patients who were receiving anti-VEGF therapy, we observed the amount of two proteins decreased with the duration of the anti-VEGF treatment time.
Conclusions: In this study, we successfully isolated and purified AH exosomes. Our results provide pioneering findings for the exosomal protein profile in AMD development and under therapy. These unique proteins could be the new targets for drug discovery or biological markers for evaluating therapeutic efficacy.
Keywords: Aqueous Humor, Age-related macular degeneration, Exosome
Introduction
The macula is a pigmented area of the retina that is responsible for sharp, clear, straight-ahead vision. As people get older, structure and blood flow changes in the macula cause age-related macular degeneration (AMD). AMD is one of the leading causes of irreversible vision loss worldwide [1]. The most significant risk factor of AMD is aging. Almost all patients with advanced AMD are over the age of 60 [2]. Genetic factors are also important, and polymorphisms in over 30 genes have been reported to be associated with increased risk of AMD [3]. Others risk factors such as smoking, uncontrolled hypertension, body mass index above 25 have been reported in previous studies [4-7].
AMD begins with the formation of drusen, which are lipoprotein-rich deposits in the macula. The size of the drusen is linked to the severity of AMD [8]. Patients with early AMD are usually asymptomatic or have decreased vision, distortion or blind spots in or around their central vision. Patients in advanced stages will have difficulty recognizing faces, driving, reading, or performing other daily activities dues to loss of central vision [9]. In general, advanced AMD is classified as wet and dry base on the presence or absence of abnormal blood vessels growing beneath the retina, resulting in blood or serum leakage. The disease process of dry AMD is slower than wet AMD, and visual acuity is better preserved [10]. For the pathogenesis of AMD is still largely unknown, the current treatments for AMD are aimed at delaying or controlling the disease progression. For example, wet AMD can be treated with intraocular injections of anti-VEGF drugs to slow down neovascularization and retinal thickening [11]. For the limited options for prevention and treatment of AMD, to explore new therapeutic targets and biomarkers for AMD is important.
Exosomes are made up of a lipid bilayer that contains transmembrane proteins, cytosolic proteins, and RNAs [12]. They are secreted from cells and are released into body fluids (such as blood, urine, tears, and spinal fluid), which can then be transported between cells via the circulatory system. As a result, they are regarded as critical messengers for cell-to-cell communication [13]. Aqueous humor (AH) is the clear liquid filling in the anterior and posterior chamber of the eye [14]. Lines of evidence suggest abundant exosomes are common in AH [15, 16]. Recent research has linked exosomal proteins or miRNAs to variety of ophthalmic diseases [17-20]. To examine the changes of exosomal proteins that occur with the development of AMD, we aimed to compare the individual protein profiles from patients with AMD, using liquid chromatography-tandem mass spectrometry (LC-MS/MS).
Materials and Methods
AH Samples Collection
The Research Ethics Committee of Taipei City Hospital (TCHIRB-10712118) approved the study protocol, which was carried out by the tenets of the Helsinki Declaration. Before enrolling in the study, all participants provided written informed consent. Human AH samples were collected from patients undergoing cataract surgery at the Zhongxing branch of Taipei City Hospital. The patients with AMD were confirmed by the use of fluorescein angiography. The control eyes came from senile cataract patients who did not have other ocular or systemic diseases. Patients who received three consecutive intravitreal injections of ranibizumab (Lucentis, 10 mg/ml, Novartis, Basel, Switzerland) were asked to provide their AH during anti-VEGF treatment. Approximately 100 μL of AH was collected from each patient via anterior chamber paracentesis with a needle inserted through the peripheral cornea at the start of the procedure. Undiluted AH samples were collected in sterile tubes and stored at - 80°C until further investigation.
Isolation of Exosomes from the AH
Exosomes were isolated from the AH using the ExoQuick precipitation solution (EXOQ5A-1, System Biosciences, Inc., Mountain View, CA) by the manufacturer's instruction. Approximately 100 μl of AH was centrifuged at 3000 × g for 15 minutes to remove cellular debris and collected the supernatants. PBS buffer (10010023, Thermo Fisher Scientific; Waltham, MA) was added to the supernatants to make a final volume of 250 μl. After that, 63 μl of precipitation solution was added to the supernatants and incubated overnight. The exosome pellets were separated by centrifugation at 12,000 × g for 90 minutes and suspended in 100 μl PBS. The concentration and size distribution of the vesicles was assessed by NanoSight LM10 (LM10, Malvern Instruments, Rancho Cucamonga, CA).
Proteins Purification and LC-MS/MS Analysis
To extract the exosomal proteins, exosomes were lysed by RIPA buffer (89900, Thermo Fisher Scientific; Waltham, MA). The proteins were then digested with trypsin (60109-101, SMART Digest Trypsin Kit, Thermo Fisher Scientific; Waltham, MA), desalted (Z720070, Millipore Ziptips Micro-C18; Sigma-Aldrich, Milwaukee, WI), purified (60309-001, SOLA™ SPE Plates; Thermo Fisher Scientific; Waltham, MA), and dissolved in 0.1% formic acid for LC-MS/MS analysis (LTQ Orbitrap Velos, Thermo Scientifics; Waltham, MA) (service provided by the Mass Core Facility of Genomics Research Center, Academia Sinica). The acquired proteomics raw data files were then searched against a UniProt human protein database (http://www.uniprot.org/) by using PEAKS Studio 7.5 (PEAKS Studio, Bioinformatics Solutions, Waterloo, Ontario, Canada). The following settings were used in PEAKS Studio 7.5 in conjunction with UniProt to search the protein database: enzyme set to trypsin with a maximum of two missed cleavage site precursor and fragment mass tolerance of 20 ppm and 0.8 Da, respectively. Finally, the spectral counts obtained from each peptide were normalized to the total spectral counts recorded for all peptides in a sample.
Statistical Analyses
Data are expressed as the mean ± standard error of the mean. A paired sample t-test was used to analyze the axial length and exosome concentration measurements. SPSS version 24 was used to analyze all of the data (SPSS, Inc; Chicago, IL). A P value of less than 0.05 was considered to display a statistically significant difference. The principal component analysis (PCA) plot of all samples was generated using Partek Genomics Suite 7.18 (Partek Genomics Suite, Partek Inc., St. Louis, MO). Gene ontology enrichment analysis was conducted by ShinyGO v0.75 (http://bioinformatics.sdstate.edu/go/)[21].
Results
Samples Collection and Exosomes Isolation
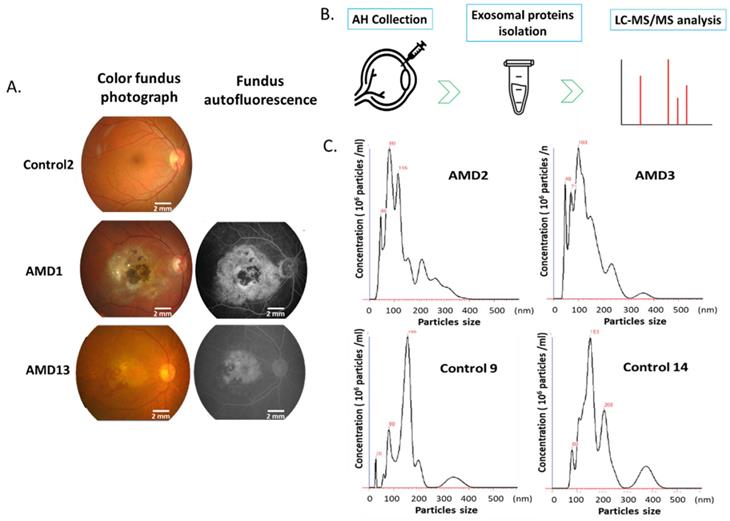
In this study, all the patients with AMD were confirmed by fluorescein angiography (Fig.1A). AH samples were collected from 28 AMD and 25 control eyes. Exosomes were isolated from each sample using the exosome purification kit. The concentration and size distribution of purified small vesicles were determined by nanoparticle tracking analysis (NTA) system. At first, the camera of NTA captures a video of all particles moving under Brownian motion. The NTA software tracks the random thermal motion of each particle to determine the diffusion coefficient which is used to calculate the size of each particle. Since NTA allows the determination of size distribution from 10 to 1000 nanometers (nm) of particles, it is one of the prominent technologies used for high-throughput analysis of individual exosomes [22]. By the analysis of NTA, most of purified exosomes were in the expected size range (Fig. 1B).
Exosomal Proteins Isolation and LC-MS/MS Analysis
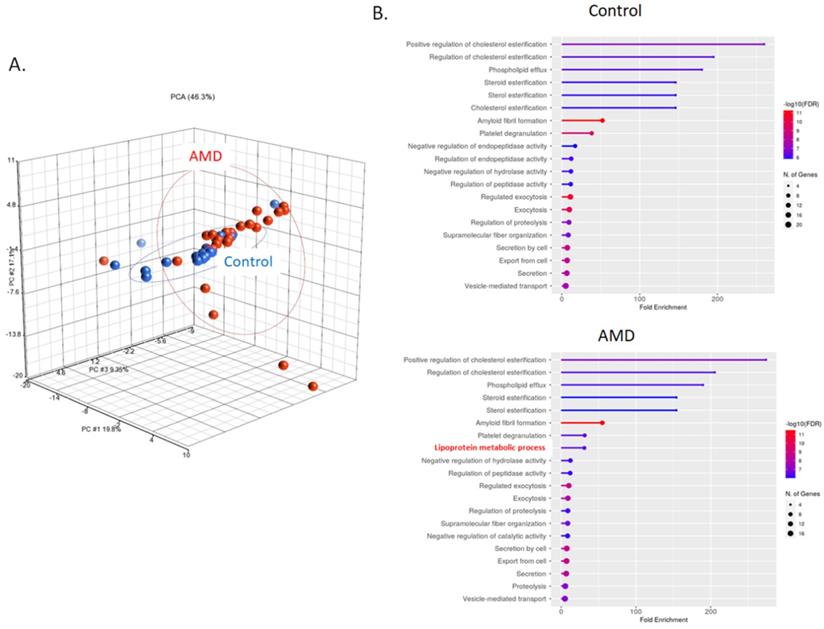
Exosomal proteins were extracted and digested with trypsin for LC-MS/MS quantitation analysis. The information about the peptides was obtained by aligning the peptide sequences to the UniProt database (www.uniprot.org). Principal component analysis (PCA) was applied to investigate internal variation between AMD and control groups (Fig. 2A). According to the PCA results, the AMD and control groups partially overlapping, but high variation was observed between individual of AMD. The analysis of gene ontology (GO) enrichment showed that most of the gene-set in the biology process were similar in the control and AMD. A gene-set, lipoprotein metabolic process, was only enriched in AMD (Fig. 2B). Since lipid metabolism is one of the important pathways in AMD pathogenesis [23]. It would be interesting to further explore their roles in cell-to-cell communication during the pathogenesis of AMD.
Ocular examination and exosomes isolation. A) Color and Fundus autofluorescence imaging for AMD eyes. B) Representative nanoparticle tracking analysis of exosomes isolated from the AH of patients with AMD and controls.
LC-MS/MS identified peptides from AMD and control. A) Common and unique identified peptides from individual AMD and control patients. B) The PCA plot of all samples was generated to assess the variability of peptide expression in myopia and control patients. (red, AMD; blue, control). C) Gene ontology enrichment analysis in biology process for AMD and control group.
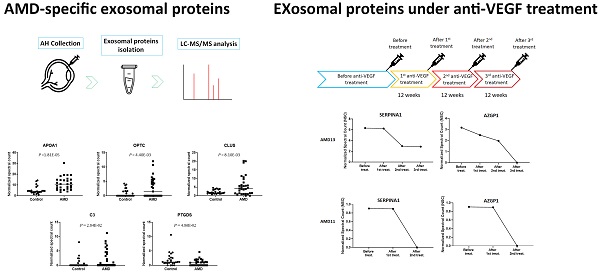
AMD-Specific Exosomal Proteins
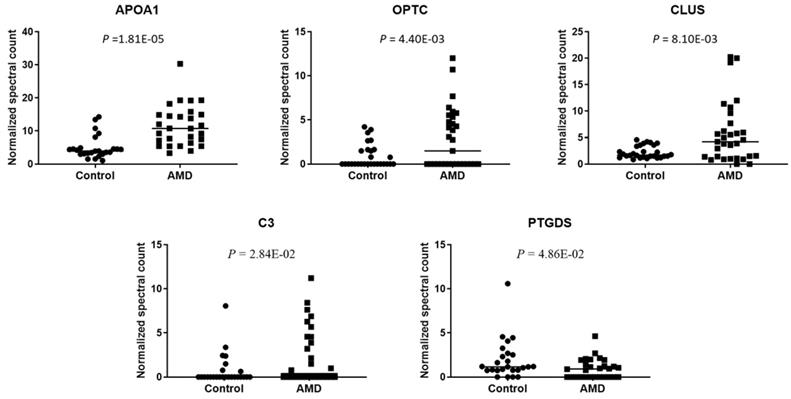
To compare the level of exosomal proteins between in AMD and control, the spectral counts obtained from each peptide were normalized to the overall spectral counts recorded for all peptides in a sample. Ten peptides were found to be significantly different between the AMD and the control (t-test, P < 0.05) (Table 1). These peptides are from 5 known proteins and 1 unidentified protein (Fig. 3). In previous studies, some of them were found to be associated with AMD. APOA1 is the main component of HDL particles [24]. The presence of APOA1 in plasma and urine has been linked to an increased risk of AMD [25-27]. CLU has been found in the drusen of patients with AMD [28]. According to the report from Kim. et. al, CLU may protect oxidative stress-induced apoptosis in retinal pigment epithelial cells [29]. C3 is one of the most important components of the complement system. Elevated plasma levels of C3 are associated with an increased risk of AMD [30, 31]. To the best of our knowledge, we are the first to identify these proteins in the exosome of AH.
Significantly changed exosomal proteins in AMD
| Protein IDs | Protein Name | Gene Name | Control (n = 25) | AMD (n = 28) | p-value |
|---|---|---|---|---|---|
| Mean ± SEM | Mean ± SEM | ||||
| P02647 | apolipoprotein A1 | APOA1 | 4.991 ± 0.683 | 11.574 ± 1.158 | 1.81E-05 |
| A0A024R3E3 | apolipoprotein A1 | APOA1 | 4.991 ± 0.683 | 11.574 ± 1.158 | 1.81E-05 |
| Q6MZU6 | Putative uncharacterized protein DKFZp686C15213 | 0.903 ± 0.211 | 3.103 ± 0.636 | 2.94E-03 | |
| Q9UBM4 | opticin | OPTC | 0.526 ± 0.171 | 2.069 ± 0.463 | 4.40E-03 |
| P10909 | clusterin | CLU | 2.032 ± 0.205 | 5.291 ± 1.101 | 8.10E-03 |
| V9HWA9 | complement C3 | C3 | 0.766 ± 0.350 | 2.420 ± 0.615 | 2.84E-02 |
| P01024 | complement C3 | C3 | 0.766 ± 0.350 | 2.420 ± 0.615 | 2.84E-02 |
| P41222 | prostaglandin D2 synthase | PTGDS | 1.938 ± 0.442 | 0.968 ± 0.211 | 4.86E-02 |
| A0A024R8G3 | prostaglandin D2 synthase | PTGDS | 1.938 ± 0.442 | 0.968 ± 0.211 | 4.86E-02 |
Normalized spectral count (NSC) of AMD-specific proteins were detected from LC-MS/MS. Scatterplot presenting the NSC of proteins detected in each patient from AMD or control. P values were calculated by paired two -tail t-tests.
Exosomal Protein profile in anti-VEGF treatment AMD
Under normal physiological conditions, the expression of VEGF is relatively low in retinal tissues. On the other hand, the expression of VEGF will increase significantly in a condition of ischemia, hypoxia, or inflammation [32]. Evaluation of VEGF level cause blood vessels growth and leakage, which is the primary cause of wet AMD [33]. Based on this theory, anti-VEGF therapies are commonly used to treat with the wet AMD [34]. The patients must be injected with the anti-VEGF drugs into the eye for every four to twelve weeks depending on their response [34]. For the exosome content is common response to external stimuli, we were interested in the change in exosomal protein profile under continuous anti-VEGF drug treatment. The AH was collected from two patients who had received continuous anti-VEGF injections of ranibizumab every 12 weeks (Fig. 4A). Comparing their exosomal proteins before and after treatment, we found that the amounts of two proteins, SERPINA1 and AZGP1, were decreased with anti-VEGF treatment time (Fig. 4B).
Discussion
In this study, we used LC-MS/MS to examine the exosomal proteins from the AH of AMD by LC-MS/MS. Comparing the protein profile of AMD and control, six proteins showed significant enrichment in patients with AMD. Three of them, APOA1, CLU, and C3, have been suggested to be linked with AMD in previous studies. To the best of our knowledge, we are the first to show that these proteins were enriched in exosome of AMD eyes. Anti-VEGF treatments are used to control the progress of wet AMD. By comparing before and after treatment, we found two exosomal proteins, SERPINA1 and AZGP1, were decreased with treatment time. As a result, we hypothesized that VEGF induced these proteins to promote AMD development. More research is needed to investigate their roles in the pathology of AMD.
Elevated HDL cholesterol levels are important in the development of AMD since drusen is composed by lipids [35]. APOA1 is one of the major components of HDL. The link between plasma level of APOA1 and the risk of AMD has been established in previous studies [25-27]. The complement system is frequently activated in many inflammatory diseases including AMD [36]. As key components of the complement cascade, genetic variations [37] and elevated plasma levels of C3 [30, 31] are associated with the risk of AMD. Our findings suggested that exosomal APOA1 and C3 are higher in AMD eyes. This raises the intriguing possibility that APOA1 and C3 in plasma could also be packaged into exosome and then cross the blood- retina barrier (BRB). BRB made up of cells that are tightly packed together to prevent uncontrolled leakage of substances such as ion, protein, and water into and out of the retinal [38]. Recently, increasing evidence suggested exosome can cross biological barriers such as the blood- brain barrier (BBB) [39]. In vivo experiments are necessary to validate our hypothesis. The pathogenesis of AMD is thought to be influenced by oxidative stress. Increased oxidative stress stimulates CLU as a physiological defense to maintain cell viability [40]. In vitro studies suggested that CLU could inhibit oxidative stress-induced caspase-3 activity, thereby protecting retinal pigment epithelial cells [29]. As a result, CLU could be considered as a preventive approach for AMD. The role of AMD-specific exosomal proteins in the pathogenesis of AMD still need to be proved. We plan to observe the changes in cell physiology and gene expression which caused by the uptake of these exosomal proteins.
Normalized spectral count (NSC) of exosomal proteins that were decreased during continuous anti-VEGF injections. A) The Flow chart of the anti-VEGF therapy process. B) The NSC of proteins detected before and after anti-VEGF therapy in different time points.
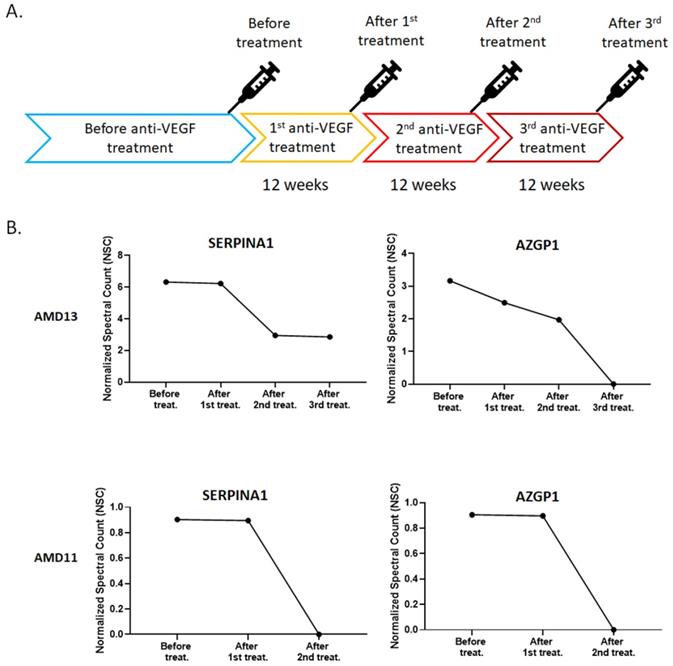
Anti-VEGF therapy is used to control the progression of wet AMD. However, some patients have a poor response or lose the efficacy to anti-VEGF agents after repeated administration [41]. Comparing the exosomal protein profile before and after anti-VEGF treatment, the level of SERPINA1 and AZGP1 in the exosome was significantly decreased. VEGF is known to stimulate endothelial cell proliferation, migration, new vessel formation, and ultimately leading to angiogenesis [42]. Previous research has shown that SERPINA1 promotes cell migration and invasion [43, 44]. AZGP1 could improve cell proliferation and epithelial-mesenchymal transition (EMT) [45, 46]. For the SERPINA1 and AZGP1 were decreased with anti-VEGF treatment time, we proposed they may under the regulation of VEGF and could be biomarkers for curative effect of anti-VEGF therapy in AMD. Due to the time-consuming for sample collection, only two patients were shown in this study. Expanding the number of samples to verify the association of SERPINA1 and AZGP1with anti-VEGF therapy is our next goal.
In conclusion, this study revealed the unique exosomal proteins in AMD patients and receiving anti-VEGF therapy patients. These proteins shed light on potential new targets for the diagnosis and treatment of AMD.
Acknowledgements
We acknowledge the technical services provided by the Center for Clinical and Biotechnological Applications of National Yang Ming Chiao Tung University. The core facility is supported by the National Core Facility for Biopharmaceuticals (NCFB), Ministry of Science and Technology. This study was supported by grants from the Ministry of Science and Technology of Taiwan (MOST 108-2314-B-532-009-) and Taipei City Hospital (TPCH-110-).
Abbreviations
AH, aqueous humor; AMD, Age-related macular degeneration; LC-MS/MS, liquid chromatography-tandem mass spectrometry; PCA, principal component analysis.
Competing Interests
The authors have declared that no competing interest exists.
References
1. GBD Blindness Vision Impairment Collaborators, Vision Loss Expert Group of the Global Burden of Disease Study. Causes of blindness and vision impairment in 2020 and trends over 30 years, and prevalence of avoidable blindness in relation to VISION 2020: the Right to Sight: an analysis for the Global Burden of Disease Study. Lancet Glob Health. 2021;9:e144-e60
2. Apte RS. Age-Related Macular Degeneration. N Engl J Med. 2021;385:539-47
3. Strunz T, Kiel C, Sauerbeck BL, Weber BHF. Learning from Fifteen Years of Genome-Wide Association Studies in Age-Related Macular Degeneration. Cells. 2020;9:2267
4. Seddon JM, Willett WC, Speizer FE, Hankinson SE. A prospective study of cigarette smoking and age-related macular degeneration in women. JAMA. 1996;276:1141-6
5. Seddon JM, Cote J, Davis N, Rosner B. Progression of age-related macular degeneration: association with body mass index, waist circumference, and waist-hip ratio. Arch Ophthalmol. 2003;121:785-92
6. Seddon JM, George S, Rosner B. Cigarette smoking, fish consumption, omega-3 fatty acid intake, and associations with age-related macular degeneration: the US Twin Study of Age-Related Macular Degeneration. Arch Ophthalmol. 2006;124:995-1001
7. Christen WG, Glynn RJ, Manson JE, Ajani UA, Buring JE. A prospective study of cigarette smoking and risk of age-related macular degeneration in men. JAMA. 1996;276:1147-51
8. Davis MD, Gangnon RE, Lee LY, Hubbard LD, Klein BE, Klein R. et al. The Age-Related Eye Disease Study severity scale for age-related macular degeneration: AREDS Report No. 17. Arch Ophthalmol. 2005;123:1484-98
9. Deng Y, Qiao L, Du M, Qu C, Wan L, Li J. et al. Age-related macular degeneration: Epidemiology, genetics, pathophysiology, diagnosis, and targeted therapy. Genes Dis. 2022;9:62-79
10. Ma HH, Liutkeviciene R. Age-Related Macular Degeneration: What Do We Know So Far? Acta Med Litu. 2021;28:36-47
11. Nashine S. Potential Therapeutic Candidates for Age-Related Macular Degeneration (AMD). Cells. 2021;10:2483
12. He C, Zheng S, Luo Y, Wang B. Exosome Theranostics: Biology and Translational Medicine. Theranostics. 2018;8:237-55
13. Colombo M, Raposo G, Thery C. Biogenesis, secretion, and intercellular interactions of exosomes and other extracellular vesicles. Annu Rev Cell Dev Biol. 2014;30:255-89
14. Goel M, Picciani RG, Lee RK, Bhattacharya SK. Aqueous humor dynamics: a review. Open Ophthalmol J. 2010;4:52-9
15. Dismuke WM, Challa P, Navarro I, Stamer WD, Liu Y. Human aqueous humor exosomes. Exp Eye Res. 2015;132:73-7
16. Wecker T, Hoffmeier K, Plotner A, Gruning BA, Horres R, Backofen R. et al. MicroRNA Profiling in Aqueous Humor of Individual Human Eyes by Next-Generation Sequencing. Invest Ophthalmol Vis Sci. 2016;57:1706-13
17. Chen CF, Hua K, Woung LC, Lin CH, Chen CT, Hsu CH. et al. Expression Profiling of Exosomal miRNAs Derived from the Aqueous Humor of Myopia Patients. Tohoku J Exp Med. 2019;249:213-21
18. Gao C, Liu X, Fan F, Yang JN, Zhou XY, Mei HJ. et al. Exosomal miR-29b found in aqueous humour mediates calcium signaling in diabetic patients with cataract. Int J Ophthalmol. 2021;14:1484-91
19. Kang GY, Bang JY, Choi AJ, Yoon J, Lee WC, Choi S. et al. Exosomal proteins in the aqueous humor as novel biomarkers in patients with neovascular age-related macular degeneration. J Proteome Res. 2014;13:581-95
20. Tsai CY, Chen CT, Lin CH, Liao CC, Hua K, Hsu CH. et al. Proteomic analysis of Exosomes derived from the Aqueous Humor of Myopia Patients. Int J Med Sci. 2021;18:2023-9
21. Ge SX, Jung D, Yao R. ShinyGO: a graphical gene-set enrichment tool for animals and plants. Bioinformatics. 2020;36:2628-9
22. Comfort N, Cai K, Bloomquist TR, Strait MD, Ferrante AW Jr, Baccarelli AA. Nanoparticle Tracking Analysis for the Quantification and Size Determination of Extracellular Vesicles. J Vis Exp. 2021;169:62447
23. Kelly UL, Grigsby D, Cady MA, Landowski M, Skiba NP, Liu J. et al. High-density lipoproteins are a potential therapeutic target for age-related macular degeneration. J Biol Chem. 2020;295:13601-16
24. van der Vorst EPC. High-Density Lipoproteins and Apolipoprotein A1. Subcell Biochem. 2020;94:399-420
25. Han X, Ong JS, Hewitt AW, Gharahkhani P, MacGregor S. The effects of eight serum lipid biomarkers on age-related macular degeneration risk: a Mendelian randomization study. Int J Epidemiol. 2021;50:325-36
26. Nordestgaard LT, Tybjaerg-Hansen A, Frikke-Schmidt R, Nordestgaard BG. Elevated Apolipoprotein A1 and HDL Cholesterol Associated with Age-related Macular Degeneration: 2 Population Cohorts. J Clin Endocrinol Metab. 2021;106:e2749-58
27. Sivagurunathan S, Selvan LDN, Khan AA, Parameswaran S, Bhattacharjee H, Gogoi K. et al. Proteomics-based approach for differentiation of age-related macular degeneration sub-types. Indian J Ophthalmol. 2021;69:647-54
28. Sakaguchi H, Miyagi M, Shadrach KG, Rayborn ME, Crabb JW, Hollyfield JG. Clusterin is present in drusen in age-related macular degeneration. Exp Eye Res. 2002;74:547-9
29. Kim JH, Kim JH, Jun HO, Yu YS, Min BH, Park KH. et al. Protective effect of clusterin from oxidative stress-induced apoptosis in human retinal pigment epithelial cells. Invest Ophthalmol Vis Sci. 2010;51:561-6
30. Scholl HP, Charbel Issa P, Walier M, Janzer S, Pollok-Kopp B, Borncke F. et al. Systemic complement activation in age-related macular degeneration. PLoS One. 2008;3:e2593
31. Sivaprasad S, Adewoyin T, Bailey TA, Dandekar SS, Jenkins S, Webster AR. et al. Estimation of systemic complement C3 activity in age-related macular degeneration. Arch Ophthalmol. 2007;125:515-9
32. Miller JW, Adamis AP, Shima DT, D'Amore PA, Moulton RS, O'Reilly MS. et al. Vascular endothelial growth factor/vascular permeability factor is temporally and spatially correlated with ocular angiogenesis in a primate model. Am J Pathol. 1994;145:574-84
33. Lu M, Adamis AP. Molecular biology of choroidal neovascularization. Ophthalmol Clin North Am. 2006;19:323-34
34. Cho YK, Park DH, Jeon IC. Medication Trends for Age-Related Macular Degeneration. Int J Mol Sci. 2021;22:11837
35. Wang L, Clark ME, Crossman DK, Kojima K, Messinger JD, Mobley JA. et al. Abundant lipid and protein components of drusen. PLoS One. 2010;5:e10329
36. Kim BJ, Mastellos DC, Li Y, Dunaief JL, Lambris JD. Targeting complement components C3 and C5 for the retina: Key concepts and lingering questions. Prog Retin Eye Res. 2021;83:100936
37. Helgason H, Sulem P, Duvvari MR, Luo H, Thorleifsson G, Stefansson H. et al. A rare nonsynonymous sequence variant in C3 is associated with high risk of age-related macular degeneration. Nat Genet. 2013;45:1371-4
38. Cunha-Vaz J, Bernardes R, Lobo C. Blood-retinal barrier. Eur J Ophthalmol. 2011;21(Suppl 6):S3-9
39. Banks WA, Sharma P, Bullock KM, Hansen KM, Ludwig N, Whiteside TL. Transport of Extracellular Vesicles across the Blood-Brain Barrier: Brain Pharmacokinetics and Effects of Inflammation. Int J Mol Sci. 2020;21:4407
40. Trougakos IP, Gonos ES. Regulation of clusterin/apolipoprotein J, a functional homologue to the small heat shock proteins, by oxidative stress in ageing and age-related diseases. Free Radic Res. 2006;40:1324-34
41. Yang S, Zhao J, Sun X. Resistance to anti-VEGF therapy in neovascular age-related macular degeneration: a comprehensive review. Drug Des Devel Ther. 2016;10:1857-67
42. Hoeben A, Landuyt B, Highley MS, Wildiers H, Van Oosterom AT, De Bruijn EA. Vascular endothelial growth factor and angiogenesis. Pharmacol Rev. 2004;56:549-80
43. Jiang L, Hu LG. Serpin peptidase inhibitor clade A member 1-overexpression in gastric cancer promotes tumor progression in vitro and is associated with poor prognosis. Oncol Lett. 2020;20:278
44. Kwon CH, Park HJ, Lee JR, Kim HK, Jeon TY, Jo HJ. et al. Serpin peptidase inhibitor clade A member 1 is a biomarker of poor prognosis in gastric cancer. Br J Cancer. 2014;111:1993-2002
45. Cao R, Ke M, Wu Q, Tian Q, Liu L, Dai Z. et al. AZGP1 is androgen responsive and involved in AR-induced prostate cancer cell proliferation and metastasis. J Cell Physiol. 2019;234:17444-58
46. Ji M, Li W, He G, Zhu D, Lv S, Tang W. et al. Zinc-alpha2-glycoprotein 1 promotes EMT in colorectal cancer by filamin A mediated focal adhesion pathway. J Cancer. 2019;10:5557-66
Author contact
![]() Corresponding author: Chian-Feng Chen, Ph.D. 155, Sec.2, Linong Street, Taipei, 112 Taiwan. E-mail: cfchenedu.tw
Corresponding author: Chian-Feng Chen, Ph.D. 155, Sec.2, Linong Street, Taipei, 112 Taiwan. E-mail: cfchenedu.tw

 Global reach, higher impact
Global reach, higher impact