Impact Factor
ISSN: 1449-1907
Int J Med Sci 2022; 19(5):858-866. doi:10.7150/ijms.71737 This issue Cite
Research Paper
Serum HBV DNA plus RNA reflecting cccDNA level before and during NAs treatment in HBeAg positive CHB patients
1. Liver disease center, Beijing YouAn Hospital, Capital Medical University, Beijing 100069, China.
2. Beijing Municipal Key Laboratory of Liver Failure and Artificial Liver Treatment & Research, Beijing YouAn Hospital, Capital Medical University, Beijing 100069, China.
3. Department of Microbiology and Infectious Disease Center, School of Basic Medical Sciences, Peking University Health Science Center, Beijing 100191, China.
4. Department of Clinical Laboratory, Shenzhen Third People's Hospital, Southern University of Science and Technology, National Clinical Research Center for Infectious Diseases, Shenzhen, 518112, PR China.
5. Academy for Advanced Interdisciplinary Studies, Peking University, Beijing 100871, China.
6. Hunan Provincial Key Laboratory of Gene Diagnostic Technology, Changsha 410205, China.
*These authors have contributed equally to this work.
Received 2022-2-2; Accepted 2022-4-20; Published 2022-5-9
Abstract
Background & Aims: Correlations between serum viral markers and intrahepatic cccDNA in patients undergoing long-term nucleos(t)ide analogues (NAs) treatment haven't been fully explored. In this study, we evaluate the correlation between intrahepatic cccDNA and other serum viral markers and intrahepatic HBV DNA in HBeAg positive chronic hepatitis B (CHB) patients during 60-month treatment with NAs.
Methods: Fifty-four HBeAg positive CHB patients received long-term NAs treatment were included in this study. Serial serum samples were regularly collected and quantitatively analyzed for HBsAg, HBV DNA, HBV RNA and HBcrAg. Histological samples from liver biopsy at baseline and month 60 were analyzed for intrahepatic HBV DNA and cccDNA.
Results: At baseline, serum HBV DNA plus RNA was positively associated with intrahepatic cccDNA in multivariate regression analysis (β=0.205, P<0.001). In the correlation analysis between cccDNA and serum viral markers, HBV DNA plus RNA had the highest correlation coefficient (r=0.698, P<0.001), followed by serum HBV DNA (r=0.641, P<0.001), HBV RNA (r=0.590, P<0.001), and HBcrAg (r=0.564, P<0.001). At month 60, correlations between these serum viral markers and cccDNA were not observed (P>0.05). Multivariate regression analysis showed that only the decreased HBV DNA plus RNA was positively associated with cccDNA decline (β=0.172, P =0.006). Changes of HBV DNA plus RNA (r=0.525, P=0.001) was better correlated with cccDNA decline as compared to HBV RNA (r=0.384, P=0.008), HBV DNA (r=0.431, P=0.003), and HBsAg (r=0.342, P=0.029).
Conclusions: Serum HBV DNA plus RNA better correlated with intrahepatic cccDNA than other viral makers before and during NAs treatment in HBeAg positive CHB patients.
Keywords: chronic hepatitis B, pregenomic RNA, covalently closed circular DNA, nucleos(t)ide analogues, hepatitis B surface antigen
Introduction
Hepatitis B virus (HBV) infection has been a global public health challenge [1]. Nucleos(t)ide analogues (NAs) suppress HBV DNA synthesis via inhibiting reverse transcription of pregenomic RNA (pgRNA) into HBV DNA, which can reverse liver fibrosis and reduce the risk of hepatocellular carcinoma [2]. Although NAs do not directly affect the covalently closed circular DNA (cccDNA), NAs could efficiently inhibit the replenishment of cccDNA pool via blocking relaxed circular DNA (rcDNA) formation [3-5]. Clinical studies also observed significant decrease of intrahepatic cccDNA after long-term NAs treatment in CHB patients [6,7], and intrahepatic cccDNA level decline is the most direct prognostic indicator of response to antiviral treatment [8]. However, the invasive procedure and potential sampling error restricted the utilization of intrahepatic cccDNA in clinical practice. Therefore, exploring noninvasive and convenient serum viral markers that indirectly reflecting intrahepatic cccDNA level has important clinical value in CHB patient receiving long-term NAs treatment.
Classical indicators, HBsAg and HBV DNA, have been thought to be positively correlated with intrahepatic cccDNA before NAs treatment, but this correlation is weak [9-11]. Besides, HBV DNA can be efficiently inhibited to an undetectable level in a majority of CHB patients after NAs treatment, and the integrated HBV DNA fragments could also generate HBsAg [12], all these factors may influence the relationship between serum HBsAg, HBV DNA and intrahepatic cccDNA level before and after NAs treatment [13,14].
Novel serum viral markers, HBV RNA [3, 14-16] and HBV core-related antigen (HBcrAg) [17-19] have been proposed as indicators to reflect intrahepatic transcriptional activity of cccDNA in CHB patients. Serum HBV RNA levels were positively correlated with intrahepatic cccDNA before NAs treatment but the correlation disappeared after 96 weeks of NAs treatment in HBeAg positive CHB patients [14]. Besides, higher serum HBV RNA/DNA ratio indicated the lower reverse transcription of pgRNA [20,21]. Huang et al. reported that serum HBV DNA plus RNA exhibited superiority than HBV RNA or HBV DNA alone in reflecting cccDNA activity in treatment-naive HBeAg positive CHB patients [21]. Recent researches have reported serum HBcrAg had a better correlation with intrahepatic cccDNA level before antiviral treatment [18, 22]. So far, it is still lacking universally acknowledged indicator to reflect cccDNA level. Whether these serum viral markers are consistently correlated with cccDNA after long-term NAs treatment remains unknown. In addition, whether the changes of these serum viral markers reflected the decline of cccDNA during NAs treatment remains to be elucidated. These studies intrigue us to conduct a head-to-head comparison of serum HBsAg, HBV RNA, HBV DNA, HBV DNA plus RNA, HBV RNA/DNA and HBcrAg levels in reflecting intrahepatic cccDNA in CHB patients treated with NAs.
In this single center, longitudinal study, we evaluate the correlation between intrahepatic cccDNA and serum HBsAg, HBV RNA, HBV DNA, HBV DNA plus RNA, HBV RNA/DNA, HBcrAg, as well as intrahepatic HBV DNA in HBeAg positive CHB patients with 60-month NAs treatment.
Methods
Patients and study design
This study was conducted using a cohort of 83 HBeAg positive CHB patients receiving NAs monotherapy. Of them, 54 patients with liver biopsy at baseline and month 60 were included in this analysis. Patients of this cohort were prospectively recruited from Beijing YouAn Hospital, Capital Medical University (Beijing, China) between June 2007 and July 2008. Eligible patients were diagnosed CHB according to the American Association for the Study of Liver Diseases guideline [23], male or female patients aged ≥ 16 years. The exclusion criteria were as follows: i) co‑infection with another viruses, including hepatitis C or D virus, Epstein-Barr virus, cytomegalovirus and human immunodeficiency viruses, the existence of autoimmune liver disease, or alcoholic liver disease; ii) with decompensated liver function (ascites, hepatic encephalopathy or upper gastrointestinal bleeding); iii) with any diseases of other major organs, such as severe heart disease or kidney disease; iv) poor compliance; v) history of a malignancy, including hepatocellular carcinoma, carcinoma in situ and atypical hyperplastic nodules; vi) with mental illness; vii) had received corticosteroids, immunosuppressants or chemotherapeutic drugs ≤6 months prior to enrollment; and viii) pregnant or breast‑feeding women.
At enrollment and 60 months on-treatment, serum specimens were collected for liver function tests, viral marker tests and HBV DNA quantification. Remaining serum samples were stored at -80 °C for subsequent research. Percutaneous liver biopsy were performed to evaluate the histology, which were diagnosed according to the modified Knodell and Ishak scoring system [24].
With the foregoing collected blood, we quantified HBsAg, HBV RNA and HBcrAg levels at the time point of baseline and month 60 of NAs treatment. Intrahepatic HBV DNA and cccDNA levels at baseline and month 60 were measured by a method as previously reported [21].
The study was conducted in compliance with the Declaration of Helsinki. Use of the research samples was approved by the Medical Ethics Review Committee of Beijing YouAn Hospital. All patients provided written informed consent authorizing us to access their medical records and to store the remaining serum specimens for research purposes.
Assays for serological HBV markers, HBV DNA, HBV RNA, and HBcrAg
Serum HBsAg, anti-HBs, HBeAg, anti-HBe, and anti-HBc were determined on a Roche Cobas e601 analyzer using an electrochemiluminescence immunoassay (Abbott Laboratories, Chicago, IL, USA). HBsAg was quantified using an Elecsys for HBsAg quantitation (Roche Diagnostics) with a lower limit of detection (LLD) of 0.05 IU/mL. The serum HBV DNA level was determined using the Cobas HBV Amplicor Monitor assay (Roche Diagnostics, Pleasanton, CA, USA), with a LLD of 50 IU/mL. Serum HBV RNA level was determined as described previously [15, 21, 25]. Briefly, HBV RNA was isolated with the nucleic acid extraction or purification kit (Sansure Biotech, Changsha, China) and treated with DNase I (Thermo Fisher Scientific, Waltham, MA, USA). The specially modified super-cis nano-magnetic beads efficiently adsorbed and enriched nucleic acids from 200 µL serum. For DNase I treatment, every reaction mixture comprised 2 µL of DNase I Reaction Buffer (10×), 2 µL of DNAse I (RNase-free), and 16 µL of total nucleic acids. The reaction was carried out at 37 °C for 30 min. Next, each mixture was incubated at 75 °C for 10 min to inactivate DNase I. Finally, DNase-I-treated HBV RNA was one-step of reverse-transcribed and real-time fluorescent quantitative PCR using the HBV pgRNA high-sensitivity quantitative kit (Sansure Biotech, Changsha, China). The LLD of the assay was 200 copies/mL. Details for HBV RNA assay could be found in Supplementary Materials. HBcrAg was determined using chemiluminescent enzyme immunoassay in automated analyzer system (Lumipulse System, Fujirebio Inc., Tokyo, Japan). The LLD was 1,000 U/mL with a linear range of 3-7 log10 U/mL.
Quantitation of intrahepatic HBV DNA and cccDNA
About 30 μm formalin fixation and paraffin embedding (FFPE) liver biopsy tissue in sections of 6 μm each was used for DNA extraction. The DNA was extracted using QIAamp FFPE DNA Mini Kit (QIAGEN, GmbH, Hilden, Germany) according to the instructions of the manufacturer. T5 Exonuclease (New England Biolabs, USA) was used to digest HBV rcDNA, replicative dsDNA and ssDNA. The reaction mixture contained 100 ng extracted DNA, 0.5 µL (10 units) T5 Exonuclease, 1 µL NEBuffer 4 (10×) with Nuclease-free H2O to a final volume of 10 μL. The digestion was carried out at 37 °C for 1 h, and stop reaction with EDTA to at least 11 mM. We combined 6.42 μL of digestion product, which was obtained in the previous step, with 7.50 μL QuantStudio™ 3D Digital PCR Master Mix, 0.06 μL of TaqMan Probe-RC-MGB (50 μM), 0.06 μL TaqMan Probe-RNAseP-VIC (50 μM), 0.24 μL primer of rc-F, 0.24 μL primer of rc-R, 0.24 μL primer of RNaseP-F and 0.24 μL primer of RNaseP-R. This sample mix 15 μL was added on each chip and loaded on ProFlex™ 2x Flat PCR System with the following program: Absolute quantification was determined using QuantStudio™ 3D Digital PCR System (Thermo Fisher Scientific Inc., Waltham, Massachusetts, USA) and analyzed with QuantStudio 3D AnalysisSuite Cloud Software. (https://china.apps.thermofisher.com/quantstudio3d/). All intrahepatic HBV cccDNA values were normalized to cell number assessed by RNase P copy number assay.
Statistical analysis
Data were analyzed using the IBM SPSS 22.0 (SPSS Inc., Chicago, IL, USA) and R software version 4.1.2 (R Foundation for Statistical Computing, Vienna, Austria). P<0.05 was considered to be statistically significant in two-tailed test. HBsAg, HBV DNA, HBV RNA, HBcrAg, intrahepatic HBV DNA and cccDNA expression were logarithmically transformed for analysis. HBV DNA plus RNA was measured using the numbers of log10 HBV DNA plus log10 HBV RNA copies/mL, the ratio between HBV RNA and DNA was also assessed using the ratio of log10 HBV RNA copies/mL to log10 HBV DNA, as previously described [21, 26]. Continuous variables with normal distribution were expressed as mean ± SD (standard deviation). Continuous variable with abnormal distribution were expressed as median and range. Dichotomy variables were expressed as counting or proportion. The comparison of continuous quantitative data between before and after treatment was performed using Wilcoxon rank sum test. Moving forward (LR) multivariate linear regression analysis was performed to determine factors associated with intrahepatic HBV cccDNA levels, and the P values of entry and removal were respectively set to 0.05 and 0.1. Correlation between two continuous variables was also calculated and visualized using the R packages 'GGally' and 'ggplot2'.
Results
Characteristics of CHB patients
A total of 54 patients with liver biopsy at baseline and month 60 were included in this study. Of them, 24 were NAs-naïve patients, 30 were previously Lamivudine exposure patients. Twenty-eight (51.85%) patients were treated with Entacavir (ETV) 0.5 mg once daily and the other 26 (48.15%) patients were treated with Adefovir dipivoxil (ADV) 10mg once daily, the differences of baseline characteristics between these two groups were not significant (all P > 0.05), detailed information were shown in Supplementary Table 1. Forty-seven (87.04%) patients were male with an average age of 36.54 ± 9.44 years. Twenty-eight patients with available genotype data were analyzed, with 20 (71.43%) were genotype C. Median inflammation and fibrosis score were 7 (range, 2-15) and 3 (range, 1-5), respectively.
During 60 months' treatment, one previous lamivudine exposure patient in ETV group was detected ETV-associated resistant variants rtV173M, rtL180M, rtM204V/I and rtT184F. Four patients in ADV group were detected ADV-associated resistant variants rtA181 V/T or rtN236T. The levels of HBsAg, HBV DNA, HBV RNA, HBV RNA plus RNA, HBcrAg, intrahepatic HBV DNA and cccDNA were all significantly decreased (P <0.001). Inflammation and fibrosis score were also significantly decreased (P <0.001), while the ratio of HBV RNA/DNA was significantly increased (P <0.001). The detailed information were shown in Table 1.
Characteristics of HBeAg positive CHB patients
| Clinical characteristics | baseline | Month 60 | P value |
|---|---|---|---|
| Sex, male/female | 47/7 | - | - |
| Age, years | 36.54±9.44 | - | - |
| Naïve/LAM treated | 24/30 | - | - |
| Therapy drug, entecavir/adefovir | 28/26 | - | - |
| BMI Kg/m2 | 23.97±3.48 | - | - |
| ALT, IU/L | 68.30 (12.60-681.90) | 22.30 (3.80-210.00) | <0.001 |
| AST, IU/L | 43.00 (10.90-358.80) | 22.20 (11.60-73.50) | <0.001 |
| AST/ALT ratio | 0.65 (0.25-2.32) | 1.02 (0.35-5.00) | <0.001 |
| TBiL, µmol/L | 15.25 (6.80-27.80) | 14.10 (7.20-37.50) | 0.090 |
| ALP, U/L | 89.70 (45.00-412.20) | 65.60 (40.60-664.60) | <0.001 |
| HBV Genotype (C/B+others)† | 20/8 | - | - |
| HBsAg (log10 IU/mL) | 3.77 (-0.07-4.95) | 3.35 (-0.33- 4.28) | <0.001 |
| HBV DNA (log10 IU/mL) | 6.60 (1.99-9.28) | 1.70 (1.70-8.70) | <0.001 |
| HBV RNA (log10 copies/mL) | 5.38 (2.30-8.01) | 2.40 (1.40-6.42) | <0.001 |
| HBV RNA/DNA ratio | 0.84 (0.34-1.62) | 1.18 (0.27-3.05) | <0.001 |
| HBV DNA plus RNA | 11.66 (4.69-16.58) | 4.21 (3.10-11.03) | <0.001 |
| HBcrAg (log10 U/mL ) | 7.40 (4.84-8.73) | 5.48 (2.95-7.86) | <0.001 |
| Intrahepatic HBV DNA (log10copies/105 cell) | 6.60 (4.42-8.50) | 4.91 (4.10-6.88) | <0.001 |
| Intrahepatic cccDNA (log10copies/105 cell) | 4.90 (2.69-7.18) | 3.34 (0.00-5.18) | <0.001 |
| Hepatic inflammation grade‡ | 7 (2-15) | 3 (1-12) | <0.001 |
| Hepatic fibrosis stage‡ | 3 (1-5) | 2(1-5) | <0.001 |
†Twenty-eight patients with available genotype data were analyzed.
‡Hepatic inflammation grade and fibrosis stage were diagnosed according to the modified knodell and Ishak scoring system respectively.
Abbreviations: ADV, Adefovir; ALP, alkaline phosphatase; ALT, alanine aminotransferase; AST, aspartate aminotransferase; BMI, body mass index; cccDNA, covalently closed circular DNA; ETV, Entecavir; HBV, hepatitis B virus; HBV RNA, hepatitis B virus ribonucleic acid; HBcrAg, hepatitis B core-related antigen; HBeAg, hepatitis B e antigen; HBsAg, hepatitis B surface antigen; LAM, Lamivudine; TBIL, total bilirubin.
Regression analysis of factors associated with intrahepatic cccDNA
At baseline, univariate linear regression analysis showed that serum HBV DNA, HBV RNA, HBV DNA plus RNA, HBcrAg, intrahepatic HBV DNA, and Ishak fibrosis score were all associated with cccDNA (all P <0.05), while HBsAg, HBV RNA/DNA and inflammation score were not associated with cccDNA (all P >0.05) Table 2. The multivariate regression analysis showed that only HBV DNA plus RNA was positively associated with cccDNA (β=0.205, 95%CI: 0.135-0.274, P <0.001).
Linear regression analysis of factors associated with intrahepatic cccDNA before and after 60 months treatment
| Variables | Baseline | Month 60 | ||||
|---|---|---|---|---|---|---|
| B | 95%CI | P value | B | 95%CI | P value | |
| Age, yrs | -0.003 | -0.030-0.024 | 0.821 | 0.015 | -0.005-0.035 | 0.141 |
| Male | -0.282 | -1.042-0.477 | 0.459 | -0.265 | -0.822-0.293 | 0.345 |
| ETV | 0.114 | -0.399-0.626 | 0.658 | 0.053 | -0.335-0.442 | 0.784 |
| Naïve | -0.332 | -0.849- 0.175 | 0.195 | 0.015 | -0.375- 0.404 | 0.940 |
| BMI Kg/m2 | -0.031 | -0.105-0.043 | 0.404 | -0.016 | -0.071-0.038 | 0.554 |
| HBV Genotype † | -0.142 | -0.528-0.245 | 0.455 | |||
| B/C/others | -0.384 | -0.891-0.122 | 0.131 | |||
| ALT, U/L | <0.001 | -0.002-0.002 | 0.787 | -0.003 | -0.009-0.003 | 0.359 |
| AST, U/L | <0.001 | -0.004-0.004 | 0.988 | -0.014 | -0.035-0.007 | 0.181 |
| AST/ALT ratio | -0.592 | -1.314-0.129 | 0.106 | 0.071 | -0.237-0.378 | 0.646 |
| TBIL, μmol/L | <0.001 | -0.048-0.049 | 0.990 | -0.005 | -0.041-0.030 | 0.763 |
| ALP, U/L | -0.002 | -0.007-0.002 | 0.308 | 0.005 | 0.001-0.011 | 0.067 |
| HBsAg (log10 IU/mL) | 0.033 | -0.234-0.300 | 0.804 | 0.069 | -0.179-0.317 | 0.576 |
| HBV DNA (log10 IU/mL) | 0.315 | 0.210-0.420 | <0.001 | 0.012 | -0.177-0.202 | 0.895 |
| HBV RNA (log10 copies/mL) | 0.327 | 0.194-0.460 | <0.001 | 0.005 | -0.128-0.138 | 0.938 |
| HBV RNA/DNA ratio | -0.554 | -1.657-0.549 | 0.317 | 0.084 | -0.225-0.392 | 0.588 |
| HBV DNA plus RNA | 0.204 | 0.143-0.266 | <0.001 | 0.022 | -0.083-0.126 | 0.680 |
| HBcrAg (log10 U/mL) | 0.477 | 0.254-0.700 | <0.001 | -0.110 | -0.325-0.105 | 0.306 |
| Intrahepatic HBV DNA (log10copies/105 cell) | 0.926 | 0.752-1.100 | <0.001 | 0.709 | 0.397-1.022 | <0.001 |
| Inflammation scores‡ | -0.033 | -0.100-0.034 | 0.329 | 0.079 | -0.077-0.235 | 0.311 |
| Fibrosis scores‡ | -0.328 | -0.575-0.082 | 0.010 | 0.305 | 0.100-0.511 | 0.004 |
†Twenty-eight patients with available genotype data were analyzed.
‡Hepatic inflammation grade and fibrosis stage were diagnosed according to the modified knodell and Ishak scoring system respectively.
Abbreviations: ALP, alkaline phosphatase; ALT, alanine aminotransferase; AST, aspartate aminotransferase; BMI, body mass index; cccDNA, covalently closed circular DNA; ETV, Entecavir; HBV, hepatitis B virus; HBV RNA, hepatitis B virus ribonucleic acid; HBcrAg, hepatitis B core-related antigen; HBeAg, hepatitis B e antigen; HBsAg, hepatitis B surface antigen; LAM, Lamivudine; TBIL, total bilirubin.
At month 60 of NAs treatment, univariate linear regression analysis showed that intrahepatic HBV DNA and Ishak fibrosis score were associated with cccDNA (both P<0.05). HBsAg, HBV DNA, HBV RNA, HBV RNA/DNA, HBV DNA plus RNA, HBcrAg and inflammation score were not associated with cccDNA in our study (P >0.05), Table 2.
During 60 months' NAs treatment, univariate linear regression analysis showed that the decreased HBsAg, HBV DNA, HBV RNA, HBV DNA plus RNA, intrahepatic HBV DNA and inflammation score were associated with cccDNA decline (all P<0.05). The changed HBV RNA/DNA, HBcrAg and fibrosis score were not associated with cccDNA decline (all P >0.05) Table 3. Multivariate regression analysis showed that only decreased HBV DNA plus RNA was positively associated with cccDNA decline (β=0.172, 95%CI: 0.054-0.289, P =0.006).
Correlation analysis between cccDNA and serum biomarkers, as well as intrahepatic HBV DNA
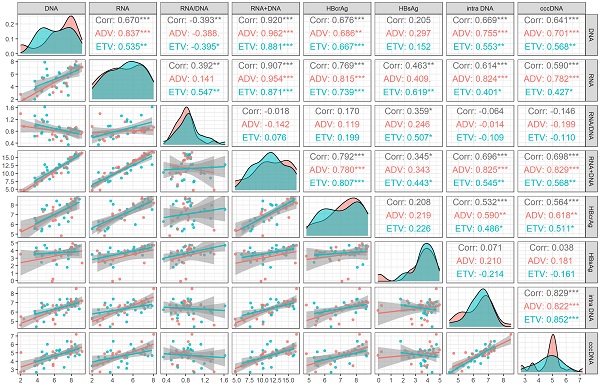
We analyzed the correlation between viral markers and cccDNA both at baseline and month 60 of NAs treatment. At baseline, HBV DNA plus RNA had the highest correlation with cccDNA (r=0.698, P<0.001), followed by HBV DNA (r=0.641, P <0.001), HBV RNA (r=0.590, P <0.001) and HBcrAg (r=0.564, P<0.001), which was lower than the correlation coefficient between intrahepatic HBV DNA and cccDNA (r=0.829, P <0.001). HBsAg and HBV RNA/DNA had no correlation with cccDNA (P = 0.804 and 0.317, respectively). The correlation of different drug subgroups were shown in Figure 1.
Based on the median value of inflammation and fibrosis score, we further studied whether the correlations between serum viral markers and cccDNA were influenced by inflammation and fibrosis. The results showed that neither inflammation score (≤7 vs. >7) nor fibrosis score (≤3 vs >3) influence the correlations between intrahepatic cccDNA and serum HBsAg, HBV DNA, HBV RNA, HBV RNA/DNA and HBV DNA plus RNA. The correlation between intrahepatic cccDNA and HBcrAg was not influenced by inflammatory (≤7 vs >7), but the correlations was influenced by fibrosis (≤3 vs >3), as shown in Table 4.
Linear regression analysis of decreased viral markers associated with intrahepatic cccDNA decline after 60 months treatment
| Variables | B | 95%CI | P value |
|---|---|---|---|
| HBsAg (log10 IU/mL) | 0.437 | 0.048-0.826 | 0.029 |
| HBV DNA (log10 IU/mL) | 0.226 | 0.079-0.373 | 0.003 |
| HBV RNA (log10 copies/mL) | 0.314 | 0.085-0.543 | 0.008 |
| HBV RNA/DNA ratio | -0.319 | -0.979-0.341 | 0.334 |
| HBV DNA plus RNA | 0.204 | 0.094-0.314 | 0.001 |
| HBcrAg (log10 U/mL) | 0.260 | -0.056-0.576 | 0.103 |
| Intrahepatic HBV DNA (log10copies/105 cell) | 1.043 | 0.775-1.311 | <0.001 |
| Inflammation scores‡ | -0.016 | -0.212- -0.021 | 0.018 |
| fibrosis scores‡ | -0.284 | -0.739-0.179 | 0.215 |
‡Hepatic inflammation grade and fibrosis stage were diagnosed according to the modified knodell and Ishak scoring system respectively.
Abbreviations: cccDNA, covalently closed circular DNA; HBV, hepatitis B virus; HBV RNA, hepatitis B virus ribonucleic acid; HBcrAg, hepatitis B core-related antigen; HBeAg, hepatitis B e antigen; HBsAg, hepatitis B surface antigen.
Correlation analysis of viral markers at baseline in HBeAg positive patients. ADV, Adefovir dipivoxil; cccDNA, covalently closed circular DNA; corr, correlation coefficient; ETV, Entacavir; HBsAg, hepatitis B surface antigen; HBcrAg, hepatitis B core-related antigen; intra DNA, intrahepatic HBV DNA; RNA+DNA, HBV RNA plus DNA; RNA/DNA, HBV RNA to HBV DNA ratio. *** indicated P < 0.001, ** indicated P < 0.01, * indicated P < 0.05.
Correlation of intrahepatic cccDNA with serum viral markers stratified by inflammation and fibrosis scores
| Parameters | cccDNA | cccDNA | ||||||
|---|---|---|---|---|---|---|---|---|
| Inflammation≤7 | Inflammation>7 | Fibrosis≤3 | Fibrosis>3 | |||||
| r | P | r | P | r | P | r | P | |
| HBsAg (log10 IU/mL) | 0.368 | 0.084 | -0.248 | 0.266 | 0.257 | 0.149 | -0.420 | 0.175 |
| HBV DNA (log10 IU/mL) | 0.656 | <0.001 | 0.632 | 0.001 | 0.526 | 0.001 | 0.880 | <0.001 |
| HBV RNA (log10 copies/mL) | 0.580 | 0.003 | 0.596 | 0.002 | 0.496 | 0.002 | 0.688 | 0.007 |
| HBV RNA/DNA | -0.401 | 0.052 | 0.068 | 0.747 | -0.017 | 0.924 | -0.519 | 0.057 |
| HBV DNA plus RNA | 0.728 | <0.001 | 0.657 | <0.001 | 0.611 | <0.001 | 0.824 | <0.001 |
| HBcrAg (log10 U/mL) | 0.567 | 0.006 | 0.519 | 0.019 | 0.592 | <0.001 | 0.436 | 0.180 |
Hepatic inflammation grade and fibrosis stage were diagnosed according to the modified knodell and Ishak scoring system respectively.
Abbreviations: cccDNA, covalently closed circular DNA; HBV, hepatitis B virus; HBV RNA, hepatitis B virus ribonucleic acid; HBcrAg, hepatitis B core-related antigen; HBeAg, hepatitis B e antigen; HBsAg, hepatitis B surface antigen.
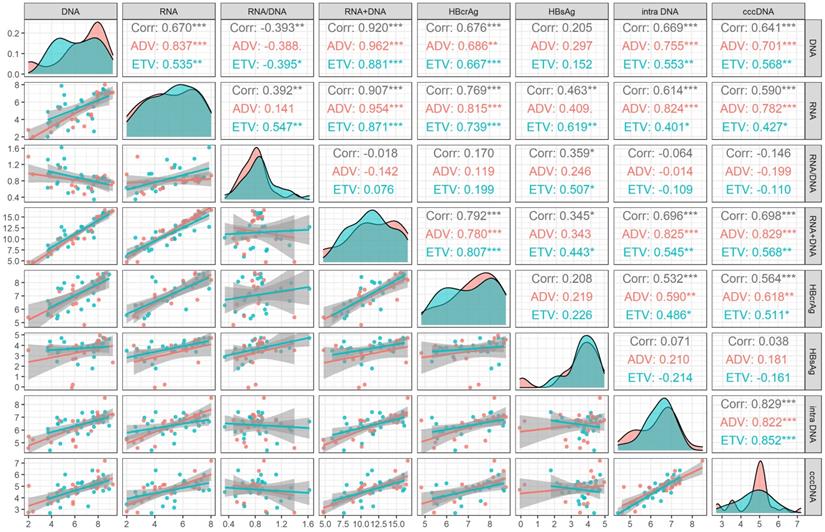
At month 60 of NAs treatment, intrahepatic HBV DNA was still significantly correlated with cccDNA level (r=0.517, P<0.001). None of the serum viral markers (HBsAg, HBV DNA, HBV RNA, HBV RNA/DNA, HBV DNA plus RNA, and HBcrAg) were correlated with cccDNA level. The correlation of different drug subgroups were shown in Figure 2.
During 60 months' treatment, the correlation coefficient between cccDNA decline and decreased serum HBV RNA plus DNA (r=0.525, P=0.001) was higher than that of HBV DNA (r=0.431, P=0.003), HBV RNA (r=0.384, P=0.008), HBsAg (r=0.342, P=0.029), HBcrAg (r=0.268, P=0.103) and HBV RNA/DNA (r=-0.159, P=0.334), only lower than the correlation coefficient between cccDNA decline and decreased intrahepatic HBV DNA (r=0.752, P<0.001), the correlation of different drug subgroups were shown in Figure 3.
Correlation coefficients changes between HBV DNA plus RNA and other serum viral markers
The correlation coefficients between HBV DNA plus RNA and HBsAg (r=0.269, P=0.112), HBV DNA (r=0.672, P<0.001) HBV RNA (r=0.843, P<0.001) and HBcrAg (r=0.620, P<0.001) at month 60 were lower than these corresponding correlation coefficients at baseline [HBsAg (r=0.345, P=0.025), HBV DNA (r=0.920, P<0.001), HBV RNA(r=0.907, P<0.001), and HBcrAg (r=0.792, P<0.001)]. On the contrary, the correlation coefficient between HBV DNA plus RNA and HBV RNA/DNA at month 60 (r=0.543, P<0.001) was higher than that at baseline (r=-0.018, P =0.905), as shown in Figure 1 and Figure 2.
Correlation analysis of viral markers at month 60 in HBeAg positive patients. ADV, Adefovir dipivoxil; cccDNA, covalently closed circular DNA; corr, correlation coefficient; ETV, Entacavir; HBsAg, hepatitis B surface antigen; HBcrAg, hepatitis B core-related antigen; intra DNA, intrahepatic HBV DNA; RNA+DNA, HBV RNA plus DNA; RNA/DNA, HBV RNA to HBV DNA ratio. *** indicated P < 0.001, ** indicated P < 0.01, * indicated P < 0.05.
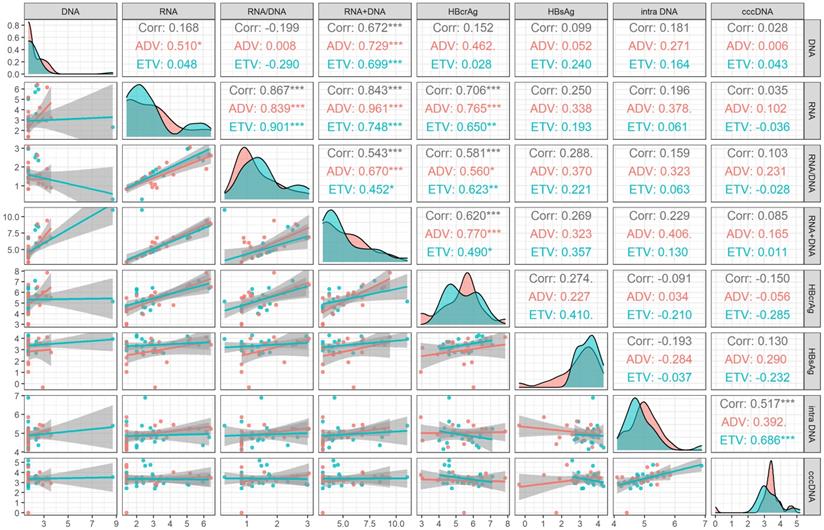
Correlation analysis of the changed value of viral markers after 60 months' treatment in HBeAg positive patients. ADV, Adefovir dipivoxil; cccDNA, covalently closed circular DNA; corr, correlation coefficient; ETV, Entacavir; HBsAg, hepatitis B surface antigen; HBcrAg, hepatitis B core-related antigen; intra DNA, intrahepatic HBV DNA; RNA+DNA, HBV RNA plus DNA; RNA/DNA, HBV RNA to HBV DNA ratio. *** indicated P < 0.001, ** indicated P < 0.01, * indicated P < 0.05.
Discussion
Based on real-life clinical practice, the present study represented a head-to-head comparison of these serum viral markers in reflecting intrahepatic cccDNA levels. We found that baseline serum HBV DNA plus RNA was better positively associated with cccDNA than HBsAg, HBV RNA, HBV DNA, HBV RNA/DNA and HBcrAg in HBeAg positive CHB patients, and this correlation were stable in patients with different inflammation and fibrosis scores. However, this association disappeared after 60 months' NAs treatment. Furthermore, the decrease of HBV DNA plus RNA was also better positively correlated with the decline of intrahepatic cccDNA than other serum viral markers during 60 months NA treatment.
According to our results, serum viral markers positively correlated with cccDNA in HBeAg positive CHB patients at baseline, but this correlation was not consistent after 60 months' NAs treatment. This was consistent with previous studies that serum HBV RNA was positively correlated with cccDNA level before NAs treatment in HBeAg positive CHB patients [14, 22], but was not correlated with cccDNA after 96 weeks of NAs treatment [14]. However, in the same study, intrahepatic cccDNA positively correlated with HBV DNA, but not with HBsAg before NAs treatment. Interestingly, after 96 weeks' NAs treatment, intrahepatic cccDNA correlated with HBsAg, but not with HBV DNA [14]. In the present study, we found that the positive correlation between cccDNA and HBV RNA plus DNA and HBcrAg lost after 60 months' NAs treatment. Consistent with previous studies [9,14], our results indicated that HBV DNA was positively correlated with intrahepatic cccDNA before NAs treatment while this correlation disappeared after NAs treatment. Besides, HBsAg was not correlated with intrahepatic cccDNA levels neither before nor after NAs treatment, the discrepancies among these studies might due to the heterogeneity of the study population.
To our knowledge, the present study was the first head-to-head comparison study of serum HBcrAg and HBV RNA plus DNA in reflecting intrahepatic cccDNA level. Previously studies have reported that serum HBcrAg may be better than HBV RNA in reflecting intrahepatic cccDNA level before treatment [22]. We did found that HBcrAg positively correlated with intrahepatic cccDNA level before NAs treatment in HBeAg positive CHB patients (r=0.564, P<0.001). However, the correlation was weaker than that between HBV RNA and cccDNA (r=0.590, P<0.001), as well as between HBV RNA plus DNA and cccDNA (r=0.698, P<0.001). Besides, HBcrAg did not show statistical significance in the multivariate linear regression analysis.
Among the studied serum viral markers (HBsAg, HBV DNA, HBV RNA and HBcrAg), HBV DNA plus RNA achieved the highest correlation with cccDNA. This may be explained by that HBsAg is produced both from cccDNA and HBV DNA integrated into the host genome [27], negatively affected the correlation between HBsAg and cccDNA. Further, serum HBV DNA or HBV RNA alone may be insufficient to reflect the intrahepatic cccDNA since reverse transcription of pgRNA could be blocked by NAs treatment. Moreover, anti-HBe antibodies are cross-reactive with HBcAg due to the amino acid sequence homology [28], interfering against the accurate measurement of HBcrAg. All these factors may potentially influence the correlation between these serum viral markers and cccDNA. Nevertheless, further research is needed to clarify the underlying mechanism.
The promising surrogate marker should also reflect the change of cccDNA. In a previous study, compared with the decrease of serum HBV RNA (r=0.28, P <0.05) and HBV DNA (r =0.35, P=0.01) levels, the decrease of serum HBsAg levels (r =0.38, P <0.01) better reflected the decrease of intrahepatic cccDNA levels after the 96 weeks' NAs treatment [14]. Wang et al. [29] have reported that the decrease of HBcrAg (r=0.282, P=0.043) correlated with the decline of cccDNA level after 96 weeks' NAs therapy in HBeAg positive patients. Our study found that the decrease of serum HBV DNA plus RNA (r=0.525, P=0.001) did the best in reflecting the decline of cccDNA among other markers, including HBV RNA (r=0.384, P=0.008), HBV DNA (r=0.431, P=0.003), and HBsAg (r=0.342, P=0.029) after 60 months' NAs treatment. While the decrease of HBcrAg was not correlated with the decline of cccDNA in our study, which was and inconsistent with previous study [29], the possible reason may be due to different NAs treatment course.
Our study has some limitations. The single-center design and limited sample size may bring bias to the study. Besides, considering the genotype of most patients was B or C, the results of this study should be carefully extrapolated for genotype A and D and for non-Chinese ethnicity. Future studies with a large sample size are needed to confirm the results of this study.
In conclusion, serum HBV DNA plus RNA was better than serum HBsAg, HBV RNA, HBV DNA, HBV RNA/DNA and HBcrAg in reflecting the intrahepatic cccDNA level at baseline and during 60 months' NAs treatment. This result deepens our knowledge and understanding of clinical significance of HBV RNA plus DNA in HBeAg positive CHB patients receiving long-term NAs treatment.
Abbreviations
ADV: Adefovir dipivoxil; ALT: alanine aminotransferase; AST: aspartate aminotransferase; BMI: body mass index; cccDNA: covalently closed circular DNA; CHB: chronic hepatitis B; HBV: hepatitis B virus; HBcrAg: hepatitis B core-related antigen; HBeAg: hepatitis B e antigen; HBsAg: hepatitis B surface antigen; HCC: hepatocellular carcinoma; LLD: lower limit of detection; NAs: nucleos(t)ide analogues; pgRNA: pregenomic RNA; rcDNA: relaxed circular DNA.
Supplementary Material
Supplementary method and table.
Acknowledgements
The authors thank the participants for their contribution to the research.
Ethics approval and consent to participate statement
The study was approved by the Medical Ethics Review Committee of Beijing YouAn Hospital.
All patients provided written informed consent authorizing us to access their medical records and to store the serum specimens for research purposes.
Availability of data and materials
The data and materials that support the findings of this study are available from the corresponding author upon reasonable request.
Funding
This study is supported in part by National Science and Technology Key Project on “Major Infectious Diseases such as HIV/AIDS, Viral Hepatitis Prevention and Treatment” (2017ZX10302201-004, 2017ZX10202203-006, 2017ZX10201201, 2017ZX10203201-005). Beijing Municipal Administration of Hospitals Clinical medicine Development of special funding support (ZYLX202125).
Author contributions
YW, YNL, HL, and ZPD equally contributed to visualization, methodology, analysis, writing and editing the manuscript. DDB, YR, GXY, YYJ and LB contributed to data curation, acquisition of data, review and editing of the manuscript. SL, ML, LZ, YC, XYC and ZPD contributed to project administration, resources, review and editing of the manuscript. FML and SJZ contributed to conceptualization, funding acquisition, designed and supervised the experiments, and critical revision of the manuscript. All authors have read and approved the final version of the manuscript to be published.
Competing Interests
The authors have declared that no competing interest exists.
References
1. Polaris Observatory C. Global prevalence, treatment, and prevention of hepatitis B virus infection in 2016: a modelling study. The lancet Gastroenterology & hepatology. 2018;3:383-403
2. Lok AS, Zoulim F, Dusheiko G, Ghany MG. Hepatitis B cure: From discovery to regulatory approval. Journal of hepatology. 2017;67:847-61
3. Lu F, Wang J, Chen X, Xu D, Xia N. Potential use of serum HBV RNA in antiviral therapy for chronic hepatitis B in the era of nucleos(t)ide analogs. Frontiers of medicine. 2017;11:502-8
4. Nguyen T, Locarnini S. Hepatitis: Monitoring drug therapy for hepatitis B-a global challenge? Nature reviews Gastroenterology & hepatology. 2009;6:565-7
5. Yang HC, Kao JH. Persistence of hepatitis B virus covalently closed circular DNA in hepatocytes: molecular mechanisms and clinical significance. Emerging microbes & infections. 2014;3:e64
6. Wong DK, Yuen MF, Ngai VW, Fung J, Lai CL. One-year entecavir or lamivudine therapy results in reduction of hepatitis B virus intrahepatic covalently closed circular DNA levels. Antiviral therapy. 2006;11:909-16
7. Werle-Lapostolle B, Bowden S, Locarnini S, Wursthorn K, Petersen J, Lau G. et al. Persistence of cccDNA during the natural history of chronic hepatitis B and decline during adefovir dipivoxil therapy. Gastroenterology. 2004;126:1750-8
8. Sung JJ, Wong ML, Bowden S, Liew CT, Hui AY, Wong VW. et al. Intrahepatic hepatitis B virus covalently closed circular DNA can be a predictor of sustained response to therapy. Gastroenterology. 2005;128:1890-7
9. Chen EQ, Feng S, Wang ML, Liang LB, Zhou LY, Du LY. et al. Serum hepatitis B core-related antigen is a satisfactory surrogate marker of intrahepatic covalently closed circular DNA in chronic hepatitis B. Scientific reports. 2017;7:173
10. Li J, Sun X, Fang J, Wang C, Han G, Ren W. Analysis of intrahepatic total HBV DNA, cccDNA and serum HBsAg level in Chronic Hepatitis B patients with undetectable serum HBV DNA during oral antiviral therapy. Clinics and research in hepatology and gastroenterology. 2017;41:635-43
11. Li W, Zhao J, Zou Z, Liu Y, Li B, Sun Y. et al. Analysis of hepatitis B virus intrahepatic covalently closed circular DNA and serum viral markers in treatment-naive patients with acute and chronic HBV infection. PloS one. 2014;9:e89046
12. Saitta C, Tripodi G, Barbera A, Bertuccio A, Smedile A, Ciancio A. et al. Hepatitis B virus (HBV) DNA integration in patients with occult HBV infection and hepatocellular carcinoma. Liver international: official journal of the International Association for the Study of the Liver. 2015;35:2311-7
13. Wang J, Du M, Huang H, Chen R, Niu J, Jiang J. et al. Reply to: "Serum HBV pgRNA as a clinical marker for cccDNA activity": Consistent loss of serum HBV RNA might predict the "para-functional cure" of chronic hepatitis B. Journal of hepatology. 2017;66:462-3
14. Gao Y, Li Y, Meng Q, Zhang Z, Zhao P, Shang Q. et al. Serum Hepatitis B Virus DNA, RNA, and HBsAg: Which Correlated Better with Intrahepatic Covalently Closed Circular DNA before and after Nucleos(t)ide Analogue Treatment? Journal of clinical microbiology. 2017;55:2972-82
15. Wang J, Shen T, Huang X, Kumar GR, Chen X, Zeng Z. et al. Serum hepatitis B virus RNA is encapsidated pregenome RNA that may be associated with persistence of viral infection and rebound. Journal of hepatology. 2016;65:700-10
16. Wang J, Yu Y, Li G, Shen C, Meng Z, Zheng J. et al. Relationship between serum HBV-RNA levels and intrahepatic viral as well as histologic activity markers in entecavir-treated patients. Journal of hepatology. 2017 S0168-8278(17): 32261-4
17. Wang ML, Chen EQ, Tao CM, Tang H. Letter: serum HBcrAg is a useful marker for disease monitoring, predicting treatment response and disease outcome of chronic hepatitis B virus infection. Alimentary pharmacology & therapeutics. 2018;47:1719-20
18. Testoni B, Lebosse F, Scholtes C, Berby F, Miaglia C, Subic M. et al. Serum hepatitis B core-related antigen (HBcrAg) correlates with covalently closed circular DNA transcriptional activity in chronic hepatitis B patients. Journal of hepatology. 2019;70:615-25
19. Mak LY, Wong DK, Cheung KS, Seto WK, Lai CL, Yuen MF. Review article: hepatitis B core-related antigen (HBcrAg): an emerging marker for chronic hepatitis B virus infection. Alimentary pharmacology & therapeutics. 2018;47:43-54
20. Wang J, Chen R, Zhang R, Ding S, Zhang T, Yuan Q. et al. The gRNA-miRNA-gRNA Ternary Cassette Combining CRISPR/Cas9 with RNAi Approach Strongly Inhibits Hepatitis B Virus Replication. Theranostics. 2017;7:3090-105
21. Huang H, Wang J, Li W, Chen R, Chen X, Zhang F. et al. Serum HBV DNA plus RNA shows superiority in reflecting the activity of intrahepatic cccDNA in treatment-naive HBV-infected individuals. Journal of clinical virology: the official publication of the Pan American Society for Clinical Virology. 2018;99-100:71-8
22. Chen EQ, Wang ML, Tao YC, Wu DB, Liao J, He M. et al. Serum HBcrAg is better than HBV RNA and HBsAg in reflecting intrahepatic covalently closed circular DNA. Journal of viral hepatitis. 2019;26:586-95
23. Lok AS, McMahon BJ. Chronic hepatitis B. Hepatology. 2007;45:507-39
24. Goodman ZD. Grading and staging systems for inflammation and fibrosis in chronic liver diseases. Journal of hepatology. 2007;47:598-607
25. Liao H, Liu Y, Li X, Wang J, Chen X, Zou J. et al. Monitoring of serum HBV RNA, HBcrAg, HBsAg and anti-HBc levels in patients during long-term nucleoside/nucleotide analogue therapy. Antiviral therapy. 2019;24:105-15
26. Tsuge M, Murakami E, Imamura M, Abe H, Miki D, Hiraga N. et al. Serum HBV RNA and HBeAg are useful markers for the safe discontinuation of nucleotide analogue treatments in chronic hepatitis B patients. Journal of gastroenterology. 2013;48:1188-204
27. Li X, Zhang J, Yang Z, Kang J, Jiang S, Zhang T. et al. The function of targeted host genes determines the oncogenicity of HBV integration in hepatocellular carcinoma. Journal of hepatology. 2014;60:975-84
28. Wang SJ, Chen ZM, Wei M, Liu JQ, Li ZL, Shi TS. et al. Specific determination of hepatitis B e antigen by antibodies targeting precore unique epitope facilitates clinical diagnosis and drug evaluation against hepatitis B virus infection. Emerging microbes & infections. 2021;10:37-50
29. Wang L, Cao X, Wang Z, Gao Y, Deng J, Liu X. et al. Correlation of HBcrAg with Intrahepatic Hepatitis B Virus Total DNA and Covalently Closed Circular DNA in HBeAg-Positive Chronic Hepatitis B Patients. Journal of clinical microbiology. 2019 57
Author contact
![]() Corresponding authors: Prof. Sujun Zheng, Artificial Liver Center, Beijing YouAn Hospital, Capital Medical University, 8 Xitoutiao, Youwai Street, Beijing 100069, China. Tel.: 086 10 63291007; Fax: 086 10 63295285; E-mail: zhengsujunedu.cn; and Prof. Fengmin Lu, Department of Microbiology and Infectious Disease Center, Peking University Health Science Center, 38 Xueyuan Road, Haidian 22 District, Beijing 100191, China. Tel.: 086 10 82805136; Fax: 86 10 82805136; E-mail: lu.fengminpku.edu.cn.
Corresponding authors: Prof. Sujun Zheng, Artificial Liver Center, Beijing YouAn Hospital, Capital Medical University, 8 Xitoutiao, Youwai Street, Beijing 100069, China. Tel.: 086 10 63291007; Fax: 086 10 63295285; E-mail: zhengsujunedu.cn; and Prof. Fengmin Lu, Department of Microbiology and Infectious Disease Center, Peking University Health Science Center, 38 Xueyuan Road, Haidian 22 District, Beijing 100191, China. Tel.: 086 10 82805136; Fax: 86 10 82805136; E-mail: lu.fengminpku.edu.cn.

 Global reach, higher impact
Global reach, higher impact