Impact Factor
ISSN: 1449-1907
Int J Med Sci 2022; 19(1):132-141. doi:10.7150/ijms.61798 This issue Cite
Research Paper
Spatiotemporal Expression Patterns of Critical Genes Involved in FGF Signaling During Morphogenesis and Odontogenesis of Deciduous Molars in Miniature Pigs
1. Beijing Laboratory of Oral Health; Capital Medical University School of Stomatology, Beijing, China.
2. Department of Oral Pathology, Peking University School and Hospital of Stomatology, Beijing, China.
3. Department of Oral Basic Science, School of Stomatology, Dalian Medical University, Dalian, China.
4. Department of Biochemistry and Molecular Biology, Capital Medical University School of Basic Medical Sciences, Beijing, China.
5. Department of Oral and Maxillofacial Surgery, Xiangya Hospital, Central South University, Changsha, China.
6. Academician Workstation for Oral-Maxillofacial Regenerative Medicine, Central South University, Changsha, China.
*These authors (Wenwen Guo and Xiaoyu Lin) contribute equally to this work.
Received 2021-4-19; Accepted 2021-11-2; Published 2022-1-1
Abstract
The fibroblast growth factor (FGF) pathway plays an important role in epithelial-mesenchymal interactions during tooth development. Nevertheless, how the ligands, receptors, and antagonists of the FGF pathway are involved in epithelial-mesenchymal interactions remains largely unknown. Miniature pigs exhibit tooth anatomy and replacement patterns like those in humans and hence can serve as large animal models. The present study investigated the spatiotemporal expression patterns of critical genes encoding FGF ligands (FGF3, FGF4, FGF7, and FGF9), antagonists (SPRY2 and SPRY4) and receptors (FGFR1, FGFR2, and FGFR3) in the third deciduous molars of miniature pigs at the cap (embryonic day 40, E40), early bell (E50), and late bell (E60) stages. The results of in situ hybridization (ISH) with tyramide signal amplification and of qRT-PCR analysis revealed increased expression of FGF7, FGFR1, FGFR2, and SPRY4 in dental epithelium and of FGF7 and FGFR1 in mesenchyme from E40 to E50. In contrast, the results revealed decreased expression of FGF3, FGF4, FGF9, and FGFR3 in dental epithelium and of FGF4, FGF9, FGFR2, and FGFR3 in the mesenchyme from E40 to E60. Mesenchyme signals of FGF3, FGF4, FGF7, SPRY2, FGFR2, and FGFR3 were concentrated in the odontoblast layer from E50 to E60. The distinct expression patterns of these molecules indicated elaborate regulation during dental morphogenesis. Our results provide a foundation for further investigation into fine-tuning dental morphogenesis and odontogenesis by controlling interactions between dental epithelium and mesenchyme, thus promoting tooth regeneration in large mammals.
Keywords: miniature pig, tooth development, deciduous molar, fibroblast growth factor, dental epithelium, dental mesenchyme
Introduction
Tooth development begins when the local oral epithelium thickens to form a placode called the dental lamina, which invaginates to surround the underlying mesenchyme and form a follicle that enters the bud stage. At the cap stage, the tip of the bud invaginates and the dental epithelium differentiates into the enamel organ, which includes inner (IEE) and outer (OEE) enamel epithelium, stellate reticulum (SR), and an intermediate cell layer. The apical part of the IEE and the underlying dental mesenchyme (DM) differentiate into ameloblasts and odontoblasts at the late bell stage [1]. Reciprocal interactions between the ectoderm-derived epithelium and underlying neural crest-derived mesenchyme occur at every step of development [1, 2].
Proteins in the fibroblast growth factor (FGF) family mediate inductive interactions between dental epithelium (DE) and the DM during successive stages of tooth formation [3]. Based on sequence similarity, receptor specificity, and binding affinity, FGFs can be subdivided into the following subfamilies: FGF1, FGF4, FGF7, FGF8, FGF9, FGF11, and FGF15 [4-7]. Most FGFs mediate their biological responses as extracellular proteins that bind to and activate cell surface tyrosine kinase FGF receptors (fibroblast growth receptors; FGFRs) 1-4 [4].
Fibroblast growth factor signaling is crucial during the development of tooth epithelium and mesenchyme [8]. The initial expression of Fgf4, Fgf8, and Fgf9 in the dental epithelium and later in the enamel knot affects cell proliferation during tooth initiation and subsequent morphogenesis that regulates the establishment of tooth shape [9-14]. Furthermore, Fgf3, Fgf7, and Fgf10 are expressed in the DM of mice [15].
Sprouty proteins act downstream of FGF signal activation by interfering with the RAS-MAPK pathway. Sprouty RTK signaling antagonist (SPRY) genes were originally identified as antagonists of FGFR-mediated signaling [16, 17]. The expression of the Spry1, Spry2, and Spry4 genes encoding antagonists is required for appropriate molar cusp patterning throughout enamel development [13, 18, 19]. However, whether joint FGF and SPRY expression regulate tooth development by modulating epithelial-mesenchymal interactions in large mammals remains unclear.
Tooth development and the cascade initiation of permanent molars have been investigated in the third deciduous molars (DM3) of miniature pigs [20-22]. Here, we characterized the dynamic expression profiles of genes associated with the FGF signaling pathway in DM3 of miniature pigs at the cap, early, and late bell (secretory) stages, with focus on associations with cusp patterning and tooth mineralization. Our findings clarified interactions between the epithelium and mesenchyme during tooth development, which might be applicable to human tooth regeneration.
Results
Morphology of DM3 across developmental stages in miniature pigs
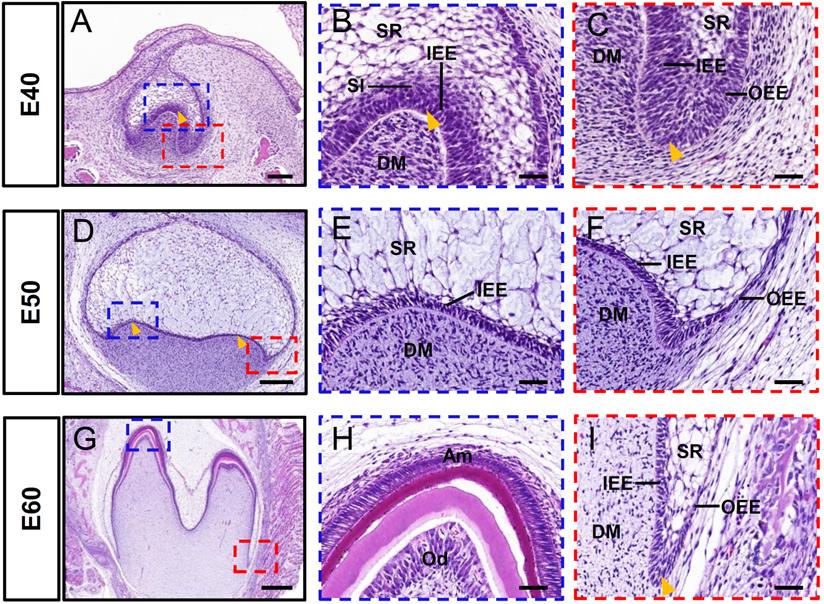
The cap, early bell, and late bell (secretory) stages of DM3 development occurred in miniature pigs at embryonic day 40 (E40), E50, and E60, respectively. The tooth bud of DM3 entered the cap stage at E40 (Figure 1A). This was characterized by formation of the primary enamel knot, a central region of epithelium (arrowhead) that comprises the signaling center involved in regulating tooth shape (Figure 1B). Furthermore, the epithelial bud underwent specific folding (arrowhead) and was composed of IEE and OEE, SR, and stratum intermedium (Figure 1B-C). At this stage, the cervical loop was formed by the IEE and OEE, together with the SR (Figure 1C). A typical feature of E50 was the formation of secondary enamel knots (arrowheads) and the start of transformation into a bell shape (Figure 1D-F). The DM3 transitioned to the later bell stage at E60 (Figure 1G). The cells at the tip of the IEE differentiated into ameloblasts and began to form pre-enamel prisms, whereas the cells at the tip of the DM differentiated into odontoblasts (Figure 1H). Furthermore, the cervical loop (arrowheads), formed by the IEE and OEE, with a minor contribution from the SR, elongated toward the direction of the future root (Figure 1I). Thus, the analysis of DM3 development at stages E40-E60 revealed information about the processes of cusp shaping, crown mineralization, and root morphogenesis in miniature pigs.
Dynamic expression of genes encoding FGF ligands and antagonists during morphogenesis of DM3
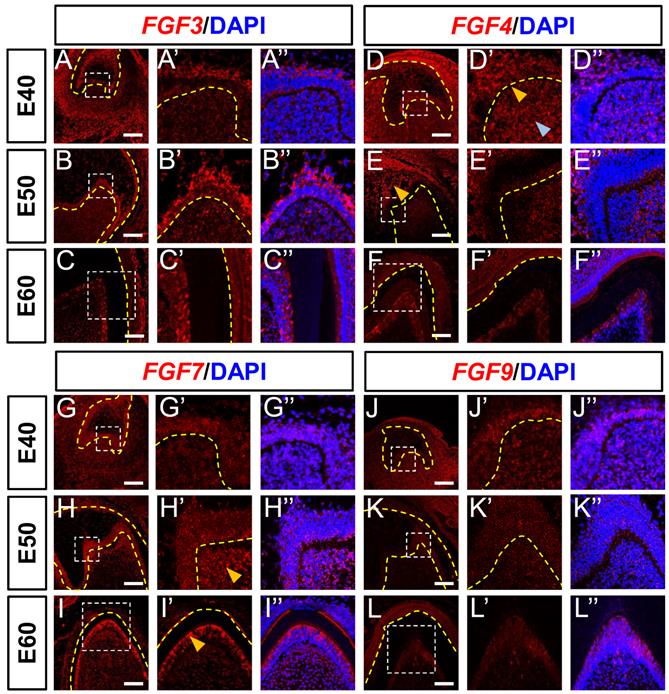
The Fgf3, Fgf4, Fgf7 and Fgf9 mRNAs are central components of the typical FGF signaling pathway, whereas Spry2 and Spry4 are key antagonists [13]. Therefore, we examined the expression of these genes in the epithelium and mesenchyme of DM3 at E40, E50, and E60 using in situ hybridization (ISH) and immunofluorescence staining. We identified FGF3 mRNA mainly in the IEE, SR, and OEE at E40 (Figure 2A), and the apical site of the IEE became more obvious at E50, indicating that FGF3 plays a role in cusp patterning (Figure 2B). The expression of FGF3 in the DM was low at E40, but gradually increased from E50 to E60 and finally localized in the odontoblast cell layer at E60 (Figure 2B and C), whereas FGF3 expression in the cervical loop increased from E40 to E50 and E60 (Figure S2A-C). We found that FGF4 mRNA was expressed in the SR, enamel knot (yellow arrow), and DM (blue arrow) at E40 (Figure 2D). However, expression at E50 was increased in the SR (yellow arrow), but decreased in the mesenchyme and finally localized at the odontoblast layer at E60 (Figure 2E and F). The expression in the cervical loop increased at E50 but decreased at E60 (Figure S2D-F).
Morphology of DM3 across developmental stages in miniature pigs. (A-I) HE staining at different stages during DM3 tooth germ morphogenesis. Boxed regions in A, D, G are magnified in B-C, E-F, H-I. (A-C) Tooth germ progresses to cap stage at embryonic day 40 (E40). Arrowheads, primary enamel knot in A and B, and cervical loop in C. (D-F) At E50, DM3 reaches early bell stage and secondary enamel knots (arrowheads) begin to form; (G-I) At E60, DM3 germ developed into late bell (secretory stage), with elongated cervical loop (arrowhead). Parts of IEE and dental papilla cells at cusp tip have differentiated into ameloblasts and odontoblasts. Am, ameloblasts; DM, dental mesenchyme; DM3, third deciduous molars; HE, hematoxylin and eosin; IEE, inner enamel epithelium; Od, odontoblasts. OEE, outer enamel epithelium; SR, stellate reticulum; SI, stratum intermedium. Scale bars: 100, 200, and 500 μm in A, D, and G, respectively, and 50 μm in B-C, E-F, H-I.
We detected FGF7 mRNA in both DE and DM at the cap stage (E40; Figure 2G). Mesenchymal expression of FGF7 mRNA was elevated apically at E50 (yellow arrow) and concentrated in the odontoblast layer (yellow arrow) at E60 (Figure 2H and I). The apical restriction and concentration indicated that FGF7 regulates odontoblast differentiation. In addition, FGF7 expression in the cervical loop was reduced from E40 to E60 (Figure S2G-I). Although FGF9 mRNA was mainly expressed in the IEE and DM at E40 (Figure 2J), the abundance was obviously reduced at E50 and E60 (Figure 2J-L). We found FGF9 mRNA in the cervical loops at E40 and E50 (Figure S2J-L). In summary, FGF3 and FGF7 mRNA expression was upregulated and concentrated in the mesenchyme from E40 to E60. In contrast, FGF4 and FGF9 mRNA expression in the mesenchyme decreased from E40 to E60.
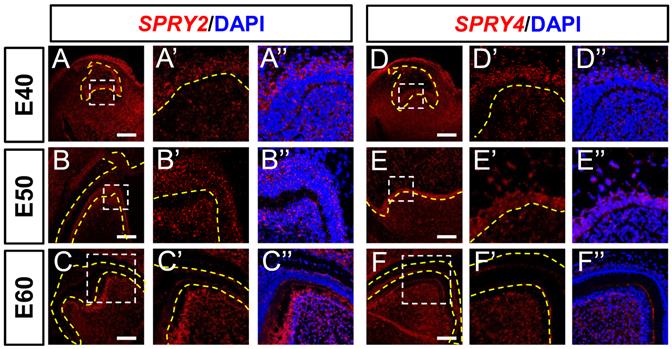
We explored the expression of FGF signaling antagonists. The expression of SPRY2 was abundant in the DE at E40, increased at E50, and decreased at E60 (Figure 3A-C). The expression of SPRY2 was increased in the DM, and concentrated in odontoblasts from E40 to E60 (Figure 3A-C). The expression of SPRY4 was detected in the intermediate cell layer at E40, and increased at E50, but decreased in the IEE at E60 (Figure 3D-F). We detected SPRY4 mRNA in DM from E40 to E60 (Figure 3E-F). Moreover, SPRY2 and SPRY4 mRNAs were expressed in the cervical loop during the three stages (Figure S3A-C and S3D-F). In summary, SPRY2 and SPRY4 were upregulated in DE, whereas only SPRY2 was upregulated in the DM during odontogenesis.
Distribution of FGF ligand proteins during morphogenesis of DM3
We examined the distribution of several FGF proteins at the three stages of DM3 development using immunofluorescence staining to confirm the results of ISH. We found that FGF3 protein was expressed mainly in the DE at E40, in both the DE and DM at E50, and in the ameloblast and odontoblast layer at E60 (Figure 4A-C). The expression of FGF3 protein was abundant in the cervical loop at E50 and E60 (Figure S4A-C). The expression patterns of FGF3 protein and mRNA were similar.
Although FGF4 protein was expressed mainly in the SR (Figure 4D) at E40, FGF4 mRNA was simultaneously expressed in the enamel knot and mesenchyme (Figure 2D). By E50, FGF4 was expressed mainly in DE (Figure 4E), whereas FGF4 mRNA was found primarily in the SR and mesenchyme, but not in the DE at E50 (Figure 2E). At E60, FGF4 protein expression was considerably reduced (Figure 4F), which was similar to the mRNA profile (Figure 2F). The FGF4 protein and FGF4 mRNA were similarly expressed in the cervical loop from E40 to E60 (Figures S4D-F and S2D-F).
The expression of FGF7 protein and FGF7 mRNA gradually increased from E40 to E60 in the DE and DM (Figures 4G-I and 2G-I), respectively. The expression of FGF7 protein and FGF7 mRNA in the cervical loop was weak from E40 to E60 (Figures S4G-I and S2G-I).
The expression level of FGF9 protein (Figure 4J-L) gradually decreased like that of FGF9 mRNA (Figure 2J-L) in the DE and DM from E40 to E60. The FGF9 protein (Figure S4J-L) and FGF9 mRNA (Figure S2J-L) were both expressed in the cervical loop from E40 to E60. The different expression levels of mRNA and protein between FGF4 and FGF7 indicated protein translocation between the epithelium and mesenchyme during tooth development.
Dynamic expression of genes encoding FGF ligands during morphogenesis of DM3. (A-L) In situ hybridization (ISH) shows mRNA expression of FGF ligands (red) and nuclei stained with DAPI (blue) from E40 to E60. White boxed regions in A-L are magnified in A'-L', and DAPI-stained nuclei are overlaid in A”-L”. Expression of FGF3 (A-C) and FGF4 (D-F) mRNA from E40 to E60. Yellow and blue arrowheads, enamel knot and mesenchyme, respectively in D'. Yellow arrowhead, SR in E. (G-I) Expression of FGF7 mRNA from E40 to E60; Yellow arrowheads, apical mesenchyme in H', and odontoblasts in I'. (J-L) Expression of FGF9 mRNA E40 to E60. Yellow dotted line, boundary of tooth epithelium and mesenchyme. Scale bar, 100 μm. E, epithelium; SR, stellate reticulum.
Dynamic expression of genes encoding FGF antagonists during morphogenesis of DM3. (A-F) In situ hybridization (ISH) shows mRNA expression of FGF antagonists (red) and nuclei stained with DAPI (blue) from E40 to E60. White boxed regions in A-F are magnified in A'-F', and DAPI staining is overlaid in A''-F”. Expression of SPRY2 (A-C) and SPRY4 (D-F) mRNA from E40 to E60. Yellow dotted line, boundary of tooth epithelium and mesenchyme. Scale bar, 100 μm.
Protein expression of FGF ligands during morphogenesis of DM3. (A-F) Immunofluorescence (IF) staining shows protein expression of FGF ligands (red) and nuclei stained with DAPI (blue) from E40 to E60. White boxed regions in A-L are magnified in A'-L', and overlaid with DAPI staining in A'-L”. Expression of (A-C) FGF3, (D-F) FGF4, (G-I) FGF7 and (J-L) FGF9 from E40 to E60. Yellow dotted line, boundary of tooth epithelium and mesenchyme. Scale bar, 50 μm.
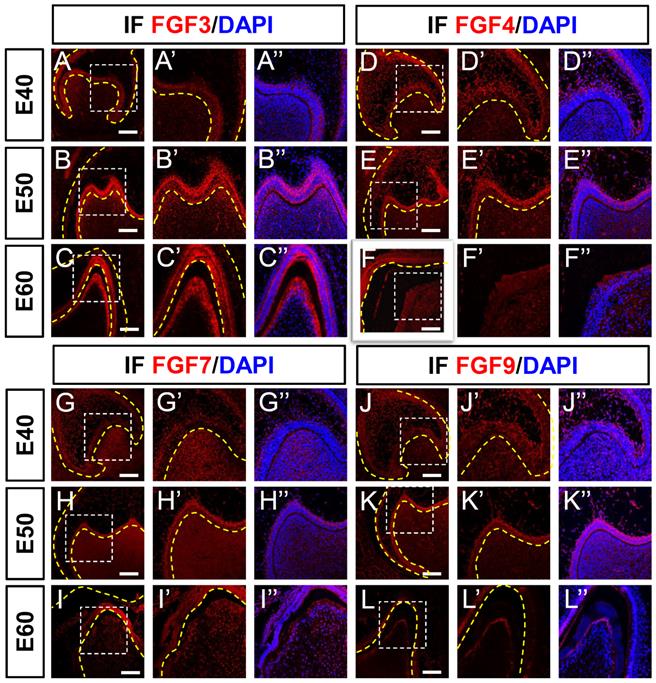
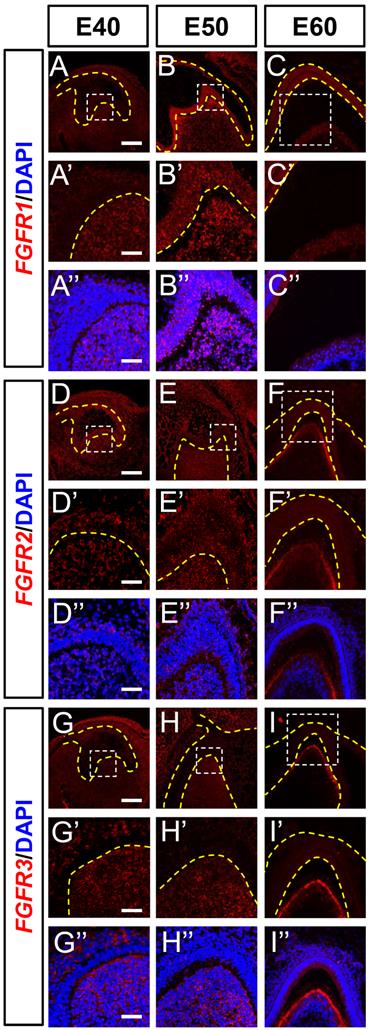
Dynamic expression of genes encoding FGF receptors during morphogenesis of DM3. (A-I) In situ hybridization (ISH) shows mRNA expression of FGF receptors (red) and nuclei stained with DAPI (blue) from E40 to E60. White boxed regions in A-I are magnified in A'-I', and DAPI staining is overlaid in A'-I”. Expression of (A-C) FGFR1, (D-F) FGFR2, and (G-I) FGFR3 mRNA from E40 to E60. Yellow dotted line, boundary of tooth epithelium and mesenchyme. Scale bar, 100 μm.
Dynamic expression of genes encoding FGF receptors during morphogenesis of DM3
Fibroblast growth factor ligands bind to FGF receptors on the surfaces of target cells to transfer signals to nuclei. We examined FGFR1, FGFR2, and FGFR3 mRNA expression levels in DM3 at the three developmental stages using ISH. The FGFR1 mRNA was expressed mainly in the DM at E40 and in both DE and DM at E50. Expression was elevated in the epithelium and mesenchyme from E40 to E50 (Figure 5A and B) but greatly reduced at E60 (Figure 5C). The expression of FGFR1 was abundant at E50 but scarce at E40 and E60 (Figure S5A-C).
Although FGFR2 was found in the DE and DM at E40 and E50, it was mainly localized in the odontoblast layer at E60 (Figure 5D-F), and FGFR3 was detected in the SR and DM at E40 (Figure 5G). The expression was restricted to the DM between E50 and E60 and limited to secretory odontoblasts at E60 (Figure 5H and I). Although FGFR2 and FGFR3 transcripts were identified in the cervical loop at E40, they were basically absent at E50 and E60 (Figure S5D-I).
Quantitative gene expression dynamics related to the FGF pathway and its antagonists in DM3
To further understand the expression dynamics of FGF-related genes and antagonists, we quantified their mRNA levels by real-time PCR and analyzed differences among the genes in DE and DM across the three stages. Tooth germs of DM3 were harvested, and epithelium including IEE, OEE, and SR was enzymatically separated from the mesenchyme using dispase II before reverse transcription [26].
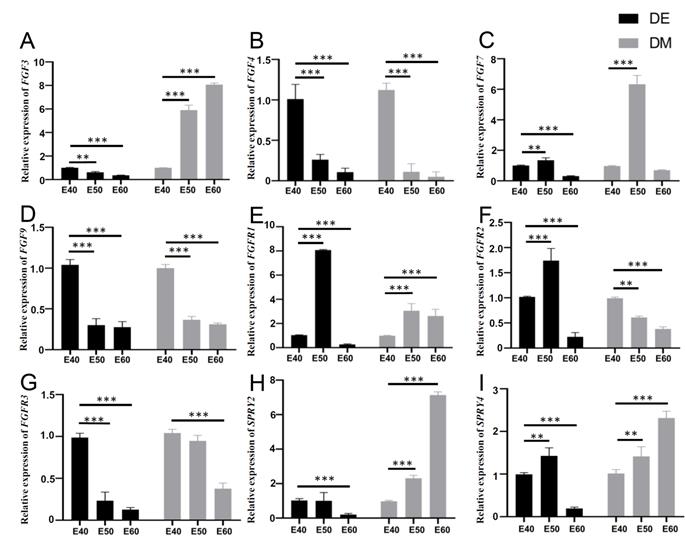
The RT-PCR results showed the expression of FGF3, FGF4, FGF9, and FGFR3 was decreased in the DE from E40 to E60. The expression of FGF7, FGFR1, FGFR2, and SPRY4 was increased in the DE from E40 to E50, but decreased in the DE from E50 to E60. The results showed the increased expression of FGF3, SPRY2, and SPRY4 and decreased expression of FGF4, FGF9, FGFR2, and FGFR3 in the DM from E40 to E60. The expression of FGF7 and FGFR1 was increased in the DM from E40 to E50, but decreased in the DM from E50 to E60 (Figure 6). The trends fit with the ISH results generally.
Discussion
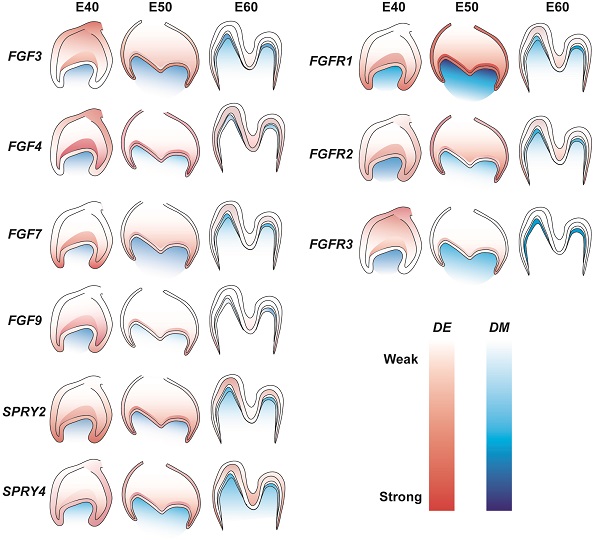
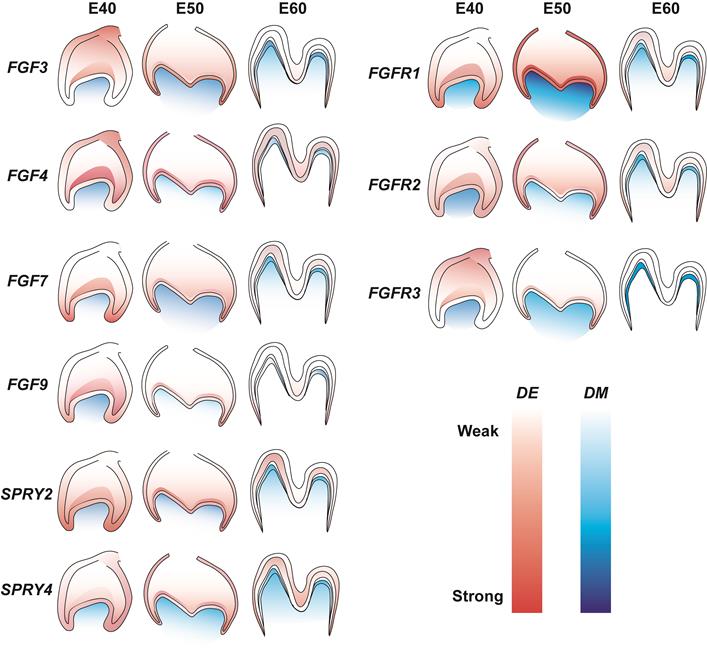
The role of FGF signaling in the tooth development of miniature pigs is not understood in detail. The present study examined the dynamic expression of representative FGF ligands and receptors in addition to DM3 antagonists at E40, E50, and E60. We found increased expression of FGF7, FGFR1, FGFR2, and SPRY4 in dental epithelium and of FGF7 and FGFR1 in mesenchyme from E40 to E50. In contrast, the results revealed decreased expression of FGF3, FGF4, FGF9, and FGFR3 in dental epithelium and of FGF4, FGF9, FGFR2, and FGFR3 in the mesenchyme from E40 to E60. The mesenchymal signals of FGF3, FGF4, FGF7, SPRY2, FGFR2, and FGFR3 were concentrated in the odontoblast layer from E50 to E60. Figure 7 shows details of related gene expression. Overall, the FGF signaling pathway and SPRY genes might communicate between the epithelium and mesenchyme to maintain tooth morphology and mineralization.
A comparison of the present, with previous findings revealed similarities and differences among mice [13, 27], pigs, and humans [28] (Table 1). Fibroblast growth factor 3 plays a significant role in the proliferation and morphogenesis of tooth development [13]. In humans, FGF3 is predominantly expressed in mesenchyme, particularly in pre-odontoblast and odontoblast cell layers at the late stage [28]. Our findings were similar to these. Although FGF7 is expressed in DE and DM at the cap stage of human teeth [28], it is not involved in tooth development in mice [12]. We found remarkably abundant FGF7 expression in the DM at the early bell stage, but it was restricted to the odontoblast layer at the secretory stage in the DM3. Both FGF4 and FGF9 mRNAs are indispensable for DE in the growth of mouse teeth [3]. We detected FGF4 and FGF9 expression in the DE and the DM from the cap until the early bell stage of DM3. However, its functional role in DM remains unclear. Our results indicated that FGF3, FGF4, FGF7, and FGF9 ligands play critical roles in cusp patterning, whereas FGF3 and Fgf7 might play fundamental roles in odontoblast differentiation.
Fibroblast growth factor ligands bind to their receptors and activate intracellular signal transduction pathways by inducing receptor phosphorylation during tooth growth. In humans, FGF7 binds specifically to FGFR2, whereas FGF3 and FGF10 bind to both FGFR1 and FGFR2 [29, 30]. The frequency of supernumerary tooth formation is reduced to 60% and 0% by Fgfr1 and Fgfr2 null allele heterozygosity, respectively, in transgenic mice [31, 32]. We found increased FGFR1 expression in IEE and DM from E40 to E50, FGFR2 expression in DE and DM, and FGFR3 expression mainly in DM at the three stages of DM3. However, these receptors are associated with gene expression and function networks in miniature pigs and require further analysis.
Quantitative gene expression dynamics related to the FGF pathway and its antagonists of DM3. (A-I) Relative mRNA expression in dental epithelium (DE) and mesenchyme (DM) from E40 to E60 determined by qRT-PCR. Expression of (A-D) FGF3, FGF4, FGF7, and FGF9, and (E-G) FGFR1, FGFR2, and FGFR3 from E40 to E60. (H-I) Expression of SPRY2 and SPRY4 from E40 to E60. *p < 0.05, **p < 0.01, ***p < 0.001.
Comparison of expression of the FGF signaling pathway and antagonists between mouse, pig, and human.
| Stage | Tissues/cells | Species | ||
|---|---|---|---|---|
| Mouse [12, 26] | Pig | Human [27] | ||
| Cap stage | Enamel knot/dental epithelium | Fgf3, Fgf4, Fgf9, Fgf15, Fgf20, Fgf16, Fg17, Fgfr1Ⅲb, Fgfr1Ⅲc, Fgfr2Ⅲb, Spry2, Spry4 | FGF3, FGF4, FGF7, FGF9, FGFR1, FGFR2, FGFR3, SPRY2, SPRY4 | FGF3, FGF4, FGF7, FGF8, FGF9, FGF10, FGFR1, FGFR2, FGFR3 |
| Dental papilla | Fgf3, Fgf10, Fgf16, Fgf17, Fgf18, Fgfr1Ⅲc, Fgfr2Ⅲc, Spry4 | FGF3, FGF4, FGF7, FGF9, FGFR1, FGFR2, FGFR3, SPRY2, SPRY4 | FGF3, FGF4, FGF7, FGF8, FGF9, FGF10, FGFR1, FGFR2, FGFR3 | |
| Bell stage | Enamel knot/ dental epithelium | Fgf4, Fgf9, Fgf16, Fgf20, Fgfr1Ⅲb, Fgfr1Ⅲc, | FGF3, FGF4, FGF7, FGF9, FGFR1, FGFR2, SPRY2 | FGF3, FGF4, FGF7, FGF8, FGF9, FGF10, FGFR1, FGFR2, FGFR3 |
| Dental papilla | Fgf3, Fgf10, Fgf15, Fgfr1IIIb, Fgfr1IIIc, Fgfr2IIIc | FGF3, FGF4, FGF7, FGFR1, FGFR2, FGFR3 SPRY2, SPRY4 | FGF7, FGF8, FGF9, FGFR1 FGFR2, FGFR3 | |
| Secretory stage | Ameloblast/ dental epithelium | Fgf2, Fgf4, Fgf9, Fgf9, Fgf16, Fgfr1, Fgfr2IIIb | FGF3, FGF7, FGFR1, SPRY2 | |
| Odontoblast/pre-odontoblast | Fgfr1IIIb, Fgfr1IIIc | FGF3, FGF7, SPRY2, FGFR1, FGFR2, FGFR3, SPRY2, SPRY4 | ||
| Dental papilla | Fgf15 | FGF3, FGFR1, FGFR2, SPRY2, SPRY4 | ||
Gene expression dynamics of FGF pathway and antagonists of DM3. Lighter and darker shades indicate decreased and increased expression, respectively. DE, dental epithelium; DM, dental mesenchyme.
Various sprouty genes act as antagonists of FGF signaling to ensure the correct morphology and size of developing teeth. Mouse Spry2 is expressed throughout the epithelium, whereas Spry4 mRNA accumulates exclusively in the DM to prevent the development of supernumerary teeth [13]. In continuously growing mouse incisors, Spry2 and Spry4 restrict the differentiation of enamel-secreting ameloblasts to the labial side, thereby allowing asymmetric enamel deposition [33]. Here, we found that SPRY2 was initially expressed in DE, then in DE and DM, and mostly in DM from the cap to the late bell stage in miniature pigs, which differed from SPRY2 expression in mice [13]. As antagonists of FGF signaling, SPRYs might form an FGF signaling gradient to precisely regulate tooth morphogenesis and odontogenesis.
Interactions among several FGF family members in the IEE and DM regulate the differentiation of dentin and subsequent dentin formation [34]. Particularly, FGF4 and FGF9 expressed in epithelial cells are thought to maintain FGF3 expression in the DM to further regulate epithelial cell proliferation and morphogenesis [35]. Specifically, FGF3 and FGF7 stimulate the proliferation of inner and outer enamel epithelial cells, respectively [35, 36]. In contrast, the loss of SPRY2 function in the epithelium leads to the formation of diastema tooth buds because Fgf3 expression in the mesenchyme is sufficient to control the expression of Shh and perhaps also Fgf4 [13]. The normal function of Spry4 in the mesenchyme is to prevent epithelial FGF ligands, including Fgf4 and Fgf9, produced in adjacent M1 tooth germ, by inducing or maintaining Fgf3 expression in the mesenchyme [13]. Based on the localization and relevant expression patterns of FGFs and SPRYs in the DE and DM of DM3, we predict that a feedback mechanism precisely regulates FGF signaling to mediate interactions between the epithelium and mesenchyme during tooth development, which should be further evaluated.
Conclusions
We determined the spatiotemporal distribution and potential gradient of FGF and sprouty molecules during odontogenesis in large mammals. Epithelial-mesenchymal interactions might be modulated by fine-tuning the FGF pathway to promote cusp patterning and dental mineralization.
Abbreviations
FGF: fibroblast growth factor; FGFRs: FGF receptors; SPRY: sprouty RTK signaling antagonist; DM3: the third deciduous molar; E40: embryonic day 40; E50: embryonic day 50; E60: embryonic day 60; DE: dental epithelium; DM: dental mesenchyme; IEE: inner enamel epithelium; OEE: outer enamel epithelium; SR: stellate reticulum; Am: ameloblasts; Od: odontoblasts; ISH: in situ hybridization; TSA: tyramide signal amplification; IF: immunofluorescence; GAPDH: glyceraldehyde-3-phosphate dehydrogenase; real-time RT-PCR: real-time reverse transcription polymerase chain reaction.
Supplementary Material
Supplementary figures and tables.
Acknowledgements
Funding
This work was supported by grants from the Chinese Research Unit of Tooth Development and Regeneration, CAMSI Innovation Fund for Medical Sciences, No. 2019-12M-5-031; National Natural Science Foundation of China (82030031 and 91649124 to S.W., and 82022014 to X.S.W.); Beijing Municipal Science & Technology Commission No. Z181100001718208; Beijing Municipal Education Commission No. 119207020201; and Beijing Hospitals Authority of Hospitals' Mission Plan, code: SML20151401; and Beijing Municipality Government grants (Beijing Scholar Program- PXM2018_014226_000021; PXM2018_193312_000006_0028S643_FCG; PXM2019_014226_000011; PXM2020_014226_000005; Z181100001718208).
Author Contributions
SLW and SXW supervised this work. WWG and XYL performed the majority of experiments, with additional contribution from RZ, JYW, FW, and JSW. WWG, LH and CMZ analyzed the data and prepared the first draft of the manuscript, which was further edited by SLW and SXW. The author(s) read and approved the final manuscript.
Competing Interests
The authors have declared that no competing interest exists.
References
1. Wu X, Li Y, Wang F. et al. Spatiotemporal Expression of Wnt/beta-catenin Signaling during Morphogenesis and Odontogenesis of Deciduous Molar in Miniature Pig. Int J Biol Sci. 2017;13:1082-1091
2. Zhang YD, Chen Z, Song YQ. et al. Making a tooth: growth factors, transcription factors, and stem cells. Cell Res. 2005;15:301-16
3. Nie X, Luukko K, Kettunen P. FGF signalling in craniofacial development and developmental disorders. Oral Dis. 2006;12:102-11
4. Itoh N, Ornitz DM. Evolution of the Fgf and Fgfr gene families. Trends Genet. 2004;20:563-9
5. Itoh N, Ornitz DM. Fibroblast growth factors: from molecular evolution to roles in development, metabolism and disease. J Biochem. 2011;149:121-30
6. Popovici C, Roubin R, Coulier F. et al. An evolutionary history of the FGF superfamily. Bioessays. 2005;27:849-57
7. Maddaluno L, Urwyler C, Werner S. Fibroblast growth factors: key players in regeneration and tissue repair. Development. 2017;144:4047-60
8. Liu C, Gu S, Sun C. et al. FGF signaling sustains the odontogenic fate of dental mesenchyme by suppressing beta-catenin signaling. Development. 2013;140:4375-85
9. Jernvall J, Kettunen P, Karavanova I. et al. Evidence for the role of the enamel knot as a control center in mammalian tooth cusp formation: non-dividing cells express growth stimulating Fgf-4 gene. Int J Dev Biol. 1994;38:463-9
10. Vaahtokari A, Aberg T, Jernvall J. et al. The enamel knot as a signaling center in the developing mouse tooth. Mech Dev. 1996;54:39-43
11. Kettunen P, Thesleff I. Expression and function of FGFs-4, -8, and -9 suggest functional redundancy and repetitive use as epithelial signals during tooth morphogenesis. Dev Dyn. 1998;211:256-68
12. Kettunen P, Laurikkala J, Itaranta P. et al. Associations of FGF-3 and FGF-10 with signaling networks regulating tooth morphogenesis. Dev Dyn. 2000;219:322-32
13. Klein OD, Minowada G, Peterkova R. et al. Sprouty genes control diastema tooth development via bidirectional antagonism of epithelial-mesenchymal FGF signaling. Dev Cell. 2006;11:181-90
14. Tapaltsyan V, Charles C, Hu J. et al. Identification of novel Fgf enhancers and their role in dental evolution. Evol Dev. 2016;18:31-40
15. Charles C, Lazzari V, Tafforeau P. et al. Modulation of Fgf3 dosage in mouse and men mirrors evolution of mammalian dentition. Proc Natl Acad Sci U S A. 2009;106:22364-8
16. Hanafusa H, Torii S, Yasunaga T. et al. Sprouty1 and Sprouty2 provide a control mechanism for the Ras/MAPK signalling pathway. Nat Cell Biol. 2002;4:850-8
17. Mason JM, Morrison DJ, Basson MA. et al. Sprouty proteins: multifaceted negative-feedback regulators of receptor tyrosine kinase signaling. Trends Cell Biol. 2006;16:45-54
18. Marangoni P, Charles C, Tafforeau P. et al. Phenotypic and evolutionary implications of modulating the ERK-MAPK cascade using the dentition as a model. Sci Rep. 2015;5:11658
19. Percival CJ, Marangoni P, Tapaltsyan V. et al. The interaction of genetic background and mutational effects in regulation of mouse craniofacial shape. G3 (Bethesda). 2017;7:1439-50
20. Wang S, Liu Y, Fang D. et al. The miniature pig: a useful large animal model for dental and orofacial research. Oral Dis. 2007;13:530-7
21. Xu J, Zheng Z, Fang D. et al. Early-stage pathogenic sequence of jaw osteoradionecrosis in vivo. J Dent Res. 2012;91:702-8
22. Wang F, Li Y, Wu X. et al. Transcriptome analysis of coding and long non-coding RNAs highlights the regulatory network of cascade initiation of permanent molars in miniature pigs. BMC Genomics. 2017;18:148
23. Li A, Song T, Wang F. et al. MicroRNAome and expression profile of developing tooth germ in miniature pigs. PLoS One. 2012;7:e52256
24. Wu X, Hu J, Li G. et al. Biomechanical stress regulates mammalian tooth replacement via the integrin beta1-RUNX2-Wnt pathway. EMBO J. 2020;39:e102374
25. Li XB, Yang G, Zhu L. et al. Gastric Lgr5(+) stem cells are the cellular origin of invasive intestinal-type gastric cancer in mice. Cell Res. 2016;26:838-49
26. Wang F, Wu Z, Fan Z. et al. The cell re-association-based whole-tooth regeneration strategies in large animal, Sus scrofa. Cell Prolif. 2018;51:e12479
27. Du W, Du W, Yu H. The role of fibroblast growth factors in tooth development and incisor renewal. Stem Cells Int. 2018;2018:7549160
28. Huang F, Hu X, Fang C. et al. Expression profile of critical genes involved in FGF signaling pathway in the developing human primary dentition. Histochem Cell Biol. 2015;144:457-69
29. Miki T, Bottaro DP, Fleming TP. et al. Determination of ligand-binding specificity by alternative splicing: two distinct growth factor receptors encoded by a single gene. Proc Natl Acad Sci U S A. 1992;89:246-50
30. Hoffman MP, Kidder BL, Steinberg ZL. et al. Gene expression profiles of mouse submandibular gland development: FGFR1 regulates branching morphogenesis in vitro through BMP- and FGF-dependent mechanisms. Development. 2002;129:5767-78
31. Trokovic N, Trokovic R, Mai P. et al. Fgfr1 regulates patterning of the pharyngeal region. Genes Dev. 2003;17:141-53
32. Yu K, Xu J, Liu Z. et al. Conditional inactivation of FGF receptor 2 reveals an essential role for FGF signaling in the regulation of osteoblast function and bone growth. Development. 2003;130:3063-74
33. Klein OD, Lyons DB, Balooch G. et al. An FGF signaling loop sustains the generation of differentiated progeny from stem cells in mouse incisors. Development. 2008;135:377-85
34. Koussoulakou DS, Margaritis LH, Koussoulakos SL. A curriculum vitae of teeth: evolution, generation, regeneration. Int J Biol Sci. 2009;5:226-43
35. Harada H, Toyono T, Toyoshima K. et al. FGF10 maintains stem cell compartment in developing mouse incisors. Development. 2002;129:1533-41
36. Harada H, Ohshima H. New perspectives on tooth development and the dental stem cell niche. Arch Histol Cytol. 2004;67:1-11
Author contact
![]() Corresponding authors: Songlin Wang, email: slwangedu.cn or Xiaoshan Wu, email: drwuxiaoshanedu.cn.
Corresponding authors: Songlin Wang, email: slwangedu.cn or Xiaoshan Wu, email: drwuxiaoshanedu.cn.

 Global reach, higher impact
Global reach, higher impact