3.2
Impact Factor
ISSN: 1449-1907
Int J Med Sci 2021; 18(15):3425-3436. doi:10.7150/ijms.63541 This issue Cite
Research Paper
Identification of Key Genes and Pathways associated with Endometriosis by Weighted Gene Co-expression Network Analysis
1. Department of Obstetrics and Gynecology, The Second Xiangya Hospital, Central South University, Changsha, Hunan 410011, China.
2. Department of Obstetrics and Gynecology, David Geffen School of Medicine, University of California, Los Angeles, California 90095, United States.
Abstract
Background: Endometriosis is a common gynecological disorder with high rates of infertility and pelvic pain. However, its pathogenesis and diagnostic biomarkers remain unclear. This study aimed to elucidate potential hub genes and key pathways associated with endometriosis in ectopic endometrium (EC) and eutopic endometrium (EU).
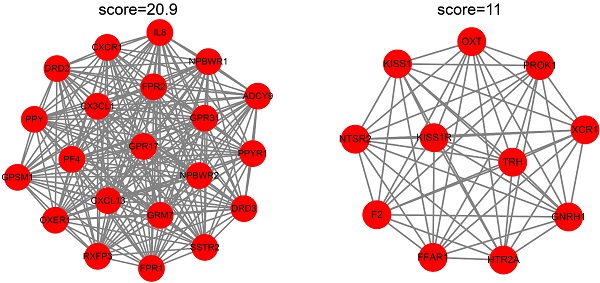
Material and Method: EC and EU-associated microarray datasets were obtained from the gene expression omnibus (GEO) database. Gene set enrichment analysis was performed to obtain further biological insight into the EU and EC-associated genes. Weighted gene co-expression network analysis (WGCNA) was performed to find clinically significant modules of highly-correlated genes. The hub genes that belong to both the weighted gene co-expression network and protein-protein interaction (PPI) network were identified using a Venn diagram.
Results: We obtained EC and EU-associated microarray datasets GSE7305 and GSE120103. Genes in the EC were mainly enriched in the immune response and immune cell trafficking, and genes in the EU were mainly enriched in stress response and steroid hormone biosynthesis. PPI networks and weighted gene co-expression networks were constructed. An EC-associated blue module and an EU-associated magenta module were identified, and their function annotations revealed that hormone receptor signaling or inflammatory microenvironments may promote EU passing through the oviducts and migrating to the ovarian surfaces, and adhesion and immune correlated genes may induce the successful ectopic implantation of the endometrium (EC). Twelve hub genes in the EC and sixteen hub genes in the EU were recognized and further validated in independent datasets.
Conclusion: Our study identified, for the first time, the hub genes and enrichment pathways in the EC and EU using WGCNA, which may provide a comprehensive understanding of the pathogenesis of endometriosis and have important clinical implications for the treatment and diagnosis of endometriosis.
Keywords: endometriosis, WGCNA, biological markers, pathway

 Global reach, higher impact
Global reach, higher impact