Impact Factor
ISSN: 1449-1907
Int J Med Sci 2022; 19(2):402-415. doi:10.7150/ijms.67815 This issue Cite
Research Paper
Potential Genes Associated with COVID-19 and Comorbidity
1. College of Medical Technology, Chengdu University of Traditional Chinese Medicine, Chengdu, Sichuan, China.
2. Non-coding RNA and Drug Discovery Key Laboratory of Sichuan Province, Chengdu Medical College, Chengdu, Sichuan, China.
3. Department of medical Laboratory, The General Hospital of Western Theater Command, Chengdu, China.
4. Chengdu Medical College, Chengdu, China.
5. School of Basic Medical Sciences, Chengdu Medical College, Chengdu, Sichuan, China.
6. Key Laboratory of Systematic Research of Distinctive Chinese Medicine Resources in Southwest China, Chengdu University of Traditional Chinese Medicine, Chengdu, China.
7. Chongqing Key Laboratory of Sichuan-Chongqing Co-construction for Diagnosis and Treatment of Infectious Diseases Integrated Traditional Chinese and Western Medicine, Chengdu University of Traditional Chinese Medicine, Chengdu, China.
8. Department of Pharmacy, Shaoxing People's Hospital, Shaoxing Hospital, Zhejiang University School of Medicine, Shaoxing, China.
*These authors contributed equally in this manuscript.
Received 2021-10-7; Accepted 2022-1-5; Published 2022-1-24
Abstract
Hypertension, diabetes mellitus, and coronary artery disease are common comorbidities and dangerous factors for infection and serious COVID-19. Polymorphisms in genes associated with comorbidities may help observe susceptibility and disease severity variation. However, specific genetic factors and the extent to which they can explain variation in susceptibility of severity are unclear. Therefore, we evaluated candidate genes associated with COVID-19 and hypertension, diabetes mellitus, and coronary artery disease. In particular, we performed searches against OMIM, NCBI, and other databases, protein-protein interaction network construction, and GO and KEGG pathway enrichment analyses. Results showed that the associated overlapping genes were TLR4, NLRP3, MBL2, IL6, IL1RN, IL1B, CX3CR1, CCR5, AGT, ACE, and F2. GO and KEGG analyses yielded 302 GO terms (q < 0.05) and 29 signaling pathways (q < 0.05), respectively, mainly including coronavirus disease-COVID-19 and cytokine-cytokine receptor interaction. IL6 and AGT were central in the PPI, with 8 and 5 connections, respectively. In this study, we identified 11 genes associated with both COVID-19 and three comorbidities that may contribute to infection and disease severity. The key genes IL6 and AGT are involved in regulating immune response, cytokine activity, and viral infection. Therefore, RAAS inhibitors, AGT antisense nucleotides, cytokine inhibitors, vitamin D, fenofibrate, and vaccines regulating non-immune and immune factors could be potential strategies to prevent and cure COVID-19. The study provides a basis for further investigation of genes and pathways with predictive value for the risk of infection and prognosis and could help guide drug and vaccine development to improve treatment efficacy and the development of personalised treatments, especially for COVID-19 individuals with common comorbidities.
Keywords: COVID-19, comorbidity, bioinformatics, susceptibility gene, disease severity, SARS-CoV-2
Introduction
SARS-CoV-2, with its highly infectious property, has spread quickly worldwide, leading the WHO to officially and publicly declare COVID-19 a global pandemic [1]. Up to 6 October 2021, WHO reported 235,673,032 cases worldwide, including 4,814,651 deaths [2]. SARS-CoV-2 can degrade angiotensin-converting enzyme 2 (ACE2) on the cell membrane. Virions bind to exfoliated ACE2, resulting in a decrease of free ACE2, disrupting the balance of human ACE/ACE2, aggravating inflammatory reactions, and promoting interstitial and endothelial fibrosis as well as myocardial hypertrophy [3, 4]. The virus also causes cytokine storms, alters cytoskeleton dynamics, strengthens vasoconstriction, and causes serious complications such as acute respiratory distress syndrome (ARDS) and pulmonary fibrosis [5, 6], which seriously harm human health. At present, several vaccines for SARS-CoV-2 have been put on the market. However, the virus is prone to mutation and its genome is characterised by high diversity and frequent recombination, leading to weak protective power and insufficient duration of protection [7]. Additionally, frequent cross-species transmission further emphasises the importance of fundamental preventive and therapeutic strategies.
The envelope of SARS-CoV-2 mainly comprises four constitutive proteins, the spike (S), membrane (M), envelope (E), and nucleocapsid (N) proteins. This viral S protein can bind to the host ACE2 and split into S1 and S2 subunits by transmembrane serine protease 2 (TMPRSS2). Human ACE2 can interact with the receptor-binding domain (RBD) on the S1 subunit, and the fusion peptide on the S2 subunit enters the infected host cell membrane [8, 9]. The RBD region of the S1 subunit can be used as a vital target for developing drugs and vaccines [10]. The S1/S2 subunit has a recognition site for furin, and S protein cleavage and pre-activation by furin increase viral invasiveness [11]. Compared to the S protein of SARS-CoV, the affinity of SARS-CoV-2 with human ACE2 increased at least ten times with stronger infectivity [12]. The E protein is the most highly conserved in SARS-CoV-2. It mainly participates in the formation of the viral envelope, nucleocapsid assembly, pore protein formation, selective transport of Ca2+ via pore proteins, as well as virus particle assembly and release from host cells [13]. Recombinant SARS-CoV-2 lacking E protein shows decreased viral titres and impaired viral maturation and reproduction [14], suggesting that E protein is a therapeutic target for latent infection. The M protein in SARS-CoV-2 contains three transmembrane domains and one conserved domain. It interacts with S and E proteins in the viral envelope, participating in the assembly and release of virus particles, enabling the host to produce neutral antibodies. It is expected to become a potent immunogen for treating COVID-19 [9, 15]. N protein, situated at the core of the virion, is a highly conserved phosphorylated protein involved in the replication and translation of viral RNA and formation and maintenance of the ribonucleoprotein complex, pivotal for viral protein assembly [16]. It is produced in large quantities during infection and has high immunogenicity, which is important for vaccine development [17]. The S protein binds to ACE2 and immune receptors, such as Toll-like receptors (TLRs) and NOD-like receptors (NLRs) [18]. A few studies characterised the binding relationship between host cell receptors and structural proteins other than S protein. Accordingly, it is necessary to study host-virus interactions to clarify the pathogenic mechanism for finding new ways of preventing and curing COVID-19.
During the course of the COVID-19 epidemic, differences in rates of infection and disease outcomes among populations have been observed. There are various risk factors for COVID-19, among which human genetic polymorphisms play an important role. Eduardo et al. [19] showed that the prevalence rate in men was significantly higher than that in women in most age groups in Chile. The important receptor ACE2 for SARS-CoV-2 binding is on the X chromosome. Because males are hemizygous, they have a higher risk of overexpressing ACE2 mutations, resulting in increased susceptibility or disease severity [20]. The rate of SARS-CoV-2 invasion in humans with A blood group is also significantly higher than that in the O blood group [21-23]. Two genome-wide association studies (GWAS) have shown that 3P21.31 and the 9q34 region containing ABO blood group sites are significantly associated with severe COVID-19 [24, 25]. The IFNL4 variant rs12979860 is associated with the presence of COVID-19 in the Spanish population, independent of gender, age, and disease severity, which may be related to reduced virus clearance [26]. The status of the PNPLA3 variant rs738409 and the TLL-1 variant rs17047200 is related to SARS-COV-2-induced infection [27]. The frequencies of TMPRSS2 variants rs12329760 and rs2298659 are associated with the severity of COVID-19, especially in young men and older women. It has also been predicted that rs12329760 could affect the stability of TMPRSS2 protease and have a protective effect on patients [28]. The frequency of rs12329760 is higher in East Asia than in European countries, and the elevated frequency is associated with a lower fatality rate [29]. In Madrid, a sequencing analysis of 23 cases of familial multiple sclerosis has shown that TMPRSS2 synonymous variants rs61735792 and rs61735794 were significantly associated with infection, whereas ACE2 and furin were not significantly associated with SARS-COV-2 infection [30]. However, previous studies have reported that the ACE2 variant rs41303171 is significantly more frequent in patients with COVID-19 than in controls [31]. Furthermore, another study of an Italian population has shown that the furin encoded by the PCSK3 variant rs769208985 is associated with COVID-19 infection [32]. These results suggest that different populations may have different alleles and genotypes that contribute to COVID-19 infection and severity.
Most patients with COVID-19 have comorbidities, which may be risk factors for susceptibility and severity [33, 34]. In particular, hypertensive disease, diabetes mellitus (DM), and coronary artery disease (CAD) are frequent COVID-19 comorbidities. Guan et al. [35] reported that 399 (25.1%) of the 1590 COVID-19 patients in China had comorbidities. Hypertension, DM, and CAD accounted for 16.9%, 8.2%, 3.7%, and 1.3% of these comorbidities, respectively, and 130 patients had at least two comorbidities. After adjusting for age and smoking status, DM and hypertension were compound endpoint risk factors for entering the intensive care unit, intrusive ventilation, and even death. A cohort study including 201 COVID-19 cases in Wuhan showed that hypertensive disease, DM, and CAD were the more frequent complications, accounting for 19.4%, 10.9%, and 4%, respectively. Comorbidities have been observed in 46.3% and 67.1% of critical and severe cases, respectively, compared with 37.8% of moderate COVID-19 cases [36]. Several meta-analyses have also confirmed that hypertensive disease, DM, and CAD were more frequent comorbidities of COVID-19. These comorbidities are more frequent in severe patients and are risk factors for disease progression [37-40]. However, the role and molecular mechanism underlying the association between common comorbidities and COVID-19 remain unclear. The polymorphism and mutation expression patterns of genes related to these diseases may lead to susceptibility to SARS-CoV-2 and different outcomes [41]. Further research is needed to be conducive to preventing and curing COVID-19.
Variants with increased transmissibility, including delta (B.1.617.2), lambda (C.37), gamma (P1), beta (B.1.351), and alpha (B.1.1.7), have recently appeared and spread worldwide [42-44]. The spread of COVID-19 caused by the delta variant was more rapid and severe than other genotypes [45], while viral mutations render established vaccines less protective, causing breakthrough infection [46]. The mutant strains can impact response measures, requiring preventive intervention even after vaccination. Identifying factors associated with susceptibility and severity and elucidating their biological mechanisms are crucial for preventing and treating COVID-19. In addition to meta-analyses and retrospective clinical studies showing that hypertension, DM, and CAD are common comorbidities and potential risk factors for infection and acute COVID-19, further evidence showed that DM and hypertension are risk factors for CAD, which can cause the three comorbidities to co-occur [47], placing patients at a greater risk of developing COVID-19 and suffering from worse clinical symptoms. However, studies of the mechanism underlying the association between the three comorbidities and COVID-19 are lacking. Identifying genes associated with comorbidities is a quintessential research goal to guide the detection of high-risk groups, prevent the disease, and treat COVID-19 patients with common comorbidities.
Bioinformatics approaches can be used to integrate and analyse genetic information and other data types, explore the mechanisms underlying specific pathological conditions, and identify new therapeutic targets. NCBI, OMIM, and other common databases were used to identify genes associated with COVID-19 as well as hypertension, DM, and CAD to predict susceptibility and severity in this study. A protein-protein interaction (PPI) network was generated using Cytoscape, and Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway enrichment analyses were performed. Nonetheless, the candidate susceptibility genes require further validation in larger studies to identify potential therapeutic targets for new adjuvant drugs and vaccines and develop personalised preventive measures and treatment plans.
Materials and methods
Collection of diseases-related genes
Disease-related genes were identified using the search terms “hypertension”, “diabetes mellitus”, “coronary artery disease”, and “COVID-19.” Four databases were used to screen for genes related to hypertension, DM, CAD, and COVID-19, including OMIM (http://www.omim.org/), KMDB/MutationView (http://mutationview.jp/MutationViewV2Server/), DisGeNET database (https://www.disgenet.org/), and NCBI database [48]. OMIM is an online catalogue of human genes and their related mutations based on human Mendelian genetics and diseases. KMDB/MutationView provides human disease-related gene mutations covering the eye, ear, heart, tumor, autoimmune disease, muscle, and blood disease genes. The DisGeNET database collects information for associations between mutation sites and diseases. The NCBI database includes information for COVID-19-related genes.
Identification of overlapping genes related to COVID-19 and comorbidities
The related genes between COVID-19 and three comorbidities were cross-analysed using Bioinformatics (http://www.bioinformatics.com.cn/). An online platform [49] was used to draw Venn diagrams of common genes associated with the diseases.
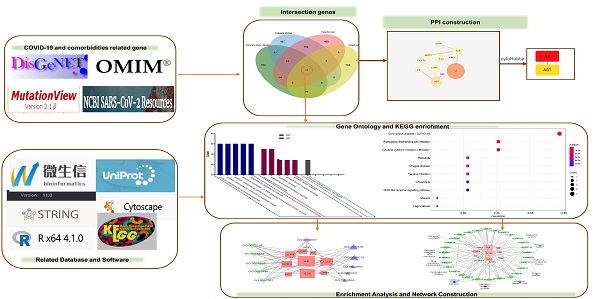
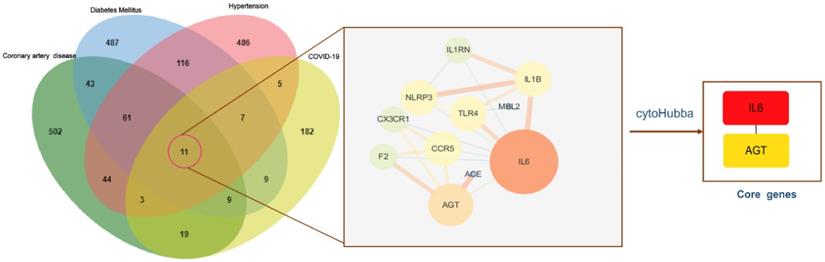
Venn diagram of the intersection genes to the three comorbidities of COVID-19. Notes: The 11 overlapping genes of comorbidities of hypertension, DM, CAD and COVID-19 were TLR4, NLRP3, MBL2, IL6, IL1RN, IL1B, CX3CR1, CCR5, AGT, ACE, F2.
Functional enrichment analysis and network visualization of intersecting genes
R (version 4.1.0) was used to load the Bioconductor (https://www.bioconductor.org/) packages “stringi,” “DOSE,” “apeglm,” “BiocManager,” “clusterProfiler,” and “pathview” to carry out GO analysis for common disease genes, KEGG analysis, and visual graphic display. The screening criteria for enrichment were p < 0.01 and q < 0.01. The p-values were estimated using the hypergeometric test and corrected for multiple testing with the Benjamini-Hochberg method. Additionally, q-values were estimated to control for the false discovery rate. The R script is provided in Supplementary File S1. A network graph of intersecting genes was constructed using Cytoscape (version 3.7.0).
Construction of a PPI network and determination of core targets
The STRING database was used to visualise the PPI network, which reveals the strength of relationships and connections between proteins corresponding to intersecting genes. Each node in the PPI represents a protein, and the connections between nodes illustrate interactions. The list “Multiple proteins” and the organism “Homo sapiens” were selected as the screening conditions. In basic settings, high confidence of 0.700 was chosen [50]. Cytoscape was used to build and visualise PPI networks. The topological parameters were analysed using Cytoscape, and the key genes were searched using cytoHubba. Genes with a degree value greater than the mean were defined as key genes.
Results
Disease-related mutations and intersecting genes
To identify genes associated with hypertension, DM, CAD, three databases (DisGeNET, OMIM, and KMDB) were used. After removing duplicate data, 733, 743, and 692 genes associated with hypertension, DM and CAD were identified, respectively. After superposition and the deletion of duplicate data for 150 genes related to COVID-19 from the SARS-CoV-2 data in NCBI and 108 genes from the gene map of OMIM, 245 genes were obtained. Then, a Venn diagram of overlapping genes was drawn using an online Bioinformatics platform. Eleven common genes (TLR4, NLRP3, MBL2, IL6, IL1RN, IL1B, CX3CR1, CCR5, AGT, ACE, and F2) were identified (Figure 1). Polymorphisms of these genes may be predictors of susceptibility and severity of COVID-19. Disease-related mutations genes are listed in Supplementary Table S1.
Enrichment analysis of intersecting genes
GO analysis of intersecting genes
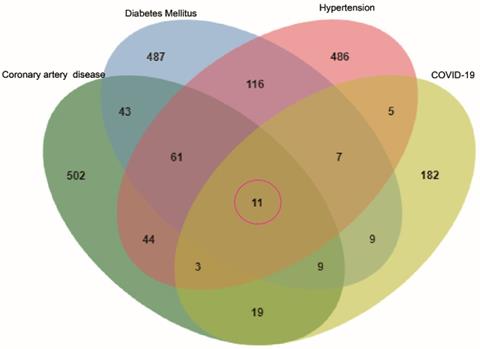
We performed a GO enrichment analysis of 11 genes associated with COVID-19 and common comorbidities. The results showed that genes were enriched in 286 biological processes (BP), including lipopolysaccharide, bacterial molecules, biological stimulation, acute inflammation, regulation of transcription factors, cytokine activity, JAK-STAT receptor signaling pathway, immunity, and metabolic regulation. The lateral plasma membrane was the only enriched cellular component (CC). The genes were associated with 15 molecular functions (MF), including cytokine binding and activity, chemokine binding and receptor activity, and cell receptor-ligand binding. The top five BP, MF, and CC terms are shown in Figure 2A, and a network map of common intersecting gene-related GO terms was constructed using Cytoscape (Figure 2B). In Table 1, detailed results of the GO enrichment analysis are provided. The results suggested that three comorbidities of COVID-19 were mainly related to metabolic and immune system disorders, insufficient or excessive response to external stimuli, and changes in the expression and activity of cytokines, resulting in changes in susceptibility and severity.
GO analysis of intersection genes and network visualization. Notes: (A) GO analysis of intersection genes. The value on the Y-axis represents counting the intersection genes in each GO term. Blue represents BP, red represents MF, and green represents CC. The X-axis is the GO term, respectively the top 5 BP, MF and the only cell component enriched according to the count value. (B) Network visualization of gene and GO term. Red represents the common gene, green represents BP, purple represents CC, blue represents MF, the larger the shape means the more connections in the whole network.
Enriched GO-terms of intersection genes
| ID | Description | Gene | Count | p | q |
|---|---|---|---|---|---|
| GO:0071222 | cellular response to lipopolysaccharide | TLR4/NLRP3/IL6/IL1B/CX3CR1/CCR5 | 6 | 4.48E-07 | 1.32E-07 |
| GO:0051091 | positive regulation of DNA-binding transcription factor activity | TLR4/NLRP3/IL6/IL1B/CX3CR1/AGT | 6 | 7.63E-07 | 2.26E-07 |
| GO:0071219 | cellular response to molecule of bacterial origin | TLR4/NLRP3/IL6/IL1B/CX3CR1/CCR5 | 6 | 4.48E-07 | 1.32E-07 |
| GO:0032496 | response to lipopolysaccharide | TLR4/NLRP3/IL6/IL1B/CX3CR1/CCR5 | 6 | 1.84E-06 | 5.44E-07 |
| GO:0071216 | cellular response to biotic stimulus | TLR4/NLRP3/IL6/IL1B/CX3CR1/CCR5 | 6 | 5.74E-07 | 1.70E-07 |
| GO:0009897 | external side of plasma membrane | TLR4/CX3CR1/CCR5/ACE/F2 | 5 | 2.26E-05 | 9.52E-06 |
| GO:0048018 | receptor ligand activity | IL6/IL1RN/IL1B/AGT/F2 | 5 | 0.000196 | 6.96E-05 |
| GO:0030546 | signaling receptor activator activity | IL6/IL1RN/IL1B/AGT/F2 | 5 | 0.000196 | 6.96E-05 |
| GO:0008083 | growth factor activity | IL6/AGT/F2 | 3 | 0.000753 | 0.000268 |
| GO:0019955 | cytokine binding | IL1RN/CX3CR1/CCR5 | 3 | 0.000753 | 0.000268 |
| GO:0070851 | growth factor receptor binding | IL6/IL1RN/IL1B | 3 | 0.000753 | 0.000268 |
Notes: Count value represents the number of target genes that core genes belong to in the term, and p was corrected by Benjamini-Hochberg method. And q value was estimated for false discovery rate control. The values p < 0.01 and q < 0.01 were considered statistically significant.
KEGG pathway analysis of intersection genes
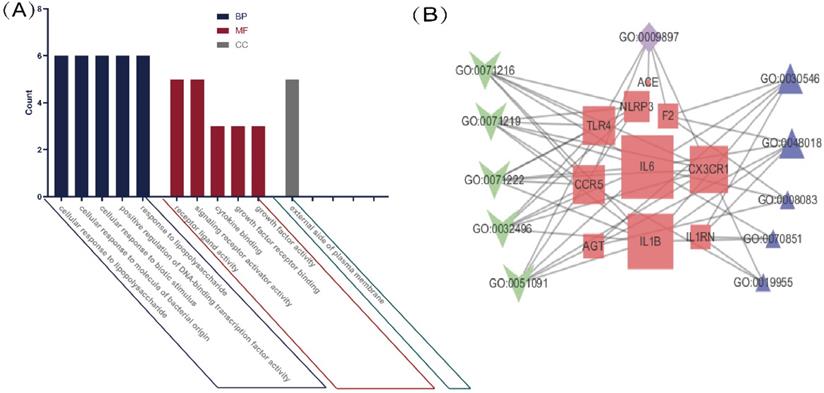
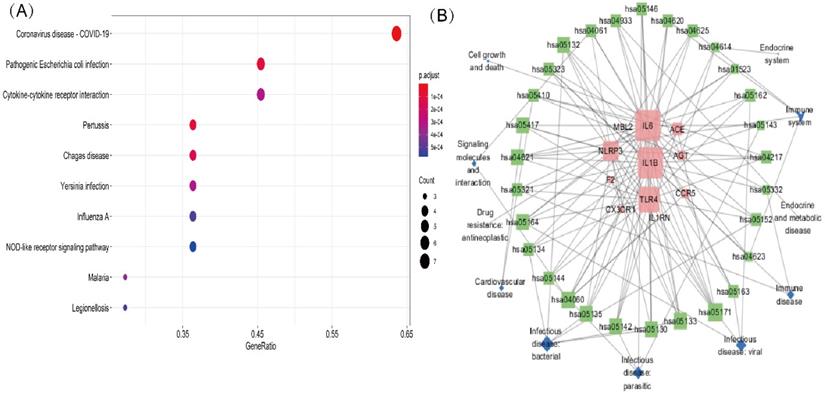
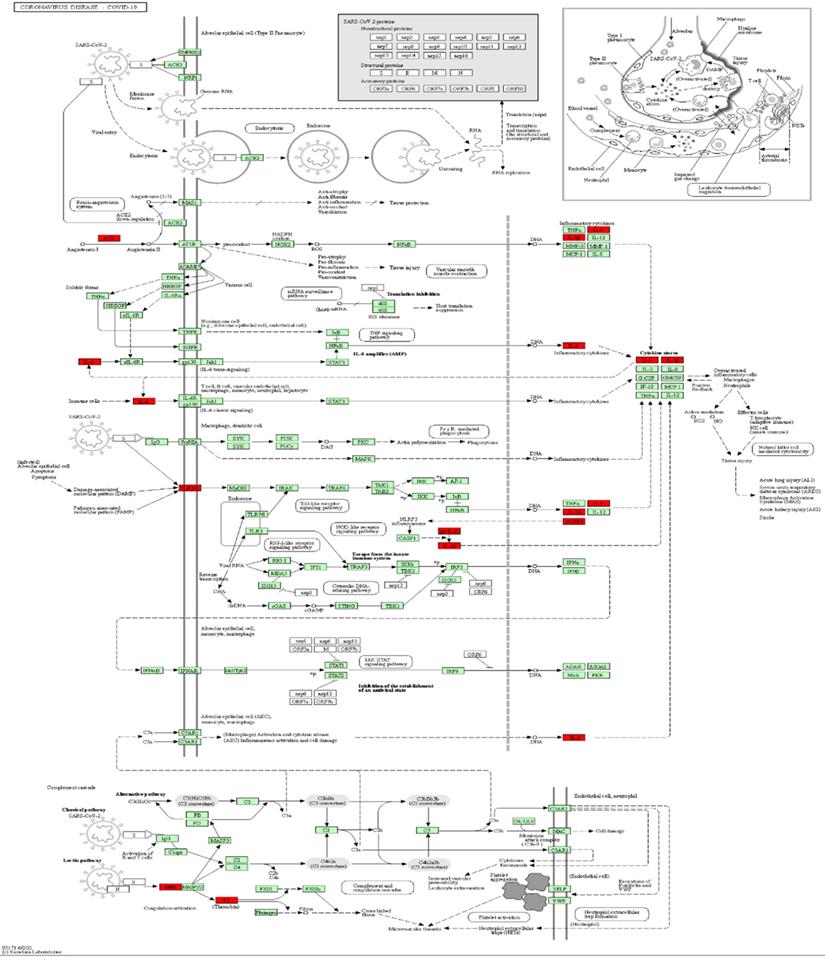
A KEGG pathway analysis was performed to identify pathways involving genes associated with COVD-19 and common comorbidities. In total, 29 KEGG pathways were enriched, including pathways related to human diseases, biological systems, environmental information processing, cellular processes, including viruses, bacteria, parasitic infectious diseases, endocrine metabolic diseases, cardiovascular diseases, immune diseases, signaling molecules and interactions, cell growth and death, immune system, and endocrine system. The top ten pathways were coronavirus disease-COVID-19, cytokine-cytokine receptor interaction, pertussis, Chagas disease, Yersinia infection, influenza A, lipid and atherosclerosis, Salmonella and Escherichia coli infection, and NLR pathway (Figure 3A). A network diagram of the intersection gene-KEGG pathways was constructed using Cytoscape (Figure 3B). The common genes were most highly enriched in coronavirus disease-COVID-19, with the highest GeneRatio value. A map of the overlapping genes in coronavirus disease-COVID-19 pathway was obtained by KEGG mapping online tool (Figure 4). These common genes were mainly involved in the inflammatory process caused by COVID-19-related cytokines, such as IL6 and IL1β. F2 was involved in the coagulation process and TLR4-mediated immune recognition against virus infection. In addition, interactions between viral proteins and cytokines, inflammatory diseases, diabetic complications, hypertrophic cardiomyopathy, the renin-angiotensin system, and other pathways were also identified in the enrichment analysis. The results, summarised in Table 2, suggested that the genes were involved in infectious and metabolic diseases, mainly by affecting the interactions between viral proteins and cytokine receptors, regulating immune and inflammatory responses. Detailed KEGG and GO analysis results are provided in Supplementary Table S2.
KEGG analysis of intersection genes and network visualization. Notes: (A) KEGG analysis of intersection genes. A higher GeneRatio value indicates a higher enrichment level; The bigger circle indicates more target genes in the pathway. The color ranges from blue to red of circles indicating that the adjusted p-values increases from small to large and the enrichment results are more significant. (B) Network visualization of intersecting genes and enriched KEGG pathways. Red represents the common gene, green represents the KEGG pathway and blue represents the upper-level classification of the KEGG pathway. The larger the shape, the more connections in the entire network.
Enriched KEGG pathways of intersection genes
| ID | Description | Gene | Count | p | q |
|---|---|---|---|---|---|
| hsa05171 | Coronavirus disease -COVID-19 | TLR4/NLRP3/MBL2/IL6/IL1B/ACE/F2 | 7 | 3.52E-07 | 1.49E-07 |
| hsa04060 | Cytokine-cytokine receptor interaction | IL6/IL1RN/IL1B/CX3CR1/CCR5 | 5 | 0.000325 | 0.000138 |
| hsa05130 | Pathogenic Escherichia coli infection | TLR4/NLRP3/IL6/IL1B/F2 | 5 | 9.03E-05 | 0.000038 |
| hsa05133 | Pertussis | TLR4/NLRP3/IL6/IL1B | 4 | 9.03E-05 | 0.000038 |
| hsa05142 | Chagas disease | TLR4/IL6/IL1B/ACE | 4 | 0.000149 | 0.000063 |
| hsa05135 | Yersinia infection | TLR4/NLRP3/IL6/IL1B | 4 | 0.000325 | 0.000138 |
| hsa05164 | Influenza A | TLR4/NLRP3/IL6/IL1B | 4 | 0.000523 | 0.000222 |
| hsa04621 | NOD-like receptor signaling pathway | TLR4/NLRP3/IL6/IL1B | 4 | 0.000578 | 0.000245 |
| hsa05417 | Lipid and atherosclerosis | TLR4/NLRP3/IL6/IL1B | 4 | 0.000937 | 0.000397 |
| hsa05132 | Salmonella infection | TLR4/NLRP3/IL6/IL1B | 4 | 0.001287 | 0.000545 |
| hsa05144 | Malaria | TLR4/IL6/IL1B | 3 | 0.000412 | 0.000175 |
| hsa05134 | Legionellosis | TLR4/IL6/IL1B | 3 | 0.000523 | 0.000222 |
| hsa05321 | Inflammatory bowel disease | TLR4/IL6/IL1B | 3 | 0.000578 | 0.000245 |
| hsa05410 | Hypertrophic cardiomyopathy | IL6/AGT/ACE | 3 | 0.001287 | 0.000545 |
| hsa05323 | Rheumatoid arthritis | TLR4/IL6/IL1B | 3 | 0.001287 | 0.000545 |
| hsa04061 | Viral protein interaction with cytokine and cytokine receptor | IL6/CX3CR1/CCR5 | 3 | 0.001287 | 0.000545 |
| hsa04933 | AGE-RAGE signaling pathway in diabetic complications | IL6/IL1B/AGT | 3 | 0.001287 | 0.000545 |
| hsa05146 | Amoebiasis | TLR4/IL6/IL1B | 3 | 0.001287 | 0.000545 |
| hsa04620 | Toll-like receptor signaling pathway | TLR4/IL6/IL1B | 3 | 0.001287 | 0.000545 |
| hsa04625 | C-type lectin receptor signaling pathway | NLRP3/IL6/IL1B | 3 | 0.001287 | 0.000545 |
| hsa05162 | Measles | TLR4/IL6/IL1B | 3 | 0.002715 | 0.001150 |
| hsa04217 | Necroptosis | TLR4/NLRP3/IL1B | 3 | 0.003568 | 0.001511 |
| hsa05152 | Tuberculosis | TLR4/IL6/IL1B | 3 | 0.004729 | 0.002003 |
| hsa05163 | Human cytomegalovirus infection | IL6/IL1B/CCR5 | 3 | 0.00864 | 0.003660 |
| hsa04614 | Renin-angiotensin system | AGT/ACE | 2 | 0.001628 | 0.000690 |
| hsa01523 | Antifolate resistance | IL6/IL1B | 2 | 0.002715 | 0.001150 |
| hsa05143 | African trypanosomiasis | IL6/IL1B | 2 | 0.003568 | 0.001511 |
| hsa05332 | Graft-versus-host disease | IL6/IL1B | 2 | 0.004412 | 0.001869 |
| hsa04623 | Cytosolic DNA-sensing pathway | IL6/IL1B | 2 | 0.008833 | 0.003742 |
Notes: Count value represents the number of target genes that core genes belong to in the term, and p was corrected by Benjamini-Hochberg method. And q value was estimated for false discovery rate control. The values p < 0.01 and q < 0.01 were considered statistically significant.
Mapping of intersection genes in the Coronavirus disease-COVID-19 pathway. Notes: The genes in red are mappings of intersection genes in Coronavirus disease-COVID-19 pathway. The intersection genes were mainly mapped in cytokines involved in the inflammatory process. F2 and TLR4 mediate coagulation process and immune recognition against the viral infection process, respectively.
PPI network analysis
To understand the interactions between genes and identify key genes among the 11 common genes associated with COVID-19 and comorbidities, the STRING database was used, and the PPI network was presented using Cytoscape software (Supplementary Table S3). Lines between nodes represent interactions between proteins, where thicker lines indicate stronger interactions. IL6 and AGT were selected as core genes, with degree values (i.e., the number of connections) of 8 and 5, respectively (Figure 5), indicating that they were central in the PPI and may be more closely related to the disease state. IL6 showed the highest number of connections and was most closely related to other common genes. It was implied that IL6 may have a vital effect on the infection and pathogenesis and can be used as a key starting point for prediction and treatment.
PPI networks of intersection genes and core genes. Notes: The bigger circle represents greater degree; Lines between nodes represent interactions between proteins encoded by genes, with thicker lines indicating stronger interactions; IL6 and AGT were selected as the core genes, and the degree value was 8, 5, respectively. IL6 showed the highest number of connections.
Discussion
We constructed a gene-protein interaction network based on common genes associated with three common comorbidities and COVID-19. Eleven common genes were found, including TLR4, NLRP3, MBL2, IL6, IL1RN, IL1B, CX3CR1, CCR5, AGT, ACE, and F2. These genes may be potential risk predictors of COVID-19. GO and KEGG enrichment analyses further indicated that these 11 common genes were related to infection, inflammation, cytokine activity, and receptor-binding. Our results were similar to those of another bioinformatics study. The hub genes related to COVID-19 were enriched in the immune and inflammatory reactions, and cytokine-cytokine receptor interaction pathway [51], suggesting the key mechanisms and pathways underlying SARS-CoV-2 infection.
Previous work has demonstrated that the core gene IL6 is connected to a higher risk of SARS-CoV-2 infecting and pathopoiesia [52]. IL6 responds to tissue damage and infection and is secreted by diverse cells as a proinflammatory cytokine, such as fibroblasts, keratinocytes, and macrophages [53]. Higher IL6 levels are commonly found in the three common comorbidities, which may lead to severe COVID-19. IL6 plays a key role in exacerbating inflammatory responses after infecting SARS-CoV-2. Excessive IL6 can lead to excessive activation of Th17 cells by activating the JAK-STAT pathway. Th17 cells produce numerous effectors (IL-17, IL-1β, IL-21, IL-22) that recruit neutrophils, upregulate fibrinogen, damage tissue, and cause systemic inflammatory symptoms [5, 54-56]. A meta-analysis of COVID-19 and IL6 polymorphisms has shown that carriers of the IL6-174C allele show more severe pneumonia [57]. Another study has shown that mortality and prevalence are negatively correlated with the IL6 rs1800795G allele in COVID-19 [58]. Immune system dysfunction and IL6 overexpression are characteristics of COVID-19, and elevated IL6 levels are related to respiratory failure and death [59]. Recent research has shown that the common Asian haplotype C-T-T, represented by alleles rs1800796, rs1524107, and rs2066992, reduces the risk of severe COVID-19 via reduced IL6 expression [56]. Clinical data has shown that the majority of patients show improved hypoxaemia and CT imaging and no significant adverse reactions after receiving tocilizumab, a monoclonal antibody binding with IL6 receptor, thus inhibiting the binding of IL6 to the receptor, to avoid damaging targeted cells and reducing inflammation [60]. Considering the importance of IL6 in COVID-19-related pathomechanism, anti-IL6 therapies may be effective for severe COVID-19, including selective inhibitors of ADAM-17 or sgp130Fc, which inhibits IL6 signal transduction [61].
The core gene AGT encodes the precursor of angiotensin, which is highly produced in the liver and is the only substrate for renin. Angiotensin is converted to the active substance angiotensin (ANG) I/II in the renin-angiotensin system (RAS). It exerts physiological effects on target organs, including the kidney, blood vessels, and adrenal gland, and regulates the vascular tone, heart functions, and vascular remodelling [62]. Sequence variation in this gene is associated with the risks of hypertensive disease, heart failure, and cardiovascular disease [63, 64]. AGT is critical for ANG I /ANG II to maintain balance. The SARS-CoV-2-infected receptor ACE2 is an important member of the RAS system. Any changes in expression and function of the RAS element caused by genetic variation can lead to differences in susceptibility and severity to COVID-19 [65], highlighting the importance of AGT in COVID-19. Cafiero et al. [66] studied the genotype and allele frequency distributions of AGT rs699 and found that asymptomatic patients have a lower frequency of the T/C genotype compared to symptomatic patients with COVID-19. The AGT rs699 SNP is potentially valuable tool for predicting clinical COVID-19 outcomes. The transfusion of AGT antisense nucleotides to inhibit AGT expression in male C57BL/6J mice for 14 days significantly reduced the expression of TMPRSS2 mRNA in the lungs [67]. It was suggested that suppression of AGT may have a protective effect against viral entry.
A relationship between ACE polymorphisms and COVID-19 has also been reported. ACE encodes an enzyme involved in regulating blood pressure and electrolyte balance, which catalyses transforming ANG I into ANG II with biologically active and participates in the imbalance of ACE2/ACE in the pathogenic process of SARS-CoV-2. The intron 16 of ACE is inserted/deleted 287bp to cause polymorphisms, which is currently the most studied polymorphism in ACE. There is a clear relationship between the polymorphism and blood ACE level [68]. The I/D genotype is close to pathogenesis and progress of diabetic complications [69]. The frequency and mortality of COVID-19 patients are closely connected to the ACE-I/D genotype that may be detected for predicting the ponderance of COVID-19 [70]. ACE2 is the vital receptor of SARS-CoV-2 binding host cell to infect humans. The expression and function of ACE are affected by its polymorphism, which plays a vital role in regulating ACE2 expression. The deficiency or overexpression of ACE alters ANG I accumulation, which may promote virus infection [68] and may explain why ACE is a potential susceptibility gene for COVID-19.
The intersecting gene F2 encodes prothrombin. The rs1799963 and rs3136516 F2 polymorphisms are associated with increases in the prothrombin level/activity and the risk of thrombosis [71, 72]. Marker genes for arterial/venous thromboembolism include F2, and SNP genotyping of F2 will help identify the population most at risk of COVID-19-related thrombotic complications [73]. Patients with coagulation abnormalities and severe infection-related systemic coagulopathies, including DIC or thrombotic microangiopathy, in severe COVID-19 show higher death rates. An elevated D-dimer level can increase the risk of death in severe cases; accordingly, anticoagulant treatment may reduce the mortality of COVID-19, as supported by studies of low-molecular-weight heparin revealing an improved prognosis [74]. A study in Hong Kong has revealed that anticoagulants and antiplatelet drugs are related to lower hazard of critical COVID-19 [75]. As the correlation of COVID-19 coagulation abnormalities becomes clearer, the polymorphism of coagulation-related genes may be a considerable reason for the adverse progression of COVID-19 patients. Further studies of prothrombin F2 polymorphisms in COVID-19 are needed.
Genes involved in immune system regulation may be related to variation in COVID-19 outcomes. Pathogens proliferate in the host in chronic illnesses, such as DM, affecting innate immunity [76]. MBL2 encodes a vital molecule in the innate immune system, a soluble mannose-binding lectin. An MBL2 deficiency is related to the susceptibility of infectious diseases [77]. The MBL2 variant rs5030737 is positively correlated with the number of COVID-19 cases but not with the number of deaths. The MBL2 variant rs1800450 is a positive correlation with the numbers of cases and deaths. The MBL2 variant rs1800451 is a negative correlation with the number of cases. These findings suggested that MBL2 inactivation is a risk factor for SARS-COV-2 infection [78, 79]. Chemokine receptor-ligand interactions mediate inflammatory cell transport and pathogen-related immune responses. Early studies of CCR5-deficient mice have shown that the Th1 immune response is suppressed, increasing susceptibility to viral and bacterial infection [80]. The deletion of the CCR5 gene is related to a higher incidence of HCV [81]. The CCR5 Δ32 variant produces a truncated protein and observably decreases receptor formulation, suppressing the immune response [82], which may lead to increased virus susceptibility. A bioinformatics study has analysed the summary statistics of the GWAS dataset from patients with respiratory failure of COVID-19 and found that the first 10 BP involved the CCR5 [83]. In an epidemiological study, CCR5 Δ32 allele frequency positively correlated with COVID-19 mortality [84], suggesting CCR5 plays an important role in susceptibility and severity of COVID-19. Studies have shown that the number of non-classical monocytes is lower in CAD+SARS-COV-2 than in CAD-SARS-COV-2, and CX3CR1 expression on the surface was impaired. CX3CR1 mediated non-classical monocyte migration along with endothelial cells in the vascular system for antiviral immune response. Non-classical monocyte numbers and phenotypes were predictors of adverse clinical outcomes in CAD+SARS-COV-2-infected patients [85], suggesting that studies combined COVID-19 and comorbidities analysis can help detect different markers and improve therapeutic decision-making for different COVID-19 populations to efficient, personalised treatment.
Among KEGG signalling pathways identified in the study, TLR, NLR, and CLR signalling pathways play vital roles in initiating innate immune response, which can activate immune system and destroy immune tolerance state, which can disrupt immune tolerance state. It has been noted that cells expressing little or no ACE2 are still susceptible to the virus, suggesting that the S protein invades the host via receptors other than ACE2 [86]. Evidence from in vitro studies and computer simulations supports the interaction between S proteins and immune receptors, including NLRs, CLRs, and TLRs [87], consistent with the results of molecular docking study demonstrating binding between the S protein and TLRs, including TLR1, TLR4, and TLR6, in which the docking result showed that TLR4 had the highest binding energy [88]. Activating TLR4 can increase ACE2 expression, promoting viral entry to cause excessive inflammation [89]. The TLR4 pathway can produce IL6, a critical cytokine associated with cytokine storm, suggesting that TLR4 participates in SARS-CoV-2 invasion. NLRP3 inflammatory bodies composed of apoptosis-associated speck-like protein, caspase-1, and oligomeric NLRP3 contribute to severe COVID-19, and inflammatory bodies might be related to severe cytokine storms, resulting in ARDS and even death [90, 91]. Anti-inflammatory and NLRP3 inflammatory body inhibitors could be potential therapeutic drugs for COVID-19.
The immune system is out of control to excessively release inflammatory mediators, resulting in a cytokine storm, a critical feature of COVID-19 recognized as the main reason causing serious outcomes. High levels of proinflammatory cytokines were observed in patients with COVID-19, such as IL1, IL1β, IL2, IL6, and TNF-α [92, 93]. COVID-19 comorbidities, including hypertensive disease, DM, and CAD, are characterised by chronic inflammation [94]. Most of the intersecting genes were associated with both COVID-19 and these comorbidities, playing a crucial role in inflammation. Mutations in genes encoding cytokines, such as IL1B, IL1R1, and IL1RN, likely cause a cytokine storm and critical COVID-19 [95]. Supportive and symptomatic treatment is necessary to control cytokine storms by inhibiting inflammation [96]. Drugs targeting IL1, IL6, IL18, and interferon-γ, are effective in treating various cytokine storm-related syndromes [97]. Therefore, this is an important area of research aimed at COVID-19 treatment.
Non-immune and immune factors are targets for the identification of new auxiliary drugs for COVID-19. The core genes ATG and ACE of the RAS are related to the ACE/ACE2 balance. The renin-angiotensin-aldosterone system (RAAS) blocker ACE-I/ARB has a protective effect against COVID-19, and RAAS inhibitors may be associated with a lower probability of death [98, 99]. AGT antisense oligodeoxynucleotides inhibit AGT, providing another potential treatment. Interventions targeting the immune pathway are effective for preventing and curing COVID-19. A study has shown that the Lianhua Qingwen capsule can treat or interfere with the pathological process of COVID-19 via the NLR and TLR signalling pathways, NLRP33, and IL6 [100]. Additionally, metformin has anti-inflammatory activity. It can target mitochondrial electron transport and reduce IL6 levels by blocking ROS/CRAC, providing new curative pathways to weaken cytokine activity and thrombosis [101]. Recent studies have shown that 25(OH)D levels are related to mortality from COVID-19. The GC variant of rs2282679 in the gene encoding a vitamin D binding protein is associated with disease severity, and the DHCR7 rs12785878 variant associated with vitamin D deficiency has a higher prevalence in the Portuguese population than in Europe and may explain more severity of COVID-19 in the Portuguese population [102]. Some scholars have suggested that using the combination of dexamethasone and vitamin D in COVID-19 may improve the symptom of ARDS [103]. Dexamethasone, a corticosteroid, has been identified as an effective immunosuppressant that weakens the innate immune response. Moderate to severe ARDS group treated using dexamethasone has been shown to decrease mechanical ventilation and overall mortality [104]. Vitamin D promotes the production of antimicrobial peptides, enhances innate cellular immunity [105], regulates the expression of TLR2 and TLR4, and reduces IL6 levels [106, 107]. In addition, vitamin D is an effective renin inhibitor, and a vitamin D deficiency leads to increased AGT expression and the activation of RAAS [108], which reduces the risk of ARDS, myocarditis, or heart injury in patients with COVID-19 [109]. Vitamin D not only enhances host immunity to avoid viral infection but also inhibits severe cytokine storm and RAAS imbalance, suggesting that it can be an important cryptic adjuvant drug for safely and effectively preventing and curing COVID-19. A new study showed that the hyperlipidaemia drug fenofibrate reduced the stability of the SARS-CoV-2 S1 protein, reducing infection by 70% [110]. In addition, fenofibrate can inhibit inflammation and reduce fibrinogen levels and cytokine production, including TNF-α, IL6, and IL1-β [111, 112]. The antithrombotic activity of fenofibrate may reduce or prevent adverse events, with good safety and low cost, therefore, we strongly recommend clinical trials for fenofibrate treating COVID-19 patients requiring hospitalization.
Ligands are used as immunogenic adjuvants to activate TLR and NLR for enhancing the efficacy of vaccines and promoting a strong immune response [113]. Early activating immune regulation and the innate immune system to enhance antiviral ability may be important ways to prevent COVID-19. After the initiation of inflammatory bodies, NLRP3 affects cellular pressure caused by various pathogens [114]. QS-21, aluminium hydroxide, and other immune adjuvants can activate NLRP3 [115, 116]. Early TLR activation can improve antiviral immunity, and the TLR4 agonist MPL is an adjuvant used in vaccines to prevent HPV and HBV infection [117]. Various epitope-based peptide and recombinant S1 subunit vaccine components can activate TLR to protect against SARS-CoV-2 [118, 119]; however, clinical trials are lacking. A mixture of TLR4 and TLR9 agonists prepared as a vaccine was more effective than a vaccine targeting TLR4 [120]. The synergistic effects of different receptor agonists for NLPR3 and TLR can activate vaccine adjuvants, promote an innate immune response, and provide solid and lasting protection against pathogens. This may be an important method for the clinical application of vaccines to prevent infection. However, excessive activation of TLR and NLR may lead to the development of cytokine storms, leading to severe COVID-19 [121]. The role of TLR and NLR during SARS-CoV-2 and their harmful effects need to be determined in further studies.
In this study, the genes associated with both three common comorbidities and COVID-19 were screened through various databases. Candidate genes for the prediction of susceptibility and severity of COVID-19 included TLR4, NLRP3, MBL2, IL6, IL1RN, IL1B, CX3CR1, CCR5, AGT, ACE, and F2. Studies of COVID-19 have evaluated the roles of polymorphisms in ACE2, TMPRSS2, IL-6, ACE, MBL2, AGT, and CCR5. However, the roles of polymorphisms in F2, NLRP3, TLR4, CX3CR, IL1RN, and IL1B in COVID-19 have not been established. This study could provide a reference for subsequent studies of susceptibility genes and marker genes of COVID-19 severity, and suggest that there are shared genetic mechanisms underlying COVID-19 and comorbidities, and reveal potential treatments. These results are of particular value for screening high-risk groups and for improving treatment efficacy and the development of personalised treatments, especially for COVID-19 individuals with the three comorbidities.
However, this study had some limitations. Database-based genomic data only provided a preliminarily explanation of the connection between three common comorbidities and COVID-19. The results should be validated in large population studies as well as functional assays. Additionally, this study focused on three common comorbidities. Other potential factors, such as smoking, obesity, and region, were not included in this study, and more mechanisms underlying associations between multiple factors and COVID-19 need to be explored. Basic and clinical research on related genes and drugs is required to provide more strategies for preventing and curing COVID-19.
Abbreviations
GO: Gene Ontology; KEGG: Kyoto Encyclopedia of Genes and Genomes; NLRP: NOD-like receptor protein; PPI: protein-protein interaction; TMPRSS2: transmembrane serine protease 2; TLR: Toll-like receptor; RBD: receptor-binding domain; ROS: reactive oxygen species; CRAC: Ca2+ release-activated Ca2+ channels; RAAS: renin-angiotensin-aldosterone system; ACE: angiotensin-converting enzyme; ARDS: acute respiratory distress syndrome.
Supplementary Material
Supplementary file.
Supplementary table 1.
Supplementary table 2.
Supplementary table 3.
Acknowledgements
We acknowledge all the participants in the Non-coding RNA and Drug Discovery Key Laboratory for helpful discussions and Editage for English language editing.
Author Contributions
Xu Jia, Jinlin Guo, Fengling Qiao, and Lin Zhang designed the study. All authors contributed to their thoughts and advice. Shanshan Feng and Fuqiang Song wrote the manuscript. The manuscript has been reviewed by Xianqin Zhang. All authors participated in the final version of the article and supported this submission.
Data availability statement
The authors confirm that some data supporting the study's findings are available in the article and supplementary materials.
Competing Interests
The authors have declared that no competing interest exists.
References
1. Cucinotta D, Vanelli M. WHO Declares COVID-19 a Pandemic. Acta Biomed. 2020;91:157-60
2. Organization WH. World Health Organization Weekly Operational Update on COVID-19. 2021.
3. Levi M, Thachil J, Iba T, Levy JH. Coagulation abnormalities and thrombosis in patients with COVID-19. The Lancet Haematology. 2020;7:e438-e440
4. Verdecchia P, Cavallini C, Spanevello A, Angeli F. The pivotal link between ACE2 deficiency and SARS-CoV-2 infection. Eur J Intern Med. 2020;76:14-20
5. Ragab D, Salah Eldin H, Taeimah M, Khattab R, Salem R. The COVID-19 Cytokine Storm; What We Know So Far. Front Immunol. 2020;11:1446
6. Pollard CA MM, Nestor-Kalinoski AL. The COVID-19 pandemic: a global health crisis. Physiol Genomics. Physiol Genomics. 2020;52:549-57
7. Hunt AC, Case JB, Park Y-J, Cao L, Wu K, Walls AC. et al. Multivalent designed proteins protect against SARS-CoV-2 variants of concern. bioRxiv. 2021. 2021 07.07.451375
8. Hoffmann M, Kleine-Weber H, Schroeder S, Kruger N, Herrler T, Erichsen S. et al. SARS-CoV-2 Cell Entry Depends on ACE2 and TMPRSS2 and Is Blocked by a Clinically Proven Protease Inhibitor. Cell. 2020;181:271-280.e8
9. Arya R, Kumari S, Pandey B, Mistry H, Bihani SC, Das A. et al. Structural insights into SARS-CoV-2 proteins. J Mol Biol. 2021;433:166725
10. Tai W, He L, Zhang X, Pu J, Voronin D, Jiang S. et al. Characterization of the receptor-binding domain (RBD) of 2019 novel coronavirus: implication for development of RBD protein as a viral attachment inhibitor and vaccine. Cell Mol Immunol. 2020;17:613-20
11. Walls AC, Park Y-J, Tortorici MA, Wall A, McGuire AT, Veesler D. Structure, Function, and Antigenicity of the SARS-CoV-2 Spike Glycoprotein. Cell. 2020;181:281-292.e6
12. Wrapp D WN, Corbett KS, Goldsmith JA, Hsieh CL, Abiona O, Graham BS, McLellan JS. Cryo-EM structure of the 2019-nCoV spike in the prefusion conformation. Science. 2020;367:1260-3
13. Mukherjee S, Bhattacharyya D, Bhunia A. Host-membrane interacting interface of the SARS coronavirus envelope protein: Immense functional potential of C-terminal domain. Biophys Chem. 2020;266:106452
14. Ortego J, Ceriani JE, Patiño C, Plana J, Enjuanes L. Absence of E protein arrests transmissible gastroenteritis coronavirus maturation in the secretory pathway. Virology. 2007;368:296-308
15. Wu F, Zhao S, Yu B, Chen Y-M, Wang W, Song Z-G. et al. A new coronavirus associated with human respiratory disease in China. Nature. 2020;579:265-9
16. Lan J, Ge J, Yu J, Shan S, Zhou H, Fan S. et al. Structure of the SARS-CoV-2 spike receptor-binding domain bound to the ACE2 receptor. Nature. 2020;581:215-20
17. Padron-Regalado E. Vaccines for SARS-CoV-2: Lessons from Other Coronavirus Strains. Infect Dis Ther. 2020:1-20
18. Gadanec LK, McSweeney KR, Qaradakhi T, Ali B, Zulli A, Apostolopoulos V. Can SARS-CoV-2 Virus Use Multiple Receptors to Enter Host Cells? International journal of molecular sciences. 2021;22:992
19. Undurraga EA, Chowell G, Mizumoto K. COVID-19 case fatality risk by age and gender in a high testing setting in Latin America: Chile, March-August 2020. Infect Dis Poverty. 2021;10:11
20. Gemmati D, Bramanti B, Serino ML, Secchiero P, Zauli G, Tisato V. COVID-19 and Individual Genetic Susceptibility/Receptivity: Role of ACE1/ACE2 Genes, Immunity, Inflammation and Coagulation. Might the Double X-chromosome in Females Be Protective against SARS-CoV-2 Compared to the Single X-Chromosome in Males? Int J Mol Sci. 2020 21
21. Li J, Wang X, Chen J, Cai Y, Deng A, Yang M. Association between ABO blood groups and risk of SARS-CoV-2 pneumonia. Br J Haematol. 2020;190:24-7
22. Zhao J YY, Huang H, Li D, Gu D, Lu X, Zhang Z, Liu L, Liu T, Liu Y, He Y, Sun B, Wei M, Yang G, Wang X, Zhang L, Zhou X, Xing M, Wang PG. Relationship between the ABO Blood Group and the COVID-19 Susceptibility. Clin Infect Dis. 2020;73:328-331
23. Ellinghaus D, Degenhardt F, Bujanda L, Buti M, Albillos A, Invernizzi P. et al. The ABO blood group locus and a chromosome 3 gene cluster associate with SARSCoV-2 respiratory failure in an Italian-Spanish genome-wide association analysis. medRxiv. 2020
24. Severe Covid GG, Ellinghaus D, Degenhardt F, Bujanda L, Buti M, Albillos A. et al. Genomewide Association Study of Severe Covid-19 with Respiratory Failure. N Engl J Med. 2020;383:1522-34
25. Shelton JF, Shastri AJ, Ye C, Weldon CH, Filshtein-Somnez T, Coker D. et al. Trans-ethnic analysis reveals genetic and non-genetic associations with COVID-19 susceptibility and severity. MedRxiv. 2020
26. Saponi-Cortes JMR, Rivas MD, Calle-Alonso F, Sanchez JF, Costo A, Martin C. et al. IFNL4 genetic variant can predispose to COVID-19. Sci Rep. 2021;11:21185
27. Grimaudo S, Amodio E, Pipitone RM, Maida CM, Pizzo S, Prestileo T. et al. PNPLA3 and TLL-1 Polymorphisms as Potential Predictors of Disease Severity in Patients With COVID-19. Front Cell Dev Biol. 2021;9:627914
28. Monticelli M, Hay Mele B, Benetti E, Fallerini C, Baldassarri M, Furini S. et al. Protective Role of a TMPRSS2 Variant on Severe COVID-19 Outcome in Young Males and Elderly Women. Genes (Basel). 2021 12
29. Jeon S, Blazyte A, Yoon C, Ryu H, Jeon Y, Bhak Y. et al. Regional TMPRSS2 V197M Allele Frequencies Are Correlated with COVID-19 Case Fatality Rates. Mol Cells. 2021;44:680-7
30. Torre-Fuentes L, Matias-Guiu J, Hernandez-Lorenzo L, Montero-Escribano P, Pytel V, Porta-Etessam J. et al. ACE2, TMPRSS2, and Furin variants and SARS-CoV-2 infection in Madrid, Spain. J Med Virol. 2021;93:863-9
31. Benetti E, Tita R, Spiga O, Ciolfi A, Birolo G, Bruselles A. et al. ACE2 gene variants may underlie interindividual variability and susceptibility to COVID-19 in the Italian population. Eur J Hum Genet. 2020;28:1602-14
32. Latini A, Agolini E, Novelli A, Borgiani P, Giannini R, Gravina P. et al. COVID-19 and Genetic Variants of Protein Involved in the SARS-CoV-2 Entry into the Host Cells. Genes (Basel). 2020 11
33. Ejaz H, Alsrhani A, Zafar A, Javed H, Junaid K, Abdalla AE. et al. COVID-19 and comorbidities: Deleterious impact on infected patients. J Infect Public Health. 2020;13:1833-9
34. Gasmi A, Peana M, Pivina L, Srinath S, Gasmi Benahmed A, Semenova Y. et al. Interrelations between COVID-19 and other disorders. Clin Immunol. 2021;224:108651
35. Guan WJ, Liang WH, Zhao Y, Liang HR, Chen ZS, Li YM. et al. Comorbidity and its impact on 1590 patients with COVID-19 in China: a nationwide analysis. Eur Respir J. 2020 55
36. Feng Y, Ling Y, Bai T, Xie Y, Huang J, Li J. et al. COVID-19 with Different Severities: A Multicenter Study of Clinical Features. Am J Respir Crit Care Med. 2020;201:1380-8
37. Li B, Yang J, Zhao F, Zhi L, Wang X, Liu L. et al. Prevalence and impact of cardiovascular metabolic diseases on COVID-19 in China. Clin Res Cardiol. 2020;109:531-8
38. Wang B, Li R, Lu Z, Huang Y. Does comorbidity increase the risk of patients with COVID-19: evidence from meta-analysis. Aging (Albany NY). 2020;12:6049-57
39. Yang J, Zheng Y, Gou X, Pu K, Chen Z, Guo Q. et al. Prevalence of comorbidities and its effects in patients infected with SARS-CoV-2: a systematic review and meta-analysis. Int J Infect Dis. 2020;94:91-5
40. Zhao J, Li X, Gao Y, Huang W. Risk factors for the exacerbation of patients with 2019 Novel Coronavirus: A meta-analysis. Int J Med Sci. 2020;17:1744-50
41. Singh MK, Mobeen A, Chandra A, Joshi S, Ramachandran S. A meta-analysis of comorbidities in COVID-19: Which diseases increase the susceptibility of SARS-CoV-2 infection? Comput Biol Med. 2021;130:104219
42. Campbell F, Archer B, Laurenson-Schafer H, Jinnai Y, Konings F, Batra N. et al. Increased transmissibility and global spread of SARS-CoV-2 variants of concern as at June 2021. Euro Surveill. 2021 26
43. Salvatore M, Bhattacharyya R, Purkayastha S, Zimmermann L, Ray D, Hazra A, et al. Resurgence of SARS-CoV-2 in India: Potential role of the B.1.617.2 (Delta) variant and delayed interventions. medRxiv. 2021: 2021.06.23.21259405
44. Wink PL, Zempulski Volpato FC, Monteiro FL, Willig JB, Zavascki AP, Luís Barth A, et al. First identification of SARS-CoV-2 Lambda (C.37) variant in Southern Brazil. medRxiv. 2021: 2021.06.21.21259241
45. Zhang M, Xiao J, Deng A, Zhang Y, Zhuang Y, Hu T. et al. Transmission Dynamics of an Outbreak of the COVID-19 Delta Variant B.1.617.2 - Guangdong Province, China, May - June 2021. China CDC Weekly. 2021;3:584-6
46. COVID-19 Vaccine Breakthrough Infections Reported to CDC - United States, January 1-April 30, 2021. MMWR Morb Mortal Wkly Rep. 2021; 70: 792-3.
47. Sundell J. Obesity and diabetes as risk factors for coronary artery disease: from the epidemiological aspect to the initial vascular mechanisms. Diabetes Obes Metab. 2005 7
48. Tatusova T, DiCuccio M, Badretdin A, Chetvernin V, Nawrocki EP, Zaslavsky L. et al. NCBI prokaryotic genome annotation pipeline. Nucleic Acids Res. 2016;44:6614-24
49. Bardou P, Mariette J, Escudié F, Djemiel C, Klopp C. jvenn: an interactive Venn diagram viewer. BMC Bioinformatics. 2014;15:293
50. von Mering C, Huynen M, Jaeggi D, Schmidt S, Bork P, Snel B. STRING: a database of predicted functional associations between proteins. Nucleic Acids Res. 2003;31:258-61
51. Auwul MR, Rahman MR, Gov E, Shahjaman M, Moni MA. Bioinformatics and machine learning approach identifies potential drug targets and pathways in COVID-19. Brief Bioinform. 2021 22
52. Ulhaq ZS SG. Interleukin-6 as a potential biomarker of COVID-19 progression. Med Mal Infect. 2020;50:382-3
53. Tanaka T, Narazaki M, Kishimoto T. IL-6 in inflammation, immunity, and disease. Cold Spring Harb Perspect Biol. 2014;6:a016295
54. Jamal M, Bangash HI, Habiba M, Lei Y, Xie T, Sun J. et al. Immune dysregulation and system pathology in COVID-19. Virulence. 2021;12:918-36
55. Wu D, Yang XO. TH17 responses in cytokine storm of COVID-19: An emerging target of JAK2 inhibitor Fedratinib. Journal of microbiology, immunology, and infection = Wei mian yu gan ran za zhi. 2020;53:368-70
56. Chen T, Lin Y-X, Zha Y, Sun Y, Tian J, Yang Z. et al. A Low-Producing Haplotype of Interleukin-6 Disrupting CTCF Binding Is Protective against Severe COVID-19. mBio. 2021;12:e0137221
57. Ulhaq ZS, Soraya GV. Anti-IL-6 receptor antibody treatment for severe COVID-19 and the potential implication of IL-6 gene polymorphisms in novel coronavirus pneumonia. Med Clin (Barc). 2020;155:548-56
58. Rajkumar RP. Genetic polymorphisms mediating behavioural and immune response to pathogens may moderate the impact of the COVID-19 pandemic: a pilot study.06.03.20120998. medRxiv. 2020
59. Liu B, Li M, Zhou Z, Guan X, Xiang Y. Can we use interleukin-6 (IL-6) blockade for coronavirus disease 2019 (COVID-19)-induced cytokine release syndrome (CRS)? J Autoimmun. 2020;111:102452
60. Xu X, Han M, Li T, Sun W, Wang D, Fu B. et al. Effective treatment of severe COVID-19 patients with tocilizumab. Proceedings of the National Academy of Sciences of the United States of America. 2020;117:10970-5
61. Du P, Geng J, Wang F, Chen X, Huang Z, Wang Y. Role of IL-6 inhibitor in treatment of COVID-19-related cytokine release syndrome. Int J Med Sci. 2021;18:1356-62
62. Yang T, Xu C. Physiology and Pathophysiology of the Intrarenal Renin-Angiotensin System: An Update. J Am Soc Nephrol. 2017;28:1040-9
63. Xu Y, Rong J, Zhang Z. The emerging role of angiotensinogen in cardiovascular diseases. J Cell Physiol. 2021;236:68-78
64. Giri A, Hellwege JN, Keaton JM, Park J, Qiu C, Warren HR. et al. Trans-ethnic association study of blood pressure determinants in over 750,000 individuals. Nat Genet. 2019;51:51-62
65. Kouhpayeh HR, Tabasi F, Dehvari M, Naderi M, Bahari G, Khalili T. et al. Association between angiotensinogen (AGT), angiotensin-converting enzyme (ACE) and angiotensin-II receptor 1 (AGTR1) polymorphisms and COVID-19 infection in the southeast of Iran: a preliminary case-control study. Transl Med Commun. 2021;6:26
66. Cafiero C, Rosapepe F, Palmirotta R, Re A, Ottaiano MP, Benincasa G. et al. Angiotensin System Polymorphisms' in SARS-CoV-2 Positive Patients: Assessment Between Symptomatic and Asymptomatic Patients: A Pilot Study. Pharmgenomics Pers Med. 2021;14:621-9
67. Wu C, Ye D, Mullick AE, Li Z, Danser AHJ, Daugherty A. et al. Effects of Renin-Angiotensin Inhibition on ACE2 (Angiotensin-Converting Enzyme 2) and TMPRSS2 (Transmembrane Protease Serine 2) Expression: Insights Into COVID-19. Hypertension. 2020;76:e29-e30
68. Ghafouri-Fard S, Noroozi R, Omrani MD, Branicki W, Pośpiech E, Sayad A. et al. Angiotensin converting enzyme: A review on expression profile and its association with human disorders with special focus on SARS-CoV-2 infection. Vascul Pharmacol. 2020;130:106680
69. Yoshida H, Kuriyama S, Atsumi Y, Tomonari H, Mitarai T, Hamaguchi A. et al. Angiotensin I converting enzyme gene polymorphism in non-insulin dependent diabetes mellitus. Kidney Int. 1996;50:657-64
70. Yamamoto N, Ariumi Y, Nishida N, Yamamoto R, Bauer G, Gojobori T. et al. SARS-CoV-2 infections and COVID-19 mortalities strongly correlate with ACE1 I/D genotype. Gene. 2020;758:144944
71. Jadaon MM. Epidemiology of Prothrombin G20210A Mutation in the Mediterranean Region. Mediterr J Hematol Infect Dis. 2011;3:e2011054
72. Demirci FYK, Dressen AS, Kammerer CM, Barmada MM, Kao AH, Ramsey-Goldman R. et al. Functional polymorphisms of the coagulation factor II gene (F2) and susceptibility to systemic lupus erythematosus. J Rheumatol. 2011;38:652-7
73. Abu-Farha M, Al-Sabah S, Hammad MM, Hebbar P, Channanath AM, John SE. et al. Prognostic Genetic Markers for Thrombosis in COVID-19 Patients: A Focused Analysis on D-Dimer, Homocysteine and Thromboembolism. Front Pharmacol. 2020;11:587451
74. Tang N, Bai H, Chen X, Gong J, Li D, Sun Z. Anticoagulant treatment is associated with decreased mortality in severe coronavirus disease 2019 patients with coagulopathy. Journal of thrombosis and haemostasis: JTH. 2020;18:1094-9
75. Zhou J, Lee S, Guo CL, Chang C, Liu T, Leung KSK. et al. Anticoagulant or antiplatelet use and severe COVID-19 disease: A propensity score-matched territory-wide study. Pharmacol Res. 2021;165:105473
76. Pal R, Bhansali A. COVID-19, diabetes mellitus and ACE2: The conundrum. Diabetes Res Clin Pract. 2020;162:108132
77. Shen W, Xiao L, Li Y, Zhou D, Zhang W. Association between polymorphisms in mannose-binding lectin 2 gene with pulmonary tuberculosis susceptibility. Hereditas. 2020;157:33
78. Medetalibeyoglu A, Bahat G, Senkal N, Kose M, Avci K, Sayin GY. et al. Mannose binding lectin gene 2 (rs1800450) missense variant may contribute to development and severity of COVID-19 infection. Infect Genet Evol. 2021;89:104717
79. Monticelli M, Mele BH, Andreotti G, Cubellis MV, Riccio G. Why does SARS-CoV-2 hit in different ways? Host genetic factors can influence the acquisition or the course of COVID-19. Eur J Med Genet. 2021;64:104227
80. Dawson TC, Beck MA, Kuziel WA, Henderson F, Maeda N. Contrasting effects of CCR5 and CCR2 deficiency in the pulmonary inflammatory response to influenza A virus. Am J Pathol. 2000;156:1951-9
81. Ahlenstiel G, Woitas RP, Rockstroh J, Spengler U. CC-chemokine receptor 5 (CCR5) in hepatitis C-at the crossroads of the antiviral immune response? J Antimicrob Chemother. 2004;53:895-8
82. Mummidi S, Ahuja SS, McDaniel BL, Ahuja SK. The human CC chemokine receptor 5 (CCR5) gene. Multiple transcripts with 5'-end heterogeneity, dual promoter usage, and evidence for polymorphisms within the regulatory regions and noncoding exons. J Biol Chem. 1997;272:30662-71
83. Oh JH, Tannenbaum A, Deasy JO. Identification of biological correlates associated with respiratory failure in COVID-19. BMC Med Genomics. 2020;13:186
84. Panda AK, Padhi A, Prusty BAK. CCR5 Δ32 minorallele is associated with susceptibility to SARS-CoV-2 infection and death: An epidemiological investigation. Clinica chimica acta; international journal of clinical chemistry. 2020;510:60-1
85. Mueller KAL, Langnau C, Günter M, Pöschel S, Gekeler S, Petersen-Uribe Á. et al. Numbers and phenotype of non-classical CD14dimCD16+ monocytes are predictors of adverse clinical outcome in patients with coronary artery disease and severe SARS-CoV-2 infection. Cardiovasc Res. 2021;117:224-39
86. Zhao Y, Zhao Z, Wang Y, Zhou Y, Ma Y, Zuo W. Single-Cell RNA Expression Profiling of ACE2, the Receptor of SARS-CoV-2. Am J Respir Crit Care Med. 2020;202:756-9
87. Gao C, Zeng J, Jia N, Stavenhagen K, Matsumoto Y, Zhang H. et al. SARS-CoV-2 Spike Protein Interacts with Multiple Innate Immune Receptors. bioRxiv. 2020. 2020 07.29.227462
88. Brandão SCS, Ramos JdOX, Dompieri LT, Godoi ETAM, Figueiredo JL, Sarinho ESC. et al. Is Toll-like receptor 4 involved in the severity of COVID-19 pathology in patients with cardiometabolic comorbidities? Cytokine Growth Factor Rev. 2021;58:102-10
89. Aboudounya MM, Heads RJ. COVID-19 and Toll-Like Receptor 4 (TLR4): SARS-CoV-2 May Bind and Activate TLR4 to Increase ACE2 Expression, Facilitating Entry and Causing Hyperinflammation. Mediators Inflamm. 2021;2021:8874339
90. Saeedi-Boroujeni A, Mahmoudian-Sani M-R, Bahadoram M, Alghasi A. COVID-19: A Case for Inhibiting NLRP3 Inflammasome, Suppression of Inflammation with Curcumin? Basic Clin Pharmacol Toxicol. 2021;128:37-45
91. Hertanto DM, Sutanto H, Wiratama BS, Wungu CDK. Modulating the host immune response to fight against COVID-19: Where are we in 2021? Virulence. 2021;12:1732-6
92. Huang C, Wang Y, Li X, Ren L, Zhao J, Hu Y. et al. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. The Lancet. 2020;395:497-506
93. Naqvi AAT, Fatima K, Mohammad T, Fatima U, Singh IK, Singh A. et al. Insights into SARS-CoV-2 genome, structure, evolution, pathogenesis and therapies: Structural genomics approach. Biochim Biophys Acta Mol Basis Dis. 2020;1866:165878
94. Ovsyannikova IG, Haralambieva IH, Crooke SN, Poland GA, Kennedy RB. The role of host genetics in the immune response to SARS-CoV-2 and COVID-19 susceptibility and severity. Immunol Rev. 2020;296:205-19
95. Fricke-Galindo I, Falfán-Valencia R. Genetics Insight for COVID-19 Susceptibility and Severity: A Review. Frontiers in immunology. 2021;12:622176
96. Nile SH, Nile A, Qiu J, Li L, Jia X, Kai G. COVID-19: Pathogenesis, cytokine storm and therapeutic potential of interferons. Cytokine Growth Factor Rev. 2020;53:66-70
97. Ye Q, Wang B, Mao J. The pathogenesis and treatment of the `Cytokine Storm' in COVID-19. The Journal of infection. 2020;80:607-13
98. Gao C, Cai Y, Zhang K, Zhou L, Zhang Y, Zhang X. et al. Association of hypertension and antihypertensive treatment with COVID-19 mortality: a retrospective observational study. Eur Heart J. 2020;41:2058-66
99. Baral R, White M, Vassiliou VS. Effect of Renin-Angiotensin-Aldosterone System Inhibitors in Patients with COVID-19: a Systematic Review and Meta-analysis of 28,872 Patients. Curr Atheroscler Rep. 2020;22:61
100. Yan H, Zou C. Mechanism and material basis of Lianhua Qingwen capsule for improving clinical cure rate of COVID-19: a study based on network pharmacology and molecular docking technology. Nan Fang Yi Ke Da Xue Xue Bao. 2021;41:20-30
101. Menendez JA. Metformin and SARS-CoV-2: mechanistic lessons on air pollution to weather the cytokine/thrombotic storm in COVID-19. Aging (Albany NY). 2020;12:8760-5
102. Freitas AT, Calhau C, Antunes G, Araújo B, Bandeira M, Barreira S. et al. Vitamin D-related polymorphisms and vitamin D levels as risk biomarkers of COVID-19 disease severity. Sci Rep. 2021 11
103. Kumar R, Rathi H, Haq A, Wimalawansa SJ, Sharma A. Putative roles of vitamin D in modulating immune response and immunopathology associated with COVID-19. Virus Res. 2021;292:198235
104. Villar J, Ferrando C, Martínez D, Ambrós A, Muñoz T, Soler JA. et al. Dexamethasone treatment for the acute respiratory distress syndrome: a multicentre, randomised controlled trial. Lancet Respir Med. 2020;8:267-76
105. Hata TR, Audish D, Kotol P, Coda A, Kabigting F, Miller J. et al. A randomized controlled double-blind investigation of the effects of vitamin D dietary supplementation in subjects with atopic dermatitis. J Eur Acad Dermatol Venereol. 2014;28:781-9
106. Sadeghi K, Wessner B, Laggner U, Ploder M, Tamandl D, Friedl J. et al. Vitamin D3 down-regulates monocyte TLR expression and triggers hyporesponsiveness to pathogen-associated molecular patterns. Eur J Immunol. 2006;36:361-70
107. Labudzynskyi D, Shymanskyy I, Veliky M. Role of vitamin D3 in regulation of interleukin-6 and osteopontin expression in liver of diabetic mice. Eur Rev Med Pharmacol Sci. 2016;20:2916-9
108. Chandel N, Ayasolla K, Wen H, Lan X, Haque S, Saleem MA. et al. Vitamin D receptor deficit induces activation of renin angiotensin system via SIRT1 modulation in podocytes. Exp Mol Pathol. 2017;102:97-105
109. Peng M-Y, Liu W-C, Zheng J-Q, Lu C-L, Hou Y-C, Zheng C-M. et al. Immunological Aspects of SARS-CoV-2 Infection and the Putative Beneficial Role of Vitamin-D. International journal of molecular sciences. 2021 22
110. Davies SP, Mycroft-West CJ, Pagani I, Hill HJ, Chen Y-H, Karlsson R. et al. The Hyperlipidaemic Drug Fenofibrate Significantly Reduces Infection by SARS-CoV-2 in Cell Culture Models. Frontiers in Pharmacology. 2021 12
111. Stolarz AJ, Farris RA, Wiley CA, O'Brien CE, Price ET. Fenofibrate Attenuates Neutrophilic Inflammation in Airway Epithelia: Potential Drug Repurposing for Cystic Fibrosis. Clin Transl Sci. 2015;8:696-701
112. Undas A, Celinska-Löwenhoff M, Löwenhoff T, Szczeklik A. Statins, fenofibrate, and quinapril increase clot permeability and enhance fibrinolysis in patients with coronary artery disease. Journal of thrombosis and haemostasis: JTH. 2006;4:1029-36
113. Maisonneuve C, Bertholet S, Philpott DJ, De Gregorio E. Unleashing the potential of NOD- and Toll-like agonists as vaccine adjuvants. Proceedings of the National Academy of Sciences of the United States of America. 2014;111:12294-9
114. Malik G, Zhou Y. Innate Immune Sensing of Influenza A Virus. Viruses. 2020 12
115. Lacaille-Dubois M-A. Updated insights into the mechanism of action and clinical profile of the immunoadjuvant QS-21: A review. Phytomedicine. 2019;60:152905
116. Reinke S, Thakur A, Gartlan C, Bezbradica JS, Milicic A. Inflammasome-Mediated Immunogenicity of Clinical and Experimental Vaccine Adjuvants. Vaccines (Basel). 2020;8:554
117. Reed SG, Hsu F-C, Carter D, Orr MT. The science of vaccine adjuvants: advances in TLR4 ligand adjuvants. Curr Opin Immunol. 2016;41:85-90
118. Kim E, Erdos G, Huang S, Kenniston TW, Balmert SC, Carey CD. et al. Microneedle array delivered recombinant coronavirus vaccines: Immunogenicity and rapid translational development. EBioMedicine. 2020;55:102743
119. Bhattacharya M, Sharma AR, Patra P, Ghosh P, Sharma G, Patra BC. et al. Development of epitope-based peptide vaccine against novel coronavirus 2019 (SARS-COV-2): Immunoinformatics approach. Journal of medical virology. 2020;92:618-31
120. Ulloa-Montoya F, Louahed J, Dizier B, Gruselle O, Spiessens B, Lehmann FF. et al. Predictive gene signature in MAGE-A3 antigen-specific cancer immunotherapy. J Clin Oncol. 2013;31:2388-95
121. Sallenave J-M, Guillot L. Innate Immune Signaling and Proteolytic Pathways in the Resolution or Exacerbation of SARS-CoV-2 in Covid-19: Key Therapeutic Targets? Frontiers in immunology. 2020;11:1229
Author contact
![]() Corresponding authors: Xu Jia, E-mail: jiaxuedu.cn; Ling Zhang, E-mail: zhanglinfudanedu.cn; Jinlin Guo, E-mail: guo596edu.cn; Fengling Qiao: 969939206com.
Corresponding authors: Xu Jia, E-mail: jiaxuedu.cn; Ling Zhang, E-mail: zhanglinfudanedu.cn; Jinlin Guo, E-mail: guo596edu.cn; Fengling Qiao: 969939206com.

 Global reach, higher impact
Global reach, higher impact