Impact Factor
ISSN: 1449-1907
Int J Med Sci 2022; 19(4):762-768. doi:10.7150/ijms.69868 This issue Cite
Research Paper
Visfatin Polymorphisms, Lifestyle Risk Factors and Risk of Oral Squamous Cell Carcinoma in a Cohort of Taiwanese Males
1. School of Dentistry, China Medical University, Taichung 40402, Taiwan.
2. Department of Dentistry, China Medical University Hospital, Taichung 40447, Taiwan.
3. School of Medicine, Chung Shan Medical University, Taichung 40201, Taiwan.
4. Department of Psychiatry, Chung Shan Medical University Hospital, Taichung 40201, Taiwan.
5. School of Medicine, China Medical University, Taichung 40402, Taiwan.
6. Division of Hematology and Oncology, Department of Internal Medicine, China Medical University Hospital, Taichung 404332, Taiwan.
7. Institute of Medicine, Chung Shan Medical University, Taichung 40201, Taiwan.
8. Department of Medical Research, Chung Shan Medical University Hospital, Taichung 40201, Taiwan.
9. Graduate Institute of Biomedical Science, China Medical University, Taichung 40402, Taiwan.
10. Chinese Medicine Research Center, China Medical University, Taichung 40402, Taiwan.
11. Department of Biotechnology, College of Health Science, Asia University, Taichung 41354, Taiwan.
Received 2021-12-8; Accepted 2022-3-18; Published 2022-4-11
Abstract
Oral cancer is the eighth greatest generally diagnosed cancer amongst males worldwide and the fourth most generally malignancy amongst Taiwanese males. The pro-inflammatory adipocytokine visfatin promotes tumor growth. Elevated plasma visfatin levels have been identified in patients with oral squamous cell carcinoma (OSCC), although the biological mechanisms underlying the involvement of visfatin in the pathogenesis of OSCC are not well understood. Moreover, no information is available regarding associations between visfatin polymorphisms and carcinogenic lifestyle factors with OSCC. This study, therefore, investigated the effects of four visfatin gene polymorphisms (rs11977021, rs61330082, rs2110385, and rs4730153) and carcinogenic lifestyle factors (betel nut chewing, alcohol consumption and cigarette smoking) on the risk of developing OSCC in 1,275 Taiwanese males with OSCC, and 1,195 healthy males (controls). We also examined the associations between these visfatin genotypes and OSCC histopathological prognostic factors (pathological stage, tumor status, lymph node status, and metastasis). We found that compared with subjects with the CC genotype of SNP rs11977021, those with the CT+TT genotype were less likely to progress OSCC. In addition, an association was found between the rs4730153 variant and lymph node metastasis in the OSCC cohort.
Keywords: genetic polymorphisms, oral squamous cell carcinoma, visfatin
Introduction
Oral cancer is the eighth most generally diagnosed cancer amongst males worldwide [1] and the fourth most common malignancy amongst Taiwanese males [2]. Of all oral cancers, oral squamous cell carcinoma (OSCC) is the greatest general (representing approximately 90% of all malignancies of the oral cavity) [3]. Major risk factors for OSCC include human papillomavirus infection [4, 5] and the habitual consumption of carcinogens such as betel nuts, tobacco and alcohol [5]. Around 86% of Taiwanese oral tumor patients are habitual betel nut chewers [2]. Oral cancer is also associated with genetic aberrations resulting from disturbances in carcinogen metabolism, DNA repair and cell cycle control [6].
The proinflammatory adipocytokine visfatin, also known as nicotinamide phosphoribosyltransferase (NAMPT), is abundantly expressed in human and mouse visceral adipose tissue [7]. Increased serum visfatin levels are positively associated with body mass index, cholesterol and insulin resistance index values, and negatively correlated with triglyceride levels in patients with coronary artery calcification [7]. Visfatin has also been detected in tumor tissue or plasma from patients with various types of cancers [8], high levels of visfatin in tissue or plasma are associated with human pancreatic ductal adenocarcinoma [9], breast tumor [10], clear cell renal cell carcinoma (ccRCC) [11] and thyroid malignancy [12]. And the previous research demonstrated that high levels of visfatin are associated with tumorigenesis through different molecular signaling pathways and genetic polymorphisms [13-15].
Polymorphisms in the visfatin gene are associated with breast cancer and coronary artery disease [7]. Visfatin expression affects the regulation of various important factors in development related to tumor progression including tumor survival rate, metastasis and resistance [8]. Although elevated plasma visfatin levels have been identified in patients with OSCC [13], the biological mechanisms underlying the involvement of visfatin in the pathogenesis of OSCC are not well understood. Moreover, no information is available regarding associations between visfatin polymorphisms and carcinogenic lifestyle factors with OSCC. This study therefore investigated the effects of visfatin gene polymorphisms and carcinogenic lifestyle factors on the risk of developing OSCC in a cohort of Taiwanese males. We also examined associations between the visfatin genotypes and OSCC histopathological prognostic factors (pathological stage, tumor status, lymph node status, and metastasis).
Materials and Methods
Study participants
1,275 male patients in this study were diagnosed with OSCC at Chung Shan Medical University Hospital, Taichung, Taiwan, between January 1, 2010 and December 31, 2019. We randomly selected 1,195 healthy controls without any history of cancer or oral precancerous disease from the Taiwan Biobank, an ongoing prospective study that has been collecting information since 2012 about genes, lifestyle, and environmental risk factors from over 150,000 individuals aged 30-70 years throughout Taiwan. Demographic data and carcinogenic lifestyle behaviors (betel nut chewing, cigarette smoking, alcohol consumption) were recorded. Daily smokers were defined as persons who had smoked at least one cigarette daily in the previous 3 months. Alcohol consumers were defined as persons who consumed more than two alcoholic beverages per day on average. OSCC was graded using the 2018 American Joint Committee on Cancer (AJCC) Cancer Staging Manual (8th edition) [16]. A pathologist rated tumor cell differentiation by AJCC classification criteria. All procedures of the study involving human participants satisfied the Declaration of Helsinki criteria. All authors had access to the study data and reviewed and approved this study. Chung Shan Medical University Hospital's Institutional Review Board approved the study investigation (IRB No.CS2-21063). Each participant provided written informed consent before enrolling into the investigation.
Selection and genotyping of SNPs
Visfatin single-nucleotide polymorphisms (SNPs) rs11977021, rs61330082, rs2110385 and rs4730153 were chosen on the basis of prior research showing that they increased the risk of various cancers [14, 17-19]. All SNPs had minor allele frequencies exceeding 5%. Genomic DNA was extracted from 3 mL peripheral blood samples using QIAamp DNA Blood Kits (Qiagen, CA, USA). Allelic discrimination of the SNPs followed previously described assessment procedures [20-22]. RNA isolation and RT-qPCR assays followed our previously described procedures [23-25].
Bioinformatics analysis
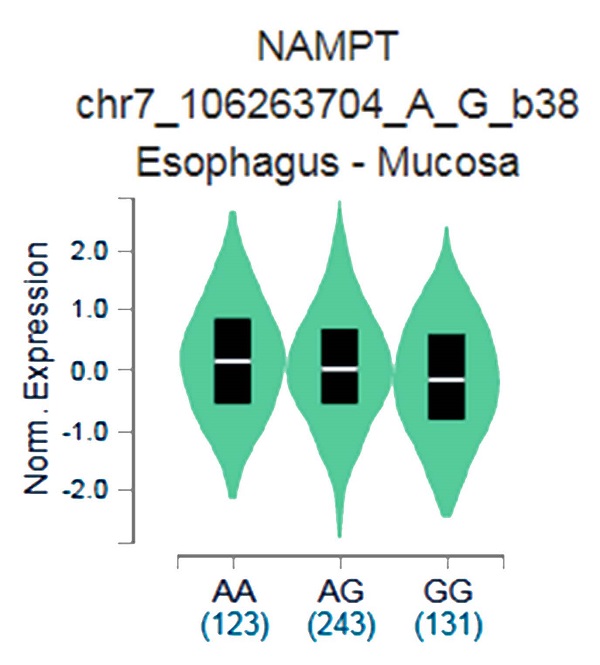
The putative functional relevance of visfatin polymorphisms was examined. Data collected from the Genotype-Tissue Expression (GTEx) database were used to recognize relationship between rs4730153 and visfatin performance in esophagus mucosal tissue [26, 27].
Statistical analysis
Previous research has reported visfatin rs11977021 frequencies for CC, CT and TT genotypes among controls of 23.2%, 54.5% and 22.3%, respectively, and corresponding visfatin rs61330082 frequencies for CC, CT and TT genotypes among controls of 39.7%, 45.1% and 15.2%, respectively [17]. Another publication has reported visfatin rs2110385 frequencies for GG, GT and TT genotypes among controls of 75.6%, 24.4% and 0%, respectively (NCBI dbSNP HapMap-HCB population), and visfatin rs4730153 frequencies for GG, GA and AA genotypes among controls of 84.1%, 15.9% and 0%, respectively [28]. The frequencies of visfatin polymorphisms in these two studies are comparable with our control samples. We only considered the rs61330082 genotype for calculating the sample size. Given a type I error (α) level of 0.05, type II error (β) level of 0.8, rs61330082 TT genotype among healthy controls of 0.15, odds ratio (OR) of 0.609, the minimum sample size required for cases is 1,195. We therefore recruited 1,195 controls and 1,275 cases for study research.
The differences between the OSCC and control groups were analyzed by the Mann-Whitney U test and the Fisher's exact test; between group differences were considered to be statistically significant if p-values were <0.05. Logistic regression was used to calculate odds ratios (ORs) and their 95% confidence intervals (CIs) for associations between genotype frequencies and the risk of OSCC. ORs were adjusted for age, betel quid chewing, cigarette smoking, and alcohol consumption. Clinical staging of OSCC disease was determined in OSCC patients with the variant rs4730153. All collective data were analyzed using Statistical Analytic System (SAS) software version 9.1 for Windows (SAS Institute Inc., CA, USA).
Results
The study enrolled 1,275 males with OSCC and 1,195 healthy, cancer-free males (controls); all were Taiwanese and living in Taichung, Taiwan. Baseline characteristics of the cases and controls, as well as clinical staging of the OSCC cohort, are shown in Table 1. Approximately half of each study group was aged less than 55 years. The mean age was 53.90 ± 10.02 years in the control group and 55.57 ± 10.76 years in the OSCC group. Compared with controls, higher proportions of patients with OSCC chewed betel nuts, were cigarette smokers and consumed alcohol (p<0.001 for all comparisons). Based on the AJCC tumor/node/metastasis (TNM) classification and staging system, 47.0% of patients had clinical stage I/II OSCC and 53.0% had clinical stage III/IV disease; the proportions of patients classified as T1+T2 or T3+T4 status were similar (50.1% and 49.9%, respectively), 65.7% were N0 lymph node status and 34.3% were N1+N2+N3 status, and nearly all patients were metastasis-free (99.2% vs 0.8% with M1 status). OSCC was moderately or poorly differentiated in 86.1% of patients and well-differentiated in 13.9%.
The results of genotyping for the four visfatin SNPs (rs11977021, rs61330082, rs2110385 and rs4730153) in the cases and controls are shown in Table 2. Having the CT+TT genotype of SNP rs11977021 diminish the risk of progressing OSCC, compared with having the CC genotype (adjusted OR [AOR] 0.801; 95% CI, 0.642 to 0.998; p=0.048). No significant associations were observed between variants rs61330082, rs2110385 and rs4730153 and OSCC. When visfatin genotypes in cases and controls were evaluated to examine the roles of carcinogenic lifestyle factors on the occurrence of OSCC, we identified that amongst study participants who consumed alcohol, those carrying the AA allele at rs4730153 were significantly less likely than those with the wild-type GG allele to develop OSCC (AOR 0.387; 95% CI, 0.071 to 2.120; p=0.046) (Table 3).
The distributions of demographical characteristics in 1,195 controls and 1,275 male patients with OSCC
| Variable | Controls (N=1,195) | Patients (N=1,275) | p value |
|---|---|---|---|
| Age (yrs) | |||
| ≤55 | 607 (50.8%) | 640 (50.2%) | p=0.766 |
| >55 | 588 (49.2%) | 635 (49.8%) | |
| Betel quid chewing | |||
| No | 996 (83.4%) | 325 (25.5%) | |
| Yes | 199 (16.6%) | 950 (74.5%) | p <0.001* |
| Daily smoker | |||
| No | 562 (47.0%) | 203 (15.9%) | |
| Yes | 633 (53.0%) | 1072 (84.1%) | p <0.001* |
| Alcohol consumer | |||
| No | 961 (80.4%) | 674 (52.9%) | |
| Yes | 234 (19.6%) | 601 (47.1%) | p <0.001* |
| Stage | |||
| I+II | 599 (47.0%) | ||
| III+IV | 676 (53.0%) | ||
| Tumor T status | |||
| T1+T2 | 639 (50.1%) | ||
| T3+T4 | 636 (49.9%) | ||
| Lymph node status | |||
| N0 | 838 (65.7%) | ||
| N1+N2+N3 | 437 (34.3%) | ||
| Metastasis | |||
| M0 | 1265 (99.2%) | ||
| M1 | 10 (0.8%) | ||
| Tumor histological grade | |||
| Well differentiated | 177 (13.9%) | ||
| Moderately or poorly differentiated | 1098 (86.1%) |
The Mann-Whitney U test or Fisher's exact test analyzed differences between healthy controls and patients with oral squamous cell carcinoma. * A p value < 0.05 was statistically significant.
Unadjusted and adjusted odds ratios (ORs) with their 95% confidence intervals (CIs) for OSCC associated with visfatin genotype frequencies
| Variable | Controls (N=1,195) (%) | Patients (N=1,275) (%) | OR (95% CI) | AOR (95% CI)a |
|---|---|---|---|---|
| rs11977021 | ||||
| CC | 319 (26.7%) | 347 (27.2%) | 1.000 (reference) | 1.000 (reference) |
| CT | 602 (50.4%) | 643 (50.4%) | 0.982 (0.813-1.185) | 0.798 (0.631-1.008) |
| TT | 274 (22.9%) | 285 (22.4%) | 0.956 (0.764-1.197) | 0.807 (0.610-1.068) |
| CT+TT | 876 (73.3%) | 928 (72.8%) | 0.974 (0.815-1.163) | 0.801(0.642-0.998)b |
| C allele | 1240 (51.9%) | 1337 (52.4%) | 1.000 (reference) | 1.000 (reference) |
| T allele | 1150 (48.1%) | 1213 (47.6%) | 0.978 (0.875-1.094) | 0.894 (0.778-1.028) |
| rs61330082 | ||||
| GG | 313 (26.2%) | 339 (26.6%) | 1.000 (reference) | 1.000 (reference) |
| GA | 602 (50.4%) | 644 (50.5%) | 0.988 (0.817-1.194) | 0.799 (0.632-1.011) |
| AA | 280 (23.4%) | 292 (22.9%) | 0.963 (0.769-1.205) | 0.814 (0.615-1.076) |
| GA+AA | 882 (73.8%) | 936 (73.4%) | 0.980 (0.819-1.172) | 0.804 (0.644-1.004) |
| G allele | 1228 (51.4%) | 1322 (51.8%) | 1.000 (reference) | 1.000 (reference) |
| A allele | 1162 (48.6%) | 1228 (48.2%) | 0.982 (0.878-1.098) | 0.899 (0.782-1.033) |
| rs2110385 | ||||
| GG | 952 (79.7%) | 1034 (81.1%) | 1.000 (reference) | 1.000 (reference) |
| GT | 231 (19.3%) | 227 (17.8%) | 0.905 (0.738-1.109) | 0.990 (0.769-1.274) |
| TT | 12 (1.0%) | 14 (1.1%) | 1.074 (0.494-2.334) | 1.418 (0.566-3.557) |
| GT+TT | 243 (20.3%) | 241(18.9%) | 0.913 (0.749-1.114) | 1.011 (0.791-1.293) |
| G allele | 2135 (89.3%) | 2295 (90.0%) | 1.000 (reference) | 1.000 (reference) |
| T allele | 255 (10.7%) | 255 (10.0%) | 0.930 (0.774-1.117) | 1.031 (0.822-1.293) |
| rs4730153 | ||||
| GG | 958 (80.2%) | 1046 (82.1%) | 1.000 (reference) | 1.000 (reference) |
| GA | 225 (18.8%) | 216 (16.9%) | 0.879 (0.715-1.081) | 0.983 (0.761-1.269) |
| AA | 12 (1.0%) | 13 (1.0%) | 0.992 (0.451-2.185) | 1.389 (0.549-3.513) |
| GA+AA | 237 (19.8%) | 229(17.9%) | 0.885 (0.723-1.083) | 1.003 (0.782-1.287) |
| G allele | 2141 (89.6%) | 2308 (90.5%) | 1.000 (reference) | 1.000 (reference) |
| A allele | 249 (10.4%) | 242 (9.5%) | 0.902 (0.748-1.086) | 1.023 (0.813-1.287) |
The ORs with their 95% CIs were estimated by logistic regression models;
a The adjusted odds ratios (AORs) with their 95% CIs were estimated by multiple logistic regression analysis controlling for age, betel quid chewing, cigarette smoking, and alcohol consumption;
b p = 0.048.
Unadjusted and adjusted odds ratios (ORs) with their 95% confidence intervals (CIs) for OSCC associated with visfatin genotype frequencies among alcohol consumers
| Variable | Controls (N=234) (%) | Patients (N=601) (%) | OR (95% CI) | AOR (95% CI)a |
|---|---|---|---|---|
| rs11977021 | ||||
| CC | 52 (22.2%) | 148 (24.6%) | 1.000 (reference) | 1.000 (reference) |
| CT | 123 (52.6%) | 319 (53.1%) | 0.911 (0.624-1.330) | 0.803 (0.515-1.252) |
| TT | 59 (25.2%) | 134 (22.3%) | 0.798 (0.514-1.239) | 0.662 (0.394-1.110) |
| CT+TT | 182 (77.8%) | 453 (75.4%) | 0.875 (0.610-1.253) | 0.756 (0.496-1.154) |
| C allele | 227 (48.5%) | 615 (51.2%) | 1.000 (reference) | 1.000 (reference) |
| T allele | 241 (51.5%) | 587 (48.8%) | 0.899 (0.726-1.113) | 0.822 (0.640-1.057) |
| rs61330082 | ||||
| GG | 51 (21.8%) | 148 (24.6%) | 1.000 (reference) | 1.000 (reference) |
| GA | 122 (52.1%) | 315 (52.4%) | 0.890 (0.608-1.302) | 0.763 (0.488-1.194) |
| AA | 61 (26.1%) | 138 (23.0%) | 0.780 (0.503-1.208) | 0.652 (0.389-1.093) |
| GA+AA | 183 (78.2%) | 453 (75.4%) | 0.853 (0.594-1.225) | 0.726 (0.474-1.110) |
| G allele | 224 (47.9%) | 611 (50.8%) | 1.000 (reference) | 1.000 (reference) |
| A allele | 244 (52.1%) | 591 (49.2%) | 0.888 (0.717-1.100) | 0.815 (0.634-1.048) |
| rs2110385 | ||||
| GG | 192 (82.1%) | 498 (82.9%) | 1.000 (reference) | 1.000 (reference) |
| GT | 37 (15.8%) | 99 (16.5%) | 1.032 (0.683-1.559) | 1.016 (0.625-1.650) |
| TT | 5 (2.1%) | 4 (0.6%) | 0.308 (0.082-1.161) | 0.468 (0.097-2.258) |
| GT+TT | 42 (17.9%) | 103 (17.1%) | 0.945 (0.637-1.404) | 0.959 (0.601-1.528) |
| G allele | 421 (90.0%) | 1095 (91.1%) | 1.000 (reference) | 1.000 (reference) |
| T allele | 47 (10.0%) | 107 (8.9%) | 0.875 (0.610-1.256) | 0.911 (0.595-1.395) |
| rs4730153 | ||||
| GG | 195 (83.3%) | 506 (84.2%) | 1.000 (reference) | 1.000 (reference) |
| GA | 34 (14.5%) | 92 (15.3%) | 1.043 (0.681-1.598) | 1.015 (0.616-1.674) |
| AA | 5 (2.2%) | 3 (0.5%) | 0.231 (0.055-0.977) | 0.387 (0.071-2.120) |
| GA+AA | 39 (16.7%) | 95 (15.8%) | 0.939 (0.624-1.411) | 0.947 (0.586-1.531) |
| G allele | 424 (90.6%) | 1104 (91.9%) | 1.000 (reference) | 1.000 (reference) |
| A allele | 44 (9.4%) | 98 (8.1%) | 0.855 (0.589-1.242) | 0.892 (0.574-1.385) |
The ORs with their 95% CIs were estimated by logistic regression models;
a Adjusted odds ratios (AORs) with their 95% CIs were estimated by multiple logistic regression analysis controlling for age, betel quid chewing, and cigarette smoking.
An investigation into the association between OSCC clinical status and allelic distributions of visfatin variant rs4730153 revealed no significant associations (Table 4). Next, we found the effect of visfatin gene polymorphisms and carcinogenic lifestyle factors on the clinical status of OSCC. Among the 950 patients who chewed betel nuts, a correlation of rs4730153 variants (GA vs GG) was found for lymph node metastasis (AOR 1.456; 95% CI, 1.022-2.075; p=0.038), but no association was found between any of the studied genotypes and clinical stage, tumor size, or OSCC differentiation (Table 5).
The GTEx data exhibited that respective carrying the AA allele of variant rs4730153 had significantly higher levels of visfatin compared with patients who had the wild-type GG homozygous genotype (p=0.0029; Fig. 1).
Genotype-Tissue Expression (GTEx) data were used to recognize relevance between visfatin expression and variant rs4730153 in esophagus mucosal tissue.
Unadjusted and adjusted odds ratios (ORs) with their 95% confidence intervals (CIs) for various clinical status categories and genotype frequencies of visfatin rs4730153 in males with OSCC (n=1,275)
| Variable | OR (95% CI) | AOR (95% CI) | ||
|---|---|---|---|---|
| Clinical Stage | ||||
| rs4730153 | Stage I+II (n=599) (%) | Stage III+IV (n=676) (%) | ||
| GG | 496 (82.8%) | 550 (81.4%) | 1.00 | 1.00 |
| GA | 97 (16.2%) | 119 (17.6%) | 1.106 (0.824-1.485) | 1.101 (0.819-1.479) |
| AA | 6 (1.0%) | 7 (1.0%) | 1.052 (0.351-3.152) | 1.045 (0.347-3.148) |
| GA+AA | 103 (17.2%) | 126 (18.6%) | 1.103 (0.828-1.470) | 1.097 (0.822-1.464) |
| Tumor size | ||||
| rs4730153 | ≤ T2 (n=639) (%) | >T2 (n=636) (%) | ||
| GG | 530 (82.9%) | 516 (81.1%) | 1.00 | 1.00 |
| GA | 102 (16.0%) | 114 (17.9%) | 1.148 (0.856-1.539) | 1.140 (0.849-1.531) |
| AA | 7 (1.1%) | 6 (0.9%) | 0.881 (0.294-2.638) | 0.836 (0.277-2.521) |
| GA+AA | 109 (17.1%) | 120 (18.9%) | 1.131 (0.849-1.505) | 1.121 (0.841-1.494) |
| Lymph node metastasis | ||||
| rs4730153 | No (n=838) (%) | Yes (n=437) (%) | ||
| GG | 693 (82.7%) | 353 (80.8%) | 1.00 | 1.00 |
| GA | 135 (16.1%) | 81 (18.5%) | 1.178 (0.869-1.596) | 1.178 (0.867-1.599) |
| AA | 10 (1.2%) | 3 (0.7%) | 0.589 (0.161-2.155) | 0.599 (0.163-2.203) |
| GA+AA | 145 (17.3%) | 84 (19.2%) | 1.137 (0.844-1.532) | 1.138 (0.844-1.536) |
| Metastasis | ||||
| rs4730153 | M0 (n=1265) (%) | M1 (n=10) (%) | ||
| GG | 1038 (82.1%) | 8 (80.0%) | 1.00 | 1.00 |
| GA | 214 (16.9%) | 2 (20.0%) | 1.213 (0.256-5.750) | 1.320 (0.276-6.312) |
| AA | 13 (1.0%) | 0 (0.0%) | - | - |
| GA+AA | 227 (17.9%) | 2 (20.0%) | 1.143 (0.241-5.419) | 1.265 (0.264-6.050) |
| Tumor histopathological grade | ||||
| rs4730153 | ≤ Grade I (n=177) (%) | > Grade I (n=1,098) (%) | ||
| GG | 150 (84.8%) | 896 (81.6%) | 1.00 | 1.00 |
| GA | 25 (14.1%) | 191 (17.4%) | 1.279 (0.814-2.008) | 1.268 (0.806-1.995) |
| AA | 2 (1.1%) | 11 (1.0%) | 0.921 (0.202-4.195) | 0.889 (0.193-4.085) |
| GA+AA | 27 (15.2%) | 202 (18.4%) | 1.252 (0.809-1.939) | 1.240 (0.799-1.924) |
Tumor histopathological grade: grade I: well differentiated; grade II: moderately differentiated; grade III: poorly differentiated;
The adjusted odds ratios (AORs) and 95% CIs were estimated by multiple logistic regression analysis controlling for age, betel quid chewing, cigarette smoking, and alcohol consumption.
Unadjusted and adjusted odds ratios (ORs) with their 95% confidence intervals (CIs) for various clinical status categories and genotype frequencies of visfatin rs4730153 in male betel quid chewers with OSCC (n=950)
| Variable | ||||
|---|---|---|---|---|
| Clinical Stage | OR (95% CI) | AOR (95% CI) | ||
| Stage I+II (n=451) (%) | Stage III+IV (n=499) (%) | |||
| rs4730153 | ||||
| GG | 381 (84.5%) | 403 (80.8%) | 1.00 | 1.00 |
| GA | 64 (14.2%) | 93 (18.6%) | 1.374 (0.970-1.945) | 1.361 (0.960-1.930) |
| AA | 6 (1.3%) | 3 (0.6%) | 0.473 (0.117-1.904) | 0.458 (0.113-1.854) |
| GA+AA | 70 (15.5%) | 96 (19.2%) | 1.297 (0.924-1.819) | 1.284 (0.914-1.804) |
| Tumor size | ||||
| rs4730153 | ≤ T2 (n=484) (%) | >T2 (n=466) (%) | ||
| GG | 399 (82.4%) | 385 (82.6%) | 1.00 | 1.00 |
| GA | 79 (16.3%) | 78 (16.7%) | 1.023 (0.726-1.442) | 1.018 (0.721-1.436) |
| AA | 6 (1.3%) | 3 (0.7%) | 0.518 (0.129-2.087) | 0.490 (0.121-1.984) |
| GA+AA | 85 (17.6%) | 81 (17.4%) | 0.988 (0.706-1.381) | 0.980 (0.700-1.372) |
| Lymph node metastasis | ||||
| rs4730153 | No (n=633) (%) | Yes (n=317) (%) | ||
| GG | 532 (84.0%) | 252 (79.5%) | 1.00 | 1.00 |
| GA | 93 (14.7%) | 64 (20.2%) | 1.453 (1.022-2.065)a | 1.456 (1.022-2.075)b |
| AA | 8 (1.3%) | 1 (0.3%) | 0.264 (0.033-2.122) | 0.260 (0.032-2.103) |
| GA+AA | 101 (16.0%) | 65 (20.5%) | 1.359 (0.961-1.920) | 1.361 (0.961-1.929) |
| Tumor histopathological grade | ||||
| rs4730153 | ≤ Grade I (n=144) (%) | > Grade I (n=806) (%) | ||
| GG | 121 (84.0%) | 663 (82.3%) | 1.00 | 1.00 |
| GA | 21 (14.6%) | 136 (16.9%) | 1.182 (0.718-1.946) | 1.177 (0.713-1.941) |
| AA | 2 (1.4%) | 7 (0.8%) | 0.639 (0.131-3.112) | 0.604 (0.123-2.972) |
| GA+AA | 23 (16.0%) | 143 (17.7%) | 1.135 (0.701-1.836) | 1.127 (0.695-1.827) |
Tumor histopathological grade: grade I: well differentiated; grade II: moderately differentiated; grade III: poorly differentiated;
The adjusted odds ratios (AORs) with their 95% CIs were estimated by multiple logistic regression analysis controlling for age, cigarette smoking, and alcohol consumption;
a p = 0.038;
b p = 0.038.
Discussion
OSCC comprises over 90% of oral cancers [29] and although medical progress has witnessed much improvement in the early detection and treatment of OSCC [30], overall 5-year survival is only around 50% [31]. Important predictors of outcome include the extent of disease at presentation, whether the patient has clinically palpable lymph nodes and histopathological findings [32]. The development of OSCC is influenced by carcinogenic lifestyle behaviors (e.g., betel nut chewing, tobacco smoking and alcohol consumption) [5]. According to literature in the Investigation of Nutritional Status and its Clinical Outcomes of Common Cancer (INSCOC) database, research has shown that malnutrition lowers quality of life and increases the risk of infection, the incidence of postoperative complications and the mortality rate [33]. The association between nutritional status with treatment outcomes in patients with OSCC should therefore be considered. Our analysis of these factors found that all three potentially facilitate the development of OSCC. This finding is echoed in recent studies that have focused on associations between gene polymorphisms and the occurrence and prognosis of oral cancer [34-36], while carcinogenic lifestyle factors specifically elicit epigenetic changes that increase rates of oral cancers [37, 38].
Visfatin is mostly (not exclusively) secreted by adipose tissue [39] and once released into the extracellular milieu, can behave in a paracrine or endocrine manner in various physiological and pathological conditions, such as cancers [39, 40]. High levels of circulating visfatin are reportedly significantly associated with the risk of cancer [41], while higher plasma visfatin levels have been reported in male patients with OSCC, independently of risk factors, which suggests that visfatin is an important contributor to the pathogenesis of OSCC [12]. Epidemiological studies have demonstrated higher alcohol consumption increases the risk of OSCC, with strong dose-response relationships [42, 43]. The risk of OSCC is three to five times higher among people who have ever consumed alcohol than people who have never consumed alcohol [44, 45]. Previous studies have described associations between other SNPs and OSCC risk: for instance, the A>G polymorphism with the rs1412115 variant increased the risk of OSCC in a Chinese Han population [46], while MDM2 SNP 309 and p53 codon 72 polymorphisms influence treatment outcomes of OSCC patients receiving postoperative irradiation [47]. In this study, we selected four visfatin gene polymorphisms (rs11977021, rs61330082, rs2110385, and rs4730153) to compare their allelic distributions between healthy, cancer-free subjects and patients with OSCC. We found that OSCC was more likely to develop in subjects who had the CC genotype of SNP rs11977021 compared with those who had the CT+TT genotype (Table 2). In our analysis of alcohol consumption, we observed that OSCC was significantly less likely to develop among study participants carrying the AA allele at rs4730153 compared with those carrying the wild-type GG allele (Table 3). This evidence indicates a critical role for alcohol consumption in association with visfatin polymorphisms and OSCC. Further research on the consequences of SNP variations at the protein level combined with the effect of alcohol consumption may provide insights into new therapeutic targets for OSCC.
Evidence has shown that visfatin promotes the growth of cancers, their metastasis and resistance to cancer therapies [8, 48]. Clinically, visfatin levels are associated with metastasis in different types of cancer. Elevated serum visfatin levels are found in patients with small cell lung cancer (SCLC) and brain metastasis [49]. In non-SCLC, elevated visfatin plasma levels are correlated with lymph node metastasis [49]. In colorectal cancer patients, upregulation of circulating serum visfatin is correlated with lymph node metastasis, inflammation and angiogenesis [50]. In patients with prostate cancer, visfatin increased the activity and expression of MMP-2 and MMP-9 [51]. Similar associations between visfatin and lymph node metastasis have been recorded in breast and gastric cancers [52, 53]. In this study, we selected visfatin rs4730153 to analyze associations between clinical status and genotypic frequencies in OSCC patients. As shown in Table 4, no differences exist between visfatin rs4730153, various clinical status categories and genotypic frequencies in patients with OSCC. We found that amongst OSCC patients who chewed betel nuts, the GA allele at rs4730153 was significantly associated with lymph node metastasis (Table 5). The result demonstrated that the role of visfatin rs4730153 in OSCC neck lymph node metastasis is probably similar with non-SCLC, prostate, breast and gastric cancers. But, the detail mechanism between the betel nuts chewing, rs4730153 variation and lymph node metastasis should be further investigation.
This study revealed significant associations between alcohol consumption and betel nut chewing and the development of OSCC in Taiwanese males with at least one visfatin polymorphism. The GTEx database has revealed a trend for higher visfatin expression amongst carriers of the AA genotype at rs4730153 compared with carriers of wild-type GG homozygous genotypes, which is mirrored by our SNP data and suggests that the SNP rs4730153 may increase visfatin expression. Limitations of this study need to be addressed. Firstly, information on underlying disease processes is not available from the Taiwan Databank, which meant that the control sample in this study could not be fully investigated regarding disease pathways. Furthermore, the long-term survival rate is also highly affected by the recall rate and will of the patient. More complex investigations using larger study samples and longer follow-up are required to examine associations between visfatin SNPs and OSCC.
In conclusion, this study discusses preliminary results of associations observed between visfatin polymorphisms and carcinogenic lifestyle behaviors in a cohort of Taiwanese males with OSCC. Compared with having the CT+TT genotype, having the CC genotype of SNP rs11977021 increased the risk of developing OSCC. Among the study participants who drank alcohol, the AA allele at rs4730153 was inversely associated with the occurrence of OSCC, suggesting a protective effect. In contrast, among those who chewed betel nuts, the GA allele at rs4730153 was significantly associated with lymph node metastasis.
Acknowledgements
We would like to thank Iona J. MacDonald from China Medical University for her editing of this manuscript.
Funding
This work was supported by grants from the Ministry of science and Technology in Taiwan (MOST 110-2320-B-039-022-MY3) and the China Medical University Hospital (DMR-111-045; DMR-111-023).
Author Contributions
Shun-Fa Yang and Chih-Hsin Tang conceived and designed the project. Chih-Hsin Tang supervised the project. The experiments and data analysis were performed by Ming-Hong Hsieh, Kwei-Jing Chen and Yen-You Lin. Reagents, materials, and analysis tools were supplied by Kwei-Jing Chen, Ming-Yu Lien, and Michael Yuan-Chien Chen. The manuscript was written by Kwei-Jing Chen and Yen-You Lin. The final manuscript was approved by all authors.
Data Availability Statement
The data presented in this study are available on request from the corresponding authors.
Competing Interests
The authors have declared that no competing interest exists.
References
1. Bray F, Ferlay J, Soerjomataram I. et al. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68:394-424
2. Vautrin A, Wesseling M, Wirix-Speetjens R. et al. Time-dependent in silico modelling of orthognathic surgery to support the design of biodegradable bone plates. J Mech Behav Biomed Mater. 2021;121:104641
3. Jemal A, Siegel R, Ward E. et al. Cancer statistics, 2008. CA Cancer J Clin. 2008;58:71-96
4. Gupta K, Metgud R. Evidences suggesting involvement of viruses in oral squamous cell carcinoma. Patholog Res Int. 2013;2013:642496
5. Scully C, Bagan J. Oral squamous cell carcinoma overview. Oral Oncol. 2009;45:301-8
6. Scully C, Field JK, Tanzawa H. Genetic aberrations in oral or head and neck squamous cell carcinoma (SCCHN): 1. Carcinogen metabolism, DNA repair and cell cycle control. Oral Oncol. 2000;36:256-63
7. Jin LW, Zheng SB, Zhou ZH. et al. Correlation between polymorphisms in the visfatin gene and its expression in the serum and coronary artery calcification. Genet Mol Res. 2016 15
8. Lin TC. The role of visfatin in cancer proliferation, angiogenesis, metastasis, drug resistance and clinical prognosis. Cancer Manag Res. 2019;11:3481-91
9. Ju HQ, Zhuang ZN, Li H. et al. Regulation of the Nampt-mediated NAD salvage pathway and its therapeutic implications in pancreatic cancer. Cancer Lett. 2016;379:1-11
10. Sharif T, Ahn DG, Liu RZ. et al. The NAD(+) salvage pathway modulates cancer cell viability via p73. Cell Death Differ. 2016;23:669-80
11. Zhang HP, Zou J, Xu ZQ. et al. Association of leptin, visfatin, apelin, resistin and adiponectin with clear cell renal cell carcinoma. Oncol Lett. 2017;13:463-8
12. Sawicka-Gutaj N, Waligorska-Stachura J, Andrusiewicz M. et al. Nicotinamide phosphorybosiltransferase overexpression in thyroid malignancies and its correlation with tumor stage and with survivin/survivin DEx3 expression. Tumour Biol. 2015;36:7859-63
13. Yu-Duan T, Chao-Ping W, Chih-Yu C. et al. Elevated plasma level of visfatin/pre-b cell colony-enhancing factor in male oral squamous cell carcinoma patients. Med Oral Patol Oral Cir Bucal. 2013;18:e180-6
14. Zhang LJ, Li XQ, Wang CD. et al. The Correlation of Visfatin and Its Gene Polymorphism with Non-Small Cell Lung Cancer. Cancer Biother Radiopharm. 2018;33:460-5
15. Li B, Zhang C, Wang J. et al. Impact of genetic variants of ABCB1, APOB, CAV1, and NAMPT on susceptibility to pancreatic ductal adenocarcinoma in Chinese patients. Mol Genet Genomic Med. 2020;8:e1226
16. Zanoni DKP, S. G. Shah, J. P.. Changes in the 8th Edition of the American Joint Committee on Cancer (AJCC) Staging of Head and Neck Cancer: Rationale and Implications. Curr Oncol Rep. 2019;21:22
17. Wu Z, Sun Y, Huang Y. et al. Genetic variant in visfatin gene promoter contributes to reduced risk of hepatocellular carcinoma in a Chinese population. Oncotarget. 2016;7:77968-77
18. Zhang C, Yan D, Wang S. et al. Genetic polymorphisms of NAMPT related with susceptibility to esophageal squamous cell carcinoma. BMC gastroenterology. 2015;15:49
19. Zhang K, Zhou B, Zhang P. et al. Genetic variants in NAMPT predict bladder cancer risk and prognosis in individuals from southwest Chinese Han group. Tumour biology: the journal of the International Society for Oncodevelopmental Biology and Medicine. 2014;35:4031-40
20. Lee HP, Chen PC, Wang SW. et al. Plumbagin suppresses endothelial progenitor cell-related angiogenesis in vitro and in vivo. Journal of Functional Foods. 2019;52:8
21. Lee HP, Wang SW, Wu YC. et al. Glucocerebroside reduces endothelial progenitor cell-induced angiogenesis. Food and Agricultural Immunology. 2019;30:12
22. Wang B, Hsu CJ, Chou CH. et al. Variations in the AURKA Gene: Biomarkers for the Development and Progression of Hepatocellular Carcinoma. Int J Med Sci. 2018;15:170-5
23. Lee HP, Wu YC, Chen BC. et al. Soya-cerebroside reduces interleukin production in human rheumatoid arthritis synovial fibroblasts by inhibiting the ERK, NF-kappa B and AP-1 signalling pathways. Food and Agricultural Immunology. 2020;31:11
24. Liu SC, Tsai CH, Wu TY. et al. Soya-cerebroside reduces IL-1β-induced MMP-1 production in chondrocytes and inhibits cartilage degradation: implications for the treatment of osteoarthritis. Food and Agricultural Immunology. 2019;30:13
25. Lee KT, Su CH, Liu SC. et al. Cordycerebroside A inhibits ICAM-1-dependent M1 monocyte adhesion to osteoarthritis synovial fibroblasts. J Food Biochem. 2022: e14108.
26. Wang CQ, Tang CH, Tzeng HE. et al. Impacts of RETN genetic polymorphism on breast cancer development. Journal of Cancer. 2020;11:2769-77
27. Consortium GT. The Genotype-Tissue Expression (GTEx) project. Nature genetics. 2013;45:580-5
28. Lai A, Chen W, Helm K. Effects of visfatin gene polymorphism RS4730153 on exercise-induced weight loss of obese children and adolescents of Han Chinese. International journal of biological sciences. 2013;9:16-21
29. Jiang X, Wu J, Wang J. et al. Tobacco and oral squamous cell carcinoma: A review of carcinogenic pathways. Tob Induc Dis. 2019;17:29
30. Messadi DV. Diagnostic aids for detection of oral precancerous conditions. Int J Oral Sci. 2013;5:59-65
31. Sim YC, Hwang JH, Ahn KM. Overall and disease-specific survival outcomes following primary surgery for oral squamous cell carcinoma: analysis of consecutive 67 patients. J Korean Assoc Oral Maxillofac Surg. 2019;45:83-90
32. Montero PH, Patel SG. Cancer of the oral cavity. Surg Oncol Clin N Am. 2015;24:491-508
33. Guo ZQ, Yu JM, Li W. et al. Survey and analysis of the nutritional status in hospitalized patients with malignant gastric tumors and its influence on the quality of life. Support Care Cancer. 2020;28:373-80
34. Shih LC, Tsai CW, Sun KT. et al. Association of Caspase-8 Genotypes With Oral Cancer Risk in Taiwan. In vivo. 2019;33:1151-6
35. Shih LC, Li CH, Sun KT. et al. Association of Matrix Metalloproteinase-7 Genotypes to the Risk of Oral Cancer in Taiwan. Anticancer Res. 2018;38:2087-92
36. Su SC, Hsieh MJ, Liu YF. et al. ADAMTS14 Gene Polymorphism and Environmental Risk in the Development of Oral Cancer. PLoS One. 2016;11:e0159585
37. Wang TH, Hsia SM, Shih YH. et al. Association of Smoking, Alcohol Use, and Betel Quid Chewing with Epigenetic Aberrations in Cancers. Int J Mol Sci. 2017 18
38. Wu CN, Chang WS, Shih LC. et al. Interaction of DNA Repair Gene XPC With Smoking and Betel Quid Chewing Behaviors of Oral Cancer. Cancer Genomics Proteomics. 2021;18:441-9
39. Wang YY, Hung AC, Lo S. et al. Adipocytokines visfatin and resistin in breast cancer: Clinical relevance, biological mechanisms, and therapeutic potential. Cancer Lett. 2021;498:229-39
40. Peiro C, Romacho T, Carraro R. et al. Visfatin/PBEF/Nampt: A New Cardiovascular Target? Front Pharmacol. 2010;1:135
41. Mohammadi M, Mianabadi F, Mehrad-Majd H. Circulating visfatin levels and cancers risk: A systematic review and meta-analysis. J Cell Physiol. 2019;234:5011-22
42. Dong J, Thrift AP. Alcohol, smoking and risk of oesophago-gastric cancer. Best Pract Res Clin Gastroenterol. 2017;31:509-17
43. Lubin JH, Cook MB, Pandeya N. et al. The importance of exposure rate on odds ratios by cigarette smoking and alcohol consumption for esophageal adenocarcinoma and squamous cell carcinoma in the Barrett's Esophagus and Esophageal Adenocarcinoma Consortium. Cancer Epidemiol. 2012;36:306-16
44. Pandeya N, Williams G, Green AC. et al. Alcohol consumption and the risks of adenocarcinoma and squamous cell carcinoma of the esophagus. Gastroenterology. 2009;136:1215-24 e1-2
45. Steevens J, Schouten LJ, Goldbohm RA. et al. Alcohol consumption, cigarette smoking and risk of subtypes of oesophageal and gastric cancer: a prospective cohort study. Gut. 2010;59:39-48
46. Ma L, Chen J, Song X. et al. Evidence that the genetic polymorphism rs1412115 on chromosome 10 is associated with risk for oral squamous cell carcinoma. Gene. 2015;560:137-9
47. Tu HF, Chen HW, Kao SY. et al. MDM2 SNP 309 and p53 codon 72 polymorphisms are associated with the outcome of oral carcinoma patients receiving postoperative irradiation. Radiother Oncol. 2008;87:243-52
48. Hung AC, Lo S, Hou MF. et al. Extracellular Visfatin-Promoted Malignant Behavior in Breast Cancer Is Mediated Through c-Abl and STAT3 Activation. Clin Cancer Res. 2016;22:4478-90
49. Liu T, Miao Z, Jiang J. et al. Visfatin Mediates SCLC Cells Migration across Brain Endothelial Cells through Upregulation of CCL2. International journal of molecular sciences. 2015;16:11439-51
50. Neubauer K, Misa IB, Diakowska D. et al. Nampt/PBEF/visfatin upregulation in colorectal tumors, mirrored in normal tissue and whole blood of colorectal cancer patients, is associated with metastasis, hypoxia, IL1beta, and anemia. Biomed Res Int. 2015;2015:523930
51. Patel ST, Mistry T, Brown JE. et al. A novel role for the adipokine visfatin/pre-B cell colony-enhancing factor 1 in prostate carcinogenesis. Peptides. 2010;31:51-7
52. Kim JG, Kim EO, Jeong BR. et al. Visfatin stimulates proliferation of MCF-7 human breast cancer cells. Mol Cells. 2010;30:341-5
53. Lu GW, Wang QJ, Xia MM. et al. Elevated plasma visfatin levels correlate with poor prognosis of gastric cancer patients. Peptides. 2014;58:60-4
Author contact
![]() Corresponding authors: Shun-Fa Yang, E-mail: ysfedu.tw; Tel.: 04-2473-0022 ext 11600. Chih-Hsin Tang, E-mail: chtangcmu.edu.tw; Tel: +886-4-22053366#7726.
Corresponding authors: Shun-Fa Yang, E-mail: ysfedu.tw; Tel.: 04-2473-0022 ext 11600. Chih-Hsin Tang, E-mail: chtangcmu.edu.tw; Tel: +886-4-22053366#7726.

 Global reach, higher impact
Global reach, higher impact