Impact Factor
ISSN: 1449-1907
Int J Med Sci 2021; 18(13):2871-2889. doi:10.7150/ijms.58191 This issue Cite
Review
Artificial Intelligence and Machine Learning in Chronic Airway Diseases: Focus on Asthma and Chronic Obstructive Pulmonary Disease
1. Department of Respiratory and Critical Care Medicine, West China Hospital, Sichuan University, Chengdu, Sichuan Province, China.
2. Department of Respiratory and Critical Care Medicine, People's Hospital of Deyang City, Affiliated Hospital of Chengdu College of Medicine, Deyang, Sichuan Province, China.
*These authors contributed equally to this work.
Received 2021-1-14; Accepted 2021-5-20; Published 2021-6-1
Abstract
Chronic airway diseases are characterized by airway inflammation, obstruction, and remodeling and show high prevalence, especially in developing countries. Among them, asthma and chronic obstructive pulmonary disease (COPD) show the highest morbidity and socioeconomic burden worldwide. Although there are extensive guidelines for the prevention, early diagnosis, and rational treatment of these lifelong diseases, their value in precision medicine is very limited. Artificial intelligence (AI) and machine learning (ML) techniques have emerged as effective methods for mining and integrating large-scale, heterogeneous medical data for clinical practice, and several AI and ML methods have recently been applied to asthma and COPD. However, very few methods have significantly contributed to clinical practice. Here, we review four aspects of AI and ML implementation in asthma and COPD to summarize existing knowledge and indicate future steps required for the safe and effective application of AI and ML tools by clinicians.
Keywords: artificial intelligence, machine learning, chronic airway diseases, asthma, chronic obstructive pulmonary disease
Introduction
Recent developments in computer operations and the rapid development of “big data” have significantly advanced artificial intelligence (AI) and machine learning (ML) technology and their applications in various fields such as medicine [1]. Medical data are difficult to capture, manage, and process using conventional tools in a timely manner because the datasets are huge, they are frequently updated, and the data come in diverse formats. Instead, imaging, genomic, proteomic and electronic health records (EHRs) data can be mined using AI/ML to extract new knowledge [2]. This development has led to rapid changes in the use of AI/ML in medicine, especially in medical imaging [3], where the techniques are used not only for rapid disease screening, but also to improve diagnostic accuracy and work efficiency [4]. Genomic data are another enormous source of complex medical information that has recently emerged. Recent studies have demonstrated that the systematic analysis of genomic data with AI/ML technology can favor precision medicine for the benefit of patients [5, 6]. Although the most widely used AI/ML technology in respiratory diseases is chest imaging, especially for the screening and diagnosis of lung nodules, the application of AI/ML tools in chronic airway diseases is attracting increasing attention [7, 8].
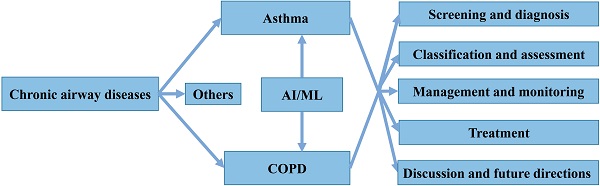
Chronic airway diseases, such as asthma, chronic obstructive pulmonary disease (COPD) and bronchiectasis, are lifelong and life-threatening pathological conditions that extensively affect people of all ages, races, and sex worldwide [9]. These diseases are characterized by airway inflammation, obstruction, and remodeling, and common symptoms include cough, sputum, and shortness of breath. Their etiology and pathogenesis are complex and not yet fully understood [10, 11]. Patients with chronic airway diseases are also prone to relapse, increasing their risk of hospitalization and death, and seriously affecting their quality of life. Among these diseases, asthma and COPD lead to the highest morbidity and socioeconomic burden worldwide [12]. Despite extensive efforts, identifying, treating and managing both disorders still face many challenges, such as under- and overdiagnosis, unclear pathogenesis, lack of uniform classification criteria for phenotypes, and high risk of death and high costs associated with exacerbations [13]. In addition, several AI/ML methods have recently been applied for both diseases, but only a few have significantly contributed to clinical practice. Thus, summarizing existing knowledge and indicating future directions is required for the safe and effective application of AI/ML tools by clinicians. Here, we systematically review the application of AI/ML technology to four different aspects of asthma and COPD: screening and diagnosis, classification and assessment, management and monitoring, as well as treatment (Figure 1). We also present the development of several models based on ML algorithms.
Structure of the present review.
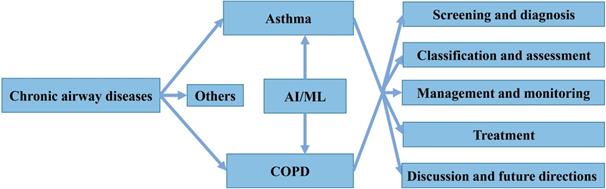
Categories of machine learning algorithms.
General concepts, terminologies and limitations of AI/ML
In order to facilitate understanding, we quickly explain the general concepts and terminologies of AI/ML that commonly appeared in this review. In addition, we summarize the evaluation indicators and current limitations of ML.
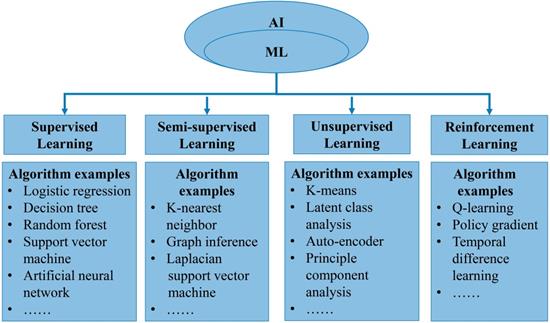
In general, AI refers to the technology that represents human intelligence through computer programs. ML is a branch of AI technology based on statistical techniques for self-learning and the development of problem-solving skills. In particular, ML uses complex algorithms to analyze large amounts of data, identify patterns, make predictions that do not require specific codes, and evolve with increasing sample size to improve learning. ML technology can be divided into supervised, semi-supervised, unsupervised and reinforcement learning [14, 15] (Figure 2). Supervised learning trains ML algorithms to labeled data. These labels, that include data types, data attributes and feature point locations, are used as expected effects to continuously modify the prediction results of the ML model. Common tasks for supervised learning are regression and classification for continuous and categorical outcome variables, respectively [15]. Semi-supervised learning can fit models to not only labeled data but also unlabeled data. When this type of ML algorithm classifies unlabeled data, it usually measures the distance/similarity between the target sample and all labeled samples [16]. Unsupervised learning aims to explore and infer potential natural connections and groupings from unlabeled data [17]. Reinforcement learning, in contrast, is a general term for ML approaches that integrate prediction and decision-making. This type of ML technology has an iterative learning approach, and can self-adapting according to the initial feedback [17]. Table 1 and Figure 3A introduce and quickly explain common ML algorithms reviewed in this article. Based on different ML algorithms, several ML models with different functions have been developed so far. It is important to note that an optimal ML model cannot be easily developed with a limited dataset. Instead, a satisfactory ML model should be constructed in two phases: the model is developed in the training phase and then its performance is assessed in the testing phase [18] (Figure 3B).
In order to evaluate the performance of trained ML models, several reasonable evaluation indicators must be used. Generally, the ML model selects different evaluation indicators according to the different classification and regression tasks. In classification, the evaluation indicators are often accuracy, false positive rate, false negative rate, sensitivity (recall), specificity, precision, F1-score, C-index (concordance index), receiver operating characteristic curve and the area underneath it (AUC). The regression tasks focus on the difference between the predicted and true value. Therefore, the evaluation indicators include mean square error, root mean square error, mean absolute error, and median absolute deviation.
Although ML technology is continuously growing in the medical field, its application is greatly limited due to issues related to the availability of adequate data (e.g. text, numbers, images), experiments and methods, and ethics [14, 15]. Inaccurate or missing data can cause serious problems, leading to incorrect model structure and biased conclusions. The imbalance and sparsity of categories in medical data can also limit ML application. Therefore, repeated experiments need to be performed and different ML methods should be explored for addressing medical challenges. Experimental design and replication, model selection, model generalization, and model interpretability are crucial aspects of applying ML techniques [19]. A good experimental design can reduce experimental errors and give more accurate conclusions. Model selection is one process of finding a solution to the research problem, but there are currently no standards to guard against model misuse or abuse. ML can also improve model generalizability to ensure more accurate prediction of future cases, but how this is best done requires further study. Interpretability of a model makes it more relevant to medical decision making, but most data-driven ML techniques remain unexplored. Another challenge of using AI/ML is to ensure ethics and eliminate prejudice during their application [20]. Ethical problems can arise due to problems with optimization, prediction, or classification, which can lead to inequality on sensitive issues or to violations of privacy. Research should not only build ML models but also resolve ethical issues associated with data use and interpretation. Despite these current limitations, AI/ML techniques are needed in the medical field due to the special ability to efficiently analyze and integrate large and heterogeneous data.
Overview of machine learning. A. Illustration of an artificial neural network algorithm. The structure of artificial neural network includes three main layers, namely input layer, hidden layer and output layer. The input layer represents the features extracted from data, which are then integrated by the hidden layer (one or more) to obtain transformed features. Finally, the transformed features are used by the output layer to predict the outcome. B. Common paths for training and testing machine learning model in medicine.
Summary of common machine learning algorithms
| Type of machine learning algorithm | Description | References describing applications |
|---|---|---|
| Natural language processing | Natural language processing is a general term for a series of technical methods. It can be divided into natural language understanding (NLU) and natural language generation (NLG). NLU focuses on how to understand text, while NLG focuses on how to generate natural text after understanding the text. | [23-25] |
| K nearest neighbor | K nearest neighbor is a type of instance-based learning algorithm, and the training process simply memorize the training data. It categorizes the sample according to the similarity. The similarity is calculated using measures such as Euclidean distance and Hamming distance. | [27,28,52], [67,78,79], [94,102,104] |
| Random forest | Random forest is an ensemble learning method. It contains multiple decision trees and integrates these decision trees to category of data. The size of trees and the number of variables usually determine the performance of model. | [27,28,30], [47,48,59], [65-67,74,79], [90,99,101], [104,107,109] |
| Support vector machine | Support vector machine is usually used for classification and regression. It learns the optimal hyperplane to classify data. Generally, it has low misclassification error and scale well to high-dimensional data. However, selecting the optimal kernel function is essential. | [28,32,35], [52,59,66-68], [75,78,79], [95,99,102], [103,104,108] |
| Artificial neural network | This is a kind of hierarchical nonlinear mapping network based on neurons and activation functions. Its structure includes three main parts, namely input layer, hidden layer and output layer. This structure is used to analyze variables in order to predict an outcome. The primary limitation is the underlying model's lack of transparency. | [32,53-56], [62,69,79], [94,102] |
| Latent class analysis | Latent class analysis is a statistically principled technique that is used in factor analysis, cluster analysis, and regression. It is to explain and estimate the association between manifest indicators by latent class variables. This method suits to classify subgroups in large and heterogeneous data. | [40-44,50], [51,82] |
| K-means | This method divides the dataset into K clusters, and each cluster is represented by the average value of all samples in the cluster, which is called the "centroid". K-means clustering is easy to interpret and computationally efficient. However, the number of clusters needs to be prespecified. | [46,84,86], [88] |
| Logistic regression | Logistic regression estimates the probability of a binary classification problem. The dependent variable of it obeys the Bernoulli distribution, and nonlinear factors are introduced through the Sigmoid function. | [47,59,60], [66-68,78,79], [95,99,102], [104,107,108] |
| Decision tree | Decision tree creates a series of decision rules to predict categorical and continuous outcomes based on input variables. It contains three main parts: a root node, leaf nodes and branches. Decision tree is easy to understanding, but unstable and prone to overfitting. | [47,49,60], [67,68,78], [79,97,100], [102] |
| Lasso regression | Lasso regression is a linear regression method using L1-regularization. L1-regularization can compress the coefficients of variables and change some coefficients to zero, so as to achieve the purpose of variable selection. | [48,59,98] |
| Naïve Bayes | Naïve Bayes is a classification algorithm based on Bayes' theorem, which is suitable for scenarios where variables are independent of each other. It is relatively simple and has good performance in the presence of noise, missing data, and irrelevant variables. | [64,67,68], [79,104] |
AI/ML and asthma
Application of AI/ML to asthma screening and diagnosis
As a heterogeneous disease, asthma is often under- or overdiagnosed, especially in poor areas. In fact, almost 20-73% of cases remain undiagnosed, while about 30-35% of people diagnosed with asthma do not actually have the condition [21, 22].
To address this issue, EHRs and Predetermined Asthma Criteria were used in a retrospective birth cohort study to develop for the first time a natural language processing algorithm for pediatric asthma diagnosis with high sensitivity (97%), specificity (95%), as well as positive (90%) and negative (98%) predictive values. The test cohort of this study consisted by 497 children, among whom the asthma prevalence was 31%. The application of the same algorithm to records from 497 children (median age, 2.3 years) at another hospital showed similar sensitivity (92%), specificity (96%), and positive (89%) and negative (97%) predictive values, confirming the algorithm's efficiency in diagnosing pediatric asthma in an external EHR system. However, the algorithm should be further validated on an adult cohort [23, 24] (Table 2). In another cross-sectional study, an ML model based on natural language processing algorithm was also developed by mining EHRs to automatically screen pediatric patients who met the Asthma Predictive Index criteria for asthma diagnosis. A total of 427 subjects with an average age of 5.3 years were enrolled in the test phase, and the sensitivity, specificity, and positive and negative predictive values of the ML model reached 86%, 98%, 88%, and 98%, respectively [25]. These results suggest that ML models based on natural language processing can be used to identify pediatric patients with undiagnosed asthma. In addition, an artificial neural network model based on 13 clinical characteristics was developed using clinical findings from EHRs, which was able to identify 100% asthma patients among 254 individuals [26].
Although spirometry and bronchial provocation tests are increasingly available, they require the full cooperation of patients and cannot confirm correct diagnosis of asthma. Therefore, the non-invasive forced oscillation technique, which does not require patient cooperation, was combined with four ML algorithms (k-nearest neighbor, random forest, decision trees, and a feature-based dissimilarity space classifier) to produce ML classifiers that serve as a useful and portable tool for diagnosing asthma airway obstruction [27]. Among the four algorithms, k-nearest neighbor led to the highest AUC of 0.91. Further research combining the forced oscillation technique with ML algorithms (k-nearest neighbor, random forest, AdaBoost with decision trees, and support vector machine) resulted in several novel classifiers that achieved AUC ≥0.9 for the differential diagnosis of patients with asthma or restrictive respiratory diseases in 97 individuals. However, the results should be further verified on an external dataset [28].
Machine learning studies on asthma
| Reference | Category | Study population | ML algorithms | Input features | Studied outcome | Results | Critical appraisal of the study |
|---|---|---|---|---|---|---|---|
| Wi CI, 2017 [23] | Screening and diagnosis | 927 children: training cohort = 430 test cohort = 497 | NLP | Clinical (EMRs) | Pediatric asthmatic subjects or not | Sensitivity = 97%, specificity = 95%, positive predictive value = 90%, negative predictive value = 98% | Pros: use of electronic medical records Cons: single internal electronic medical record system |
| Wi CI, 2018 [24] | Screening and diagnosis | 595 children: training cohort = 298 test cohort = 297 | NLP | Clinical (EHR) | Pediatric asthmatic subjects or not | Sensitivity = 92%, specificity = 96%, positive predictive value = 89%, negative predictive value = 97% | Pros: use of an external electronic medical records system Cons: has not yet been validated on an adult cohort |
| Kaur H, 2018 [25] | Screening and diagnosis | 514 children: training cohort = 87 test cohort = 427 | NLP | Clinical (EHR) | Pediatric asthmatic subjects or not | Sensitivity = 86%, specificity = 98%, positive predictive value = 88%, negative predictive value = 98% | Pros: development of the first algorithm to automatically extract patients who meet the Asthma Predictive Index criteria Cons: relatively small sample |
| Alizadeh B, 2015 [26] | Screening and diagnosis | 254 subjects: training cohort = 70% test cohort = 30% | ANN | Clinical | Asthmatic subjects or not | Accuracy = 100% | Pros: based on 13 clinical characteristics used by physicians to diagnose asthma Cons: single data source and relatively small sample |
| Amaral J, 2017 [27] | Screening and diagnosis | 75 stable asthma patients: 39 with airway obstruction and 36 without | KNN, RF, ADAB, FDSC | Forced oscillation technique parameters | Airway obstruction | KNN reached the highest accuracy range (AUC = 0.91) | Pros: use of the non-invasive forced oscillation technique Cons: the exact sensitivity and specificity values are unknown |
| Amaral J, 2020 [28] | Screening and diagnosis | 97 individuals: controls = 20 asthmatic patients = 38 restrictive patients = 39 | KNN, RF, ADAB, SVM | Forced oscillation technique parameters | Asthmatic or restrictive respiratory diseases subjects | All classifiers achieved high accuracy (AUC≥0.9) | Pros: differential diagnosis of asthma and restrictive respiratory diseases Cons: single practice site and relatively small sample |
| Zhan J, 2020 [29] | Screening and diagnosis | 355 asthma patients and 1,480 healthy individuals | Mahalanobis-Taguchi system | Routine blood biomarkers | Asthmatic subjects or not | Accuracy = 94.15% in asthma patients and 97.20% in healthy individuals | Pros: diagnosis of asthma based on routine blood biomarkers Cons: a complete blood reference space is required to more accurately identify asthma patients |
| Sinha A, 2017 [30] | Screening and diagnosis | 89 asthmatic subjects and 20 healthy controls | RF | Nuclear magnetic resonance spectra of exhaled breath condensate | Asthmatic subjects or not | Sensitivity = 80%, specificity = 75% | Pros: advocated the use of exhaled breath condensate spectral signatures Cons: did not actually measure any metabolites |
| Islam MA, 2018 [32] | Screening and diagnosis | 60 subjects: normal = 30 asthma patients = 30 | ANN, SVM | Clinical (lung sounds) | Normal or asthmatic subjects | Accuracy = 89.2(±3.87)% in ANN and 93.3(±3.10)% in SVM | Pros: used lung respiratory sound signals Cons: did not collect respiratory sounds of both upper and lower lung |
| Singh OP, 2018 [34] | Screening and diagnosis | non-asthmatic = 30 asthmatic = 43 | SVM, KNN, NB | Respired carbon dioxide waveform | Asthmatic subjects or not | Accuracy = 94.52%, sensitivity = 97.67%, and specificity = 90% in SVM | Pros: non-invasive, patient-independent method based on simple signal processing algorithm to screen for asthma Cons: relatively small sample |
| Tomita K, 2019 [35] | Screening and diagnosis | 566 adult out-patients (367 asthma patients) | SVM, DNN | Clinical, Lung function test, Bronchial challenge test | Adult asthmatic subjects or not | Accuracy = 98% in DNN and 82% in SVM | Pros: models based on symptoms, physical signs and objective tests Cons: single practice site |
| Couto M, 2015 [40] | Classification and assessment | asthmatic athletes = 150 healthy athletes = 129 athletes with other pathologic conditions = 45 | LCA | Clinical (athletes' records) | Asthmatic phenotypes | Two phenotypes: atopic asthma and sports asthma | Pros: identification of asthmatic athlete phenotypes Cons: need to be validated by larger clinical interventional trials |
| Chen Q, 2012 [41] | Classification and assessment | 689 asthma children | LCA, BIC | Clinical (questionnaire data) | Asthmatic phenotypes | Four phenotypes: never/infrequent, early-transient, early-persistent, and late-onset | Pros: identification of phenotypes based on wheeze Cons: some children could not provide precision data |
| Weinmayr G, 2013 [42] | Classification and assessment | >4,000 asthma children | LCA, BIC | Clinical (questionnaire), Bronchial hyperresponsiveness | Childhood asthma phenotypes | Seven phenotypes: one corresponding to healthy children; three related to wheeze; three related to congestion and coughed-up phlegm | Pros: identification of phenotypes according to respiratory symptoms Cons: recall bias |
| Bochenek G, 2014 [43] | Classification and assessment | 201 aspirin-exacerbated respiratory disease patients | LCA | Clinical (questionnaire, spirometry, blood eosinophilia, urinary LTE4 concentrations) | Subphenotypes within AERD phenotype | Four subphenotypes: asthma with a moderate course; asthma with a mild course; asthma with a severe course; poorly controlled asthma with frequent and severe exacerbations | Pros: identification of aspirin-exacerbated respiratory disease phenotypes Cons: LCA stability over time not established |
| Havstad S, 2014 [44] | Classification and assessment | 594 asthma children (2 years old) | LCA | Serum IgE data on 10 allergens | Atopic asthma phenotypes | Four phenotypes: low to no sensitization; highly sensitized; milk and egg dominated; peanut and inhalant(s)/no milk | Pros: examination of a more recently born, younger, and racially mixed cohort Cons: lack of additional information on lung function, cytokines, and eosinophils |
| Ross MK, 2018 [45] | Classification and assessment | 1,019 children from the CAMP study and 669 children from the ACRN/CARE dataset | PP | Clinical | Pediatric asthma phenotypes | Four phenotypes: allergic-not-obese, obese-not-allergic, allergic-and-obese, and not-obese-not-allergic | Pros: discovery of more detailed predictive features for long-term asthma control other than the current control state Cons: elimination of some features due to missing data |
| Wu W, 2019 [46] | Classification and assessment | 346 adult asthma in the Severe Asthma Research Program | Multiple-kernel k-means | Clinical, physiological, inflammatory, demographic | Asthma control state | Four phenotypes: clusters 1 and 2: young modestly corticosteroid responsive allergic asthmatics with relatively normal lung function; cluster 3: late onset asthmatics with low lung function; cluster 4: primarily young obese females with severe airflow limitation | Pros: identification of phenotypes based on corticosteroid responses Cons: limited to a single dose of systemic corticosteroid (without placebo) and a single point in time |
| Prosperi MC, 2014 [47] | Classification and assessment | 554 asthma adults | LR, RF, DT, AB | Clinical, genetic | Current asthma, wheeze, eczema | Optimal AUC = 0.84, 0.76 and 0.64 for asthma, wheeze, and eczema, respectively | Pros: integrated genomics information Cons: genetic analysis was restricted to candidate genes |
| Krautenbacher N, 2019 [48] | Classification and assessment | 260 individuals: healthy children = 43%, mild-to-moderate, allergic asthmatics = 47%, nonallergic asthmatics = 11% | Lasso regression, elastic net, RF | Genetic, immunological, environmental | Asthma phenotypes | AUC for three classes of phenotypes = 0.81 | Pros: identification of three important genes for classifying childhood asthma phenotypes: PKN2, PTK2 and ALPP Cons: should be validated in other cohort studies |
| Williams-De Vane CR, 2013 [49] | Classification and assessment | 205 individuals | DT | Clinical, genetic, demographic | Asthma endotypes | Decision tree-based methods were useful tools for identifying asthma endotypes | Pros: integrated data to identify asthma endotypes Cons: should be validated in external data |
| Siroux V, 2014 [50] | Classification and assessment | 3,001 asthmatic adults | LCA | Clinical (questionnaire data), genetic | Asthma phenotypes | Four phenotypes: inactive/mild nonallergic asthma, inactive/mild allergic asthma, active allergic asthma, and active adult-onset nonallergic asthma | Pros: large sample of asthmatic adults Cons: lack of formal replication of the genetic association signals |
| Mäkikyrö EM, 2017 [51] | Classification and assessment | 1,995 asthma subjects | LCA | Clinical (questionnaire data), asthma-related healthcare use | Asthma phenotypes | Four subtypes for women: mild asthma, moderate asthma, unknown severity, and severe asthma. Three subtypes for men: mild asthma, unknown severity, and severe asthma. | Pros: development of a simpler way to categorize asthmatic subtypes Cons: did not test the population for biomarkers and form endotypes; did not verify the subtypes with full scale lung function testing |
| Nabi FG, 2019 [52] | Classification and assessment | 55 asthma patients | Ensemble, SVM, KNN | Wheeze sounds | Asthma severity | The best positive predictive value for the mild, moderate, and severe samples were 95% (ensemble), 88% (ensemble) and 90% (SVM), respectively. | Pros: classified wheeze sounds of asthmatic patients according to severity Cons: relatively small sample |
| Moustris KP, 2012 [53] | Management and monitoring | 3,602 children | ANN | Meteorological and ambient air pollution data | Childhood asthma admissions | Index of Agreement = 0.837 Coefficient of determination = 0.528 | Pros: predicted the childhood asthma admission based on the bioclimatic and air pollution Cons: some environmental factors had not been included, such as relative humidity |
| Messinger AI, 2019 [54] | Management and monitoring | 128 asthmatic children: training set = 102 testing set = 26 | ANN | Demographic, clinical (EHR) | Respiratory score | The performance of pediatric-automated asthma severity scores was better than Pediatric Asthma Score. | Pros: pARS had the potential to help standardize acute pediatric asthma care in the PICU. Cons: incomplete data from the clinical record and sign database; a single center study |
| Xiang Y, 2020 [55] | Management and monitoring | 31,433 adult asthma patients | ANN | Clinical (EHR) | Asthma exacerbation | AUC = 0.7003 | Pros: a time-sensitive predictive model Cons: some potential risk factors for asthma exacerbations might not be recorded or might even be incorrectly recorded |
| Khatri KL, 2018 [56] | Management and monitoring | Patients of visiting emergency departments in Dallas County for respiratory diseases | ANN | Clinical, meteorological and environmental pollution data | Emergency department visits | Overall accuracy = 81.0% | Pros: can serve as useful tool for peak demand prediction in emergency departments Cons: limited number of variables; primary diagnosis may not be accurate |
| Grunwell JR, 2020 [57] | Management and monitoring | 513 asthmatic children | LCA | Clinical, demographics | Asthma exacerbation | The class of multiple sensitizations with partially reversible airflow limitation had the highest exacerbation risk (64.3%) | Pros: prediction of exacerbation in school-age children Cons: factors responsible for asthma exacerbations were not adequately addressed by the study design |
| Fitzpatrick AM, 2020 [58] | Management and monitoring | 2,593 children with mild to moderate asthma aged 5-18 years | LCA | Clinical, demographics, lung function test | Lung function and exacerbation rate | Children who had multiple sensitizations with partially reversible airflow limitation had the highest exacerbation risk (52.5%) | Pros: large sample size of diverse and representative children across the United States Cons: model selection for LCA can be subjective |
| Das LT, 2017 [59] | Management and monitoring | 2,691 asthmatic children | LR, Lasso regression, RF, SVM | Clinical (EHR) | Emergency department visits | AUC = 0.86 reached by LR | Pros: based on electronic health records (EHRs) Cons: record of emergency department visits to one medical center |
| Zhang O, 2020 [60] | Management and monitoring | 2,010 asthma patients | LR, DT, NB, perceptron algorithms | Daily monitoring data | Asthma exacerbations | AUC = 0.85, sensitivity = 90%, and specificity = 83% reached by LR | Pros: use of a large international dataset to detect severe asthma exacerbations Cons: data were collected using paper diaries, which be inaccurate or fabricated |
| Luo L, 2018 [61] | Management and monitoring | 6,813 admission records | XGBoost | Search index, air pollution data, weather data, historical admissions | Asthma admission | AUC = 0.832 | Pros: use of an easily accessible and daily updated daily search index Cons: data from a single geographical region |
| Ram S, 2015 [62] | Management and monitoring | Emergency department visits for asthma to the Children's Medical Center of Dallas (between October 2013 and December 2013) | ANN | Twitter data, Google search interests, environmental data | Emergency department visits | Accuracy = 70% | Pros: based on real-time environmental and internet-based data Cons: data of emergency department visits from one hospital |
| Finkelstein J, 2016 [64] | Management and monitoring | 7,001 records submitted by adult asthma patients | NB, BN, SVM | Daily self-monitoring reports | Asthma exacerbations | BN model reached sensitivity, specificity, and accuracy of 100% | Pros: use of home telemonitoring data Cons: the number of cases of asthma exacerbations was small |
| Huffaker MF, 2018 [65] | Management and monitoring | 33 subjects | RF | Recorded physiologic data | The time period during which onset of asthma symptoms occurred | Sensitivity = 47.2%, specificity = 96.3%, accuracy = 87.4% | Pros: showed that passive physiologic monitoring can be used in the home to assess asthma control Cons: small sample |
| Luo L, 2020 [66] | Management and monitoring | Cost data of asthmatic patients | LR, RF, SVM, classification regression tree, backpropagation neural network | Cost data | Treatment cost | AUC and sensitivity increase of 46.89% and 101.07%, respectively | Pros: use of machine learning to predict high cost Cons: lack of analysis of low-frequency comorbidities |
| Khasha R, 2019 [67] | Management and monitoring | 96 asthma patients | LR, XGBoost, RF, DT, KNN, NB, SVM | Clinical, demographics, lung function test | Control level | Optimal accuracy = 91.66% | Pros: developed a novel ensemble learning method for asthma control level detection Cons: limited factors affecting asthma control were included |
| Tsang K, 2020 [68] | Management and monitoring | 5,875 asthma patients | LR, NB, DT, SVM | mHealth data | Stable and unstable periods | Optimal sensitivity = 86.6%, optimal specificity = 72.5%, optimal AUC = 0.871 | Pros: personalized algorithms to enhance asthma management Cons: self-reported data rather than objective measures |
| Hosseini SA, 2020 [69] | Treatment | 80 patients with mild or moderate allergic asthma | ANN | Clinical, immunologic, hematologic, demographic | Low to high level of effect | Accuracy>99% | Pros: new machine learning model for the prediction of asthmatic drug effectiveness Cons: relatively small sample |
Abbreviations: AB, AdaBoost; ADAB, AdaBoost with decision trees; AERD, aspirin-exacerbated respiratory disease; ANN, artificial neural networks; AUC, area under the receiver operating characteristic curve; BN, Bayesian networks; BIC, Bayesian Information Criterion; DT, decision trees; DNN, deep neural network; EMRs, electronic medical records; EHR, electronic health records; FDSC, feature-based dissimilarity space classifier; KNN, k-nearest neighbor; LCA, latent class analysis; LR, logistic regression; NB, naïve Bayesian; NLP, natural language processing; PP, predictor pursuit; RF, random forest; SVM, support vector machine.
Despite the lack of specific biomarkers for asthma, its diagnosis can be improved by combining multiple methods and clinical data. For instance, a novel AI system (Mahalanobis-Taguch) was developed based on ML algorithm and several biomarkers determined from routine blood samples, such as platelet distribution width, white blood cell count, and eosinophil count. This system was trained using data from 319 asthmatic patients, then validated in 35 asthmatic patients with a classification accuracy of 94.15% [29]. Further confirmation of the effectiveness of this AI system in clinical practice will simplify the diagnosis of asthma. In another study, a random forest classifier based on nuclear magnetic resonance spectroscopy of exhaled breath condensate was developed using the metabolome as the biomarker source [30]. The classifier differentiated asthma patients and healthy controls with 80% sensitivity and 75% specificity. However, the sample in the study (n = 109) was relatively small, and no actual metabolites were measured, suggesting that the method requires further validation.
A recent systematic review has also suggested that automated analysis of respiratory sounds by ML algorithms can be used for effective screening and diagnosis of respiratory diseases [31]. Indeed, multichannel lung respiratory sound signals derived from 30 asthmatic patients and 30 healthy controls were combined with artificial neural network or support vector machine classifiers for the diagnosis of asthma with respective accuracies of 89.2 ± 3.87% and 93.3 ± 3.10% [32]. Interestingly, this study did not rely on the presence of the typical wheezing asthmatic symptom as a sound signal. Future studies should collect respiratory sounds of both upper and lower lung lobes for further validation of the results.
Given the usefulness of end-tidal capnography for disease diagnosis, a non-invasive, patient-independent method to process carbon dioxide waveform signals was developed based on the support vector machine classifier to differentiate 30 non-asthmatic and 43 asthmatic patients. The average accuracy, sensitivity, and specificity of the algorithm reached 94.52%, 97.67% and 90%, respectively, suggesting end-tidal capnography as an effective technique for asthma diagnosis [33, 34]. However, further validation of the results is required due to the small samples in those studies.
Recently, several classical ML algorithms, such as logistic regression analysis, support vector machine, and deep neural network, were compared for their diagnostic ability when based only on symptoms and physical signs, or when based on the combination of symptoms, physical signs, biochemical findings, lung function tests, and the bronchial provocation tests. That study included 566 adult outpatients and indicated that the deep neural network model was more accurate than other conventional ML tools, reaching an accuracy of 98% when symptoms, physical signs and objective tests were also used [35]. This study may be the first to report that AI can perform comparably to human experts for diagnosing asthma in adults. However, the results should be interpreted and generalized carefully, as different ML predictive models perform differently depending on the conditions.
Application of AI/ML to the classification and assessment of asthma
Asthma is a heterogeneous disease with multiple phenotypes and endotypes that must be properly distinguished for precise prevention and personalized treatment [36-38]. In clinical practice, spirometry and bronchial provocation tests are used to assess airflow limitation and hyperresponsiveness, allowing the identification of some asthma phenotypes, while eosinophil count analysis and fractional exhaled nitric oxide measurements can also be applied [39]. However, further research is still required to practically and accurately identify the asthma phenotypes.
Latent class analysis can generally fit a probabilistic model to a dataset of several variables such as asthma symptoms or allergy. Therefore, several recent studies have developed, verified, and applied ML-based latent class analysis for asthma classification, indicating its suitability for modelling data from symptomatic or asymptomatic asthma patients [40-44]. For instance, based on the data of athlete records that included respiratory symptoms, airway inflammation and hyperresponsiveness, allergic sensitization and lung function test, latent class analysis successfully identified two asthma phenotypes in a total of 150 elite asthmatic athletes who came from Portugal and Norway. The atopic asthma phenotype was defined by allergy symptoms, rhinitis, and high exhaled nitric oxide level, while the sports asthma phenotype was defined by exercise-induced respiratory symptoms and airway hyperresponsiveness, but no allergy [40]. The study also found that athletes who practiced water and winter sports were at higher risk of developing the sports asthma phenotype. The validation of this classification method using additional data sources or clinical interventional trials would significantly benefit the personalized treatment of asthmatic athletes.
A predictor pursuit algorithm based on clinical treatment and outcome data was also developed to analyze phenotypes of 1688 childhood asthma patients. Four phenotypes were identified with better (P < 0.001) than traditional ML methods [45]. The study also found that nedocromil was better than budesonide in controlling asthma in children with obesity and allergy. A similar classification approach was later reported that focused mainly on the response of severe asthma patients to corticosteroids. Using an unsupervised ML approach (multiple-kernel k-means clustering), four phenotypes were identified in a total of 346 asthma patients. The greatest corticosteroid responsiveness was observed for patients with late-onset, poor lung function as well as high baseline eosinophilia, while the lowest responsiveness was observed for young, obese female patients with severe airflow limitation and mild eosinophilic inflammation [46]. Applying these methods to the timely and accurate classification of asthma patients will be a valuable reference for individualized treatment, especially for difficult-to-treat asthma, while reducing the unnecessary use of corticosteroids and related complications.
Precision medicine is an emerging approach of medical science for disease diagnosis and treatment, while genomics is an important manner. In recent years, genetic data have been combined with other clinical information (e.g., demographic, laboratory, and environmental factors) within different ML algorithms to determine asthma phenotypes [47-50]. For example, 14 clinical features from 3001 adults with asthma, which included demography, medical history, respiratory symptoms, allergic characteristics, lung function test and bronchial hyperresponsiveness, were integrated with genomics data from previous analyses in order to differentiate asthma phenotypes. Four phenotypes were obtained using latent class analysis algorithm: inactive/mild nonallergic asthma (18%), inactive/mild allergic asthma (37%), active allergic asthma (27%) and active adult-onset nonallergic asthma (18%). This study also identified 15 single nucleotide polymorphisms associated with at least one these four asthma phenotypes, most of them were linked to the “active allergic asthma” phenotype [50]. Further research is needed to overcome the limitations of the in-house validation and small sample for genetic analysis, as well as to incorporate more factors and longitudinal data.
ML algorithms have also been used to classify asthma phenotypes according to the disease severity. In particular, latent class analysis was applied to questionnaire data included demographic and clinical features to classify female asthma patients into four phenotypes (“controlled, mild asthma”, “partly controlled, moderate asthma”, “uncontrolled asthma of unknown severity”, and “uncontrolled, severe asthma”) and male asthma patients into three phenotypes (“controlled, mild asthma”, “poorly controlled asthma of unknown severity”, and “partly controlled, severe asthma”) [51]. Although the study provided a simpler method for identifying asthmatic phenotypes, there are still several limitations, such as the lack of formal verification of lung function testing. In a similar study, the correlation of wheeze sounds with asthmatic severity was analyzed in 55 asthmatic patients using three ML algorithms, including the ensemble, support vector machine and k-nearest neighbor. The ensemble algorithm showed better performance, and the wheeze sound was identified as a sensitive and specific predictor of asthma severity [52].
Application of AI/ML to the monitoring and management of asthma
Asthma exacerbation and admission have a significant impact on the life quality and mortality of patients. Artificial neural networks have been extensively used to monitor and manage asthma exacerbation and admission [53-56]. For example, an artificial neural network was used to analyze clinical data and create an automated pediatric asthma severity score, which showed better performance than the pediatric asthma score and could therefore help manage pediatric asthma exacerbation in the pediatric intensive care unit [54]. Similarly, a retrospective cohort study of 31,433 adult asthma patients reported a time-sensitive predictive model based on an artificial neural network, which integrated clinical variables in the observed time window to predict asthma exacerbation [55]. In addition, a modified artificial neural network was applied to predict emergency department visits of asthma and COPD patients due to exacerbation. The developed ML model integrated several daily variables, including the number of emergency department visits as well as meteorological and environmental pollution data, reaching an overall accuracy of 81%. Nevertheless, further studies should include other variables associated with the exacerbation of these diseases [56].
In recent studies, latent class analysis has been used to predict the exacerbation risk of asthma and the decline of lung function in school-age children [57, 58]. In the latter study, a dataset consisting of 19 demographic, clinical, and laboratory variables derived from 2,593 children with mild to moderate asthma was used, and the analysis identified allergy and lung function as the main predictors of exacerbation. A similar retrospective cohort study was also performed using EHRs from 2,691 asthmatic children. Among several ML methods, the multivariable logistic regression model proved to be the most accurate, with AUC = 0.86 [59]. However, all data in that study came from a single medical center, and the multivariable logistic regression model could not be validated.
The application of four ML algorithms (logistic regression, decision tree, naïve Bayes, and perceptron algorithm) to predict severe exacerbations of asthma was recently reported based on daily monitoring data of 576 severe exacerbation events in 2,010 asthma patients. The logistic regression-based model yielded an optimal AUC of 0.85, sensitivity of 90% and specificity of 83% [60]. Given the close correlation between severe exacerbations and asthma mortality, the model may be useful to physicians as a reference, but the research data were collected from paper diaries that may be inaccurate.
In order to assess the predictability of an Internet search index for asthma admission, an ML-based prediction model (XGBoost) was developed by combining search index and data, such as air pollution, weather, and previous admission events, yielding a maximum AUC of 0.832. However, the model performance should be further validated in other geographical regions [61]. In a similar approach, an artificial neural network model was applied to predict, in real time, asthma-related emergency department visits using environmental and social media data, such Google searches and Twitter [62]. The model accuracy was 70%, making it suitable for early intervention in asthma patients to avoid exacerbation. A growing number of studies have also used electronic devices to manage, monitor, and follow asthma patients in real time [63-65]. For example, several ML algorithms (naïve Bayesian classifier, adaptive Bayesian network, and support vector machine) were used to analyze telemonitoring data from laptops at home in order to predict asthma exacerbation in a timely fashion. The sensitivity, specificity, and accuracy of the adaptive Bayesian network model reached 100%, but the study was limited by the low number of exacerbations in the collected data [64]. Another study used a contactless bed sensor to capture physiological and environmental data for the early detection of asthma exacerbation in children. Using a random forest classification model, an accuracy of 87.4% was achieved [65].
Moreover, a comorbidity portfolio model was also designed based on ML algorithms to improve the prediction of asthma treatment costs over traditional approaches [66]. The study found that a combination of cardiovascular and other respiratory diseases was a major risk for increased treatment costs in asthmatic patients. This study provides an important perspective on controlling asthma expenses in light of the high financial burden of asthma worldwide and continuing concern about treatment costs.
Successful monitoring of asthma control levels also plays a significant role in the treatment of the disease. Therefore, physicians' expertise was combined with an ensemble ML algorithm to detect asthma control. The optimal accuracy of the model was 91.66% and, although the study included relatively few factors affecting asthma control, the model could help clinicians develop timely treatment plans [67]. Based on these findings, several common supervised ML algorithms were further used to analyze asthmatic monitoring data from 5,875 patients enrolled in the Asthma Mobile Health Study. Both logistic regression and naïve Bayes-based classifiers identified the control level with high accuracy (AUC > 0.87) [68], suggesting that this method could serve as a valuable reference for the treatment of asthma in clinical practice. However, these models should be further validated using more diverse data, preferably data based on objective measures rather than self-report.
Application of AI/ML to the treatment of asthma
Despite the wide variety of studies on AI/ML implementation in asthma, very few studies have reported the application of AI/ML systems to the treatment of the disease, as such treatment is usually controlled by specific guidelines. In addition to the two aforementioned studies [45, 46], the effects of anti-inflammatory and antioxidative saffron on the treatment of mild-to-moderate allergic asthma in 80 patients were predicted using a genetic algorithm developed by modifying an artificial neural network system. The accuracy of the prediction system was greater than 99% in both the training and testing phase [69], which probably makes it suitable for predicting the treatment effect of other asthma drugs. Nevertheless, the performance of this prediction system needs to be confirmed with studies on more patients with allergic or other types of asthma.
Discussion and future directions
With the continuous improvement of computer learning and the accumulation of asthma-related data, the application of AI/ML in asthma has made great progress with good results for specific clinical research purposes [70]. Using AI/ML techniques, the mining and analysis of huge clinical, metabolomics, genomics, and other heterogeneous asthma data can help to better understand the pathogenesis and guide individual treatment of the disease. Nevertheless, some AI/ML models developed for asthma diagnosis, classification, assessment, and prediction have many limitations, such as a single data source, small sample, or lack of external confirmation, which should be overcome in future studies.
The application of AI/ML in asthma is also still limited. For example, although the current studies can be combined with different data or variables to build ML models for identifying asthma phenotypes, the integration of relative comprehensive data or variables, including demographic, environmental, medical history, symptoms, laboratory examination, pulmonary function testing, genomics, metabolomic, and imaging, is still absent. Moreover, AI/ML methods have rarely been used to identify or predict patients with severe asthma. Such studies would be of great clinical significance, because an in-depth understanding of asthma phenotypes and the reliable identification of specific subgroups could guide the use of specific drugs, such as biological-targeted agents, to bring precision medicine to asthma patients. In addition, most studies on the application of AI/ML in asthma are based on populations and datasets from developed countries, while very little research has been conducted on populations and data in developing countries, where the need for diagnosis, treatment and management is more urgent due to the weaker healthcare systems. To make the situation more challenging, the COVID-19 pandemic has severely disrupted health services for patients with chronic airway diseases. Therefore, the use of AI/ML tools to establish a management system for such patients during an infectious disease epidemic should be seriously considered.
AI/ML and COPD
Application of AI/ML to the screening and diagnosis of COPD
There are no specific symptoms related to COPD, but the disease can be diagnosed using the pulmonary function test. However, its accuracy is highly dependent on the patient's cooperation, which explains the common under- and overdiagnosis of COPD in clinical practice [71]. To address this challenge, several AI/ML techniques have been used to develop an economical, safe, and effective method for COPD diagnosis. For instance, as an AI diagnostic tool, an “expert system” was built using the following steps: questionnaire formation, WebFlex code development, expert panel pilot validation and clinical validation. The questionnaire information included demography, symptoms, environment, and diagnostic tests. In the clinical validation phase, this “expert system” reached an overall accuracy of 97.5% in 241 patients [72]. In a similar manner, a subsequent study used data from lung function tests and clinical information from 1,430 subjects to build AI-based software for the diagnosis of COPD [73]. That study showed that the developed software can reach an accuracy of 82% in 50 COPD patients significantly exceeding the diagnostic performance of pulmonologists (44.6 ± 8.7%). It is therefore clear that AI technology can considerably help clinicians make diagnostic decisions for COPD patients.
To reduce the dependence on lung function tests for early diagnosis of COPD, ML has also been used to mine and analyze transcriptomic data extracted from human bronchial epithelial cells, leading to the identification of abnormal expression of 15 genes in the disease, 10 of which had not previously been reported as COPD biomarkers. The different gene combinations were then analyzed by the random forest algorithm to distinguish non-smokers from smokers and COPD patients [74] (Table 3). Despite the remarkable diagnostic accuracy of each subgroup (65%), further studies are required to improve the model performance in distinguishing COPD patients from smokers without COPD. Given the lack of specific biomarkers for COPD diagnosis, support vector machine was also integrated with two blood biomarkers, N-acetyl-glycoprotein and lipoprotein, which were obtained by comparing 54 COPD patients with 74 normal individuals. The model achieved a diagnostic accuracy of 84.62% and an AUC of 0.90 [75], suggesting that the combination of ML algorithms with biomarkers may favor COPD diagnosis and reduce the dependence on lung function tests. However, further validation in a larger patient sample is needed.
Respiratory sounds are an important sign of the lungs and their analysis can be useful in the diagnosis of respiratory diseases [76, 77]. In a recent study, 39 features of respiratory sound were integrated with three lung function features derived from 30 COPD patients and 25 healthy subjects, and five ML classifiers were used to categorize normal individuals and COPD patients. Support vector machine and logistic regression achieved a diagnostic accuracy, sensitivity, and specificity of almost 100% [78]. In a similar approach, 22 different clinical features were extracted from each of 132 subjects. Based on this dataset, a decision support system was developed to diagnose COPD and asthma, with the random forest classifier showing the highest COPD diagnostic accuracy (97.7%) compared to other techniques. Moreover, smoking, forced expiratory volume in 1 second (FEV1), age, and forced vital capacity proved to be the main predictors [79]. However, the results of these studies should be carefully interpreted due to their small, single-center samples.
Machine learning studies on chronic obstructive pulmonary disease
| Reference | Category | Study population | ML algorithms | Input features | Studied outcome | Results | Critical appraisal of the study |
|---|---|---|---|---|---|---|---|
| Matsumura K, 2020 [74] | Screening and diagnosis | non-smokers 68 smokers = 88 COPD subjects = 48 | RF | Genetic (transcriptomic data) | Smokers or early stage of COPD | Each group with 65% accuracy. The discrimination accuracy of COPD subjects from smokers was only 29% | Pros: identification of novel genes associated with COPD Cons: limited number of patients with clear description of the smoking status |
| Zheng H, 2020 [75] | Screening and diagnosis | COPD patients = 54 normal individuals = 74 | SVM | Serum metabolic biomarkers | COPD subjects or not | Accuracy = 84.62%, AUC = 0.90 | Pros: based on serum metabolomics Cons: should be validated in a larger clinical sample |
| Haider NS, 2020 [78] | Screening and diagnosis | COPD patients = 30 healthy subjects = 25 | SVM, KNN, LR, DT, DA | Clinical (lung sound), spirometry features | COPD subjects or not | Optimal accuracy = 100% | Pros: combination of spirometry data with lung sound features for COPD diagnosis Cons: small sample and single-center data |
| Spathis D, 2019 [79] | Screening and diagnosis | 132 patients | NB, LR, ANN, SVM, KNN, DT, RF | Clinical, demographic | Asthma or COPD | Optimal accuracy = 97.7% | Pros: identification of COPD based on 22 different clinical features Cons: relatively small sample |
| Al Sallakh MA, 2018 [82] | Screening and diagnosis | Secure Anonymised Information Linkage (SAIL) Databank | LCA | Clinical (EHR) | Asthma-COPD overlap | A protocol | Pros: based on electronic health records (EHRs) Cons: incomplete information of electronic health records |
| Pikoula M, 2019 [84] | Classification and assessment | 30,961 COPD patients | K-means, hierarchical clustering | Clinical (EHR) | COPD phenotypes | Five phenotypes: anxiety/depression; non-comorbid; cardiovascular/diabetes; severe COPD/frailty; obesity/atopy | Pros: identification of phenotypes based on EHRs Cons: unclear boundaries of some clusters |
| Burgel PR, 2017 [85] | Classification and assessment | 6,060 COPD patients | CART | Clinical | COPD phenotypes | Five phenotypes: mild respiratory, moderate-to-severe respiratory, moderate-to-severe comorbid/obese, very severe respiratory, very severe comorbid | Pros: integrated respiratory characteristics and comorbidities Cons: assessment of comorbidities was based on physician diagnoses that did not consider occult conditions |
| Yoon HY, 2019 [86] | Classification and assessment | 1,195 COPD patients | K-means | Clinical (seven variables) | COPD phenotypes | Four phenotypes: putative asthma-COPD overlap, mild COPD, moderate COPD, severe COPD | Pros: demonstrated that phenotype is linked to the occurrence of acute exacerbation Cons: short follow-up duration |
| Kim WJ, 2018 [87] | Classification and assessment | 1,676 COPD patients from 13 Asian cities | Hierarchical cluster analysis | Clinical | COPD phenotypes | Three phenotypes: worse lung function and fewer symptoms, worse lung function and more symptoms. milder COPD and a preserved FEV1 and FEV1/FVC ratio | Pros: identification of COPD subgroups in a large Asian sample Cons: 90% male subjects |
| Castaldi PJ, 2014 [88] | Classification and assessment | 10,192 smokers | K-means | Clinical | COPD phenotypes | Four phenotypes: relatively resistant smokers, mild upper zone emphysema-predominant, airway disease-predominant, severe emphysema | Pros: identification of phenotypes based on airway disease and emphysema Cons: non-inclusion of biomarkers and comorbidities |
| Bodduluri S, 2020 [90] | Classification and assessment | 8980 individuals | DNN, RF | Spirometry data | Chest CT phenotypes (normal, airway predominant, emphysema predominant, and mixed emphysema/airway) | The DNN model had the highest accuracy (AUC = 0.80 and 0.91) | Pros: used spirometry data to train the model Cons: nonsmokers with and at risk for COPD were not included in the cohort |
| Gawlitza J, 2019 [94] | Classification and assessment | 75 COPD patients | KNN, XGBoost, ANN | Quantified computed tomography | Pulmonary function | KNN model with the lowest mean relative error (16%) | Pros: prediction of lung function values from quantitative computed tomography parameters Cons: small sample |
| Westcott A, 2019 [95] | Classification and assessment | 95 COPD patients | LR, SVM | Thoracic computed tomography | Lung ventilation | Accuracy = 88%, AUC = 0.82 | Pros: development of a computed tomography analysis pipeline Cons: few mild COPD patients |
| González G, 2018 [96] | Classification and assessment | 8,983 COPDGene participants and 1,672 ECLIPSE participants | Convolutional neural network | Chest computed tomography | COPD, stage, acute respiratory disease events, mortality | C-index = 0.856, accuracy = 51.1% in COPDGene cohort | Pros: based on chest computed tomography images Cons: high training computational cost and memory requirements |
| Peng J, 2020 [97] | Classification and assessment | 410 hospitalized AECOPD patients | DT | Clinical (medical records) | Mild and severe AECOPD | Accuracy = 80.3% | Pros: fast identification of the deterioration and death risk of AECOPD patients Cons: non-inclusion of interleukin and other inflammatory cytokines |
| Goto T, 2019 [98] | Management and monitoring | 44,929 hospitalized COPD patients | Lasso regression, DNN | Clinical | 30-day readmission | C-statistic = 0.61 | Pros: huge sample size and more than 1000 predictors Cons: unable to identify patients readmitted to different hospitals |
| Min X, 2019 [99] | Management and monitoring | 111,992 patients from the Geisinger Health System | LR, RF, SVM, GBDT, MLP | Medical claims data | 30-day readmission | Optimal AUC = 0.653 | Pros: combined knowledge and data driven features Cons: lack of mortality information for patients |
| Cavailles A, 2020 [100] | Management and monitoring | 143,006 patients hospitalized for AECOPD | DT | Clinical | Risk of readmission | Previous admission times was the most important risk of readmission | Pros: identification of variables associated with readmission Cons: no information on spirometry or severity |
| Chen W, 2020 [101] | Management and monitoring | 4,167 subjects | RF | Clinical, spirometry | Prebronchodilator FEV1, risk of airflow limitation | C-statistic = 0.86-0.87 | Pros: development of a personalized risk model to predict the risk of airflow limitation Cons: lack of ethnic diversity in the cohort |
| Ma X, 2020 [102] | Management and monitoring | COPD patients = 441 control subjects = 192 | KNN, LR, DT, SVM, ANN, XGBoost | Genetic, clinical | Early-stage COPD | KNN and LR had the highest precision (82%) and accuracy (81%) ANN had the highest sensitivity (100%) | Pros: identification of the association of genes and COPD development Cons: unbalanced samples from the seven centers; only nine 9 genes and five clinical features were obtained |
| Lanclus M, 2019 [103] | Management and monitoring | 62 COPD patients | SVM | Functional respiratory imaging | COPD exacerbations | Accuracy = 80.65%, positive predictive value = 82.35% | Pros: use of functional respiratory imaging for AECOPD prediction Cons: more advanced COPD patients in the cohort |
| Wang C, 2020 [104] | Management and monitoring | AECOPD patients = 135 not AECOPD patients = 168 | RF, SVM, LR, KNN, NB | Clinical (EMRs) | COPD acute exacerbations | Optimal sensitivity = 80%, specificity = 83%, positive predictive value = 81%, negative predictive value = 85%, and AUC = 0.90 from SVM | Pros: decision support for clinicians Cons: single-center cohort |
| Luo L, 2020 [107] | Management and monitoring | 780,295 hospitalizations data | LR, RF, XGBoost | Medical insurance data | High-cost COPD patients | AUC = 0.787 (LR); AUC = 0.792 (RF); AUC = 0.801 (XGBoost) | Pros: identification of high costs for COPD patients Cons: no smoking status, household income, or family population information |
| Morales DR, 2018 [108] | Management and monitoring | 54879 COPD patients | LR, SVM | Clinical | 1-year mortality | C-statistic = 0.723 | Pros: use of external data to validate models Cons: analysis of patients only with complete data |
| Moll M, 2020 [109] | Management and monitoring | 2,632 participants from COPDGene cohort and 1,268 participants from ECLIPSE cohort | RF | Clinical, spirometry, imaging | Time to death from any cause | C-index ≥ 0.7 in both cohorts | Pros: prediction of all-cause mortality Cons: cohorts were not representative of the general population |
| Orchard P, 2018 [110] | Treatment | 135 COPD patients | Sparse maximum-margin classifier, ensembles of boosted classifier, multitask neural network model | Clinical (telemonitoring data), weather | Admission and initiation of oral corticosteroid treatment | Optimal AUC = 0.74 | Pros: the model serves as a guide for corticosteroid therapy Cons: lack of a gold standard definition for exacerbation |
Abbreviations: ANN, artificial neural networks; AUC, the area under the curve; BN, bayesian networks; CART, classification and regression tree; DA, discriminant analysis; DNN, deep neural network; DT, decision trees; EMRs, electronic medical records; EHR, electronic health records; GBDT, gradient boosting decision tree; KNN, k-nearest neighbors; LCA, latent class analysis; LR, logistic regression; MLP, multi-layer perceptron; NB, naïve Bayes; RF-random forest; SVM-support vector machine.
Inequities in access to medical resources also affect the diagnosis of COPD, especially in less developed areas. Therefore, an automated telehealth AI system was recently developed and verified in 780 patients from several medical institutions [80]. The diagnostic accuracy reached 97%, and the simple equipment involved may allow its use in remote areas and in patients with less mobility.
Some patients may also have both asthma and COPD, known as asthma-COPD overlap (ACO). However, the lack of accurate diagnostic criteria has led to insufficient data on the prevalence and treatment of ACO [81]. The only relevant study of which we are aware has been registered so far as a protocol, and it aims to precisely classify COPD, asthma, and ACO patients by applying a modified latent class model to EHRs from the Secure Anonymized Information Linkage databank [82]. The analyzed data will include demographic characteristics, history of present illness, allergy, and smoking history, so the future study is expected to provide useful information.
Application of AI/ML to the classification and assessment of COPD
According to the Global Obstructive Lung Disease Initiative, COPD patients are classified into four phenotypes based on their symptomatic assessment, exacerbation and hospitalization history [83]. However, the discriminatory ability of this method is insufficient, leading to the AI/ML-based integration of additional information, including physiological features, lung function test results, comorbidities, genome, and biomarkers, for precise phenotype classification, severity assessment, and therapeutic guidance [84-89]. For example, k-means clustering was applied to analyze eight factors in 1,195 COPD patients such as physiological features, medical history, COPD assessment test score, and post-bronchodilator FEV1. Four phenotypes were identified: putative asthma-COPD overlap (cluster 1), mild COPD (cluster 2), moderate COPD (cluster 3), and severe COPD (cluster 4). Cluster 4 showed the worst post-bronchodilator FEV1 (46.7%), the shortest 6-min walking distance (365 m), and the highest COPD assessment test score (17.5), whereas cluster 1 showed the highest risk of acute exacerbation [86]. Nevertheless, the results need to be supported by a longer follow-up duration (>6 months). In another study, the variation in lung function and life quality scores among 1,676 Asian COPD patients were monitored for one year, identifying three phenotypes of COPD patients. Cluster 1 was defined by worse lung function but fewer symptoms, while cluster 3 showed mild severity but higher body mass index; cluster 2 showed severe disease and more symptoms, including the highest risk of acute exacerbation and rate of FEV1 deterioration [87]. However, one of the main study limitations was the high proportion of male subjects (90%). Moreover, using two ML methods (k-means and hierarchical clustering) and based on comorbidities and risk factors, 30,961 COPD patients were classified into five phenotypes: anxiety and depression, severe airflow limitation and weakness, cardiovascular disease and diabetes, as well as obesity/atopy and non-comorbidity [84]. Although the aforementioned studies used different ML algorithms and clinical variables and had some limitations, all supported the idea that exploring different phenotypic classification can improve individualized treatment. For similar purpose, the spirometry data of 8980 individuals (COPDGene cohort study) was used to develop a deep neural network model for the identification of four chest computed tomography imaging phenotypes (normal, airway predominant, emphysema predominant, and mixed emphysema/airway). The deep neural network model had a higher accuracy both in the classification of predominant emphysema/airway phenotypes (AUC = 0.80) and predominant emphysema/small airway phenotypes (AUC = 0.91) than FEV1/forced vital capacity, FEV1% predicted and random forest classifier. However, non-smokers with and at risk for COPD should be included in future studies [90].
The assessment of persistent airflow limitation in COPD patients depends on lung function tests. However, only some COPD patients complete these tests in clinical practice, limiting the diagnosis of airway limitation to 56% [91, 92]. Considering that it is difficult to identify FEV1 values in structured EHRs, an automatic AI tool was designed to mine FEV1 values in EHRs of 41,689 veterans with COPD. The novel AI tool showed an accuracy of 95%, serving as a helpful tool for the assessment of COPD severity in large patient population [93].
Chest computed tomography has also been widely used to detect lung texture abnormalities and assess the state of COPD. However, a large amount of image data cannot be identified with the naked eye, highlighting the need for AI/ML systems in this field [94]. In a recent prospective study, the pulmonary ventilation function of COPD was assessed using a support vector machine and logistic regression algorithms to analyze chest computed tomography images. The assessment model (quadratic support vector machine) was based on 87 image features, and its validity was tested in 27 COPD patients with an accuracy of 88% and an AUC value of 0.82 [95]. While these results are encouraging, the sample was small and most patients had moderate to severe COPD, suggesting that condign mild COPD patients should be included in future works. In another study, a convolutional neural network algorithm was used to analyze chest computed tomography images from smokers and to assess the diagnosis, stage, exacerbation, and mortality of COPD patients. Smokers in the study were divided into a training phase consisting of 8,983 participants and an evaluation phase consisting of 1,672 participants, which came from COPDGene and ECLIPSE cohorts. The algorithm yielded a c-index of 0.856 for COPD detection and an accuracy of 51.1% for the exact determination of the COPD stage in the COPDGene cohort. Moreover, the c-indices for predicting exacerbation and mortality were 0.64 and 0.72 in the COPDGene cohort, and 0.55 and 0.60 in the ECLIPSE cohort [96]. These results indicated better performance of the convolutional neural network in the COPDGene cohort, while suggesting its applicability in stage classification and risk assessment of COPD at the population level. However, this method may have limited applicability because it requires extensive training and computational resources.
Assessing the severity of hospitalized acute exacerbations of COPD (AECOPD) patients is also beneficial to clinical practice. Hence, a modified decision tree algorithm was used to analyze 28 clinical features, including demographics, medical history, and biomarkers derived from 202 inpatients with severe AECOPD and 208 inpatients with mild AECOPD. The classification of severe and mild patients was based on their admission to the intensive care unit. The overall accuracy of the developed classifier reached 80.3%, suggesting that it can be used to assess the severity of hospitalized AECOPD patients [97]; however, the patient's body mass index and other inflammatory cytokines should be included in a future prospective study.
Application of AI/ML to the management and monitoring of COPD
Persistent chronic airway inflammation and airflow limitation in COPD can induce the recurrence of acute exacerbation and readmission. In order to effectively manage COPD patients and monitor the disease, several studies have used ML-based approaches, which proved to be more effective than conventional methods [98-100]. In particular, ML algorithms, such as lasso regression and deep neural network, were used to analyze 44,929 COPD hospitalizations divided into a training (70%) and a test (30%) set. The developed models aimed to predict readmission within 30 days after discharge and showed higher prediction ability (c-statistic = 0.61) than the traditional method [98]. Similarly, several non-deep and deep ML algorithms were used to mine a database containing medical claims data of COPD patients in order to predict readmission 30 days after discharge, and the optimal AUC was 0.653 [99]. A retrospective study in France applied decision tree analysis to predict the readmission of 143,006 COPD patients older than 40 years. The study not only showed that the most relevant risk factor of readmission was the number of previous admissions, but it also assessed the cost of readmission within six months [100]. Although these studies have reported several limitations, such as the lack of important clinical features, the prediction models could be used by clinicians as a reference.
Persistent airflow limitation along with persistent respiratory symptoms make COPD a lifelong and life-threatening disease. Thus, it is particularly important to monitor variation in lung function and prevent persistent airflow limitation. A ML model based on random forest was recently developed using spirometry data obtained from 4,167 participants in order to predict individuals most likely to develop or have COPD. The primary outcome of the model was FEV1, while the secondary outcome was the risk of airflow limitation (FEV1/forced vital capacity). This model may be a useful tool for personalized risk prediction of airflow limitation and early prevention of COPD [101].
Given the irreversibility of COPD, its early detection and diagnosis are crucial. Thus, six ML models were used to predict the development of COPD based on 101 single-nucleotide polymorphisms and 5 clinical characteristics of 441 patients and 192 normal participants. Among them, 9 single-nucleotide polymorphisms were significantly associated with this disease, including 6 risk and 3 protective factors. In the test set, among the examined models, the k-nearest neighbor classifier and logistic regression showed the highest precision of 82% and accuracy of 81%, while the highest sensitivity (recall) of 100% was achieved using the multilayer perceptron classifier based on the artificial neural network algorithm [102]. Although only a few genes and clinical features were included, this model may be effective for early diagnosis of COPD, compensating for the lack of lung function testing among patients in the early disease stages.
ML algorithms were also used to analyze functional respiratory imaging for the prediction of exacerbation and early identification of AECOPD patients [103]. Similarly, a series of ML algorithms (logistic regression, random forest, naïve Bayesian, support vector machine and k-nearest neighbor) were used to mine EHRs data derived from 135 AECOPD and 168 control subjects. Further validation and comparison of the developed models indicated that the support vector machine algorithm showed the best performance (AUC = 0.90) [104]. Consequently, ML models, and especially the support vector machine model, can help physicians identify AECOPD patients and make timely decisions; however, the models' performance should be further validated using data from external sources. In another approach, a mobile telehealth system was designed to improve self-management in COPD and detect acute exacerbations of stable COPD patients in a timely manner. The system could continuously monitor the clinical information of the enrolled patients at home and warn of an acute exacerbation three days in advance [105]. Although the accuracy was only 40% and the study lasted only six months, the development of a simple, effective AI-based monitoring and warning system deserves further investigation.
The global economic burden of COPD increases every year, a trend exacerbated by the aging population. In Europe, the total cost of COPD is estimated at 56% of annual healthcare expenditure for respiratory diseases [106]. To identify and predict the costs of COPD patients in China and to provide crucial health management information, three ML algorithms (logistic regression, random forest, and extreme gradient boosting) were used to analyze 54 different demographic parameters and medical information from 780,295 hospitalizations. Although all ML models showed excellent predictive efficiency, the extreme gradient boosting model showed the highest sensitivity (71.3%) and AUC (0.801) [107], indicating that it may serve as a valuable predictive tool for patients, clinicians, insurance policy makers, and other healthcare professionals in developing countries.
Since COPD is one leading causes of death worldwide, some studies have also used AI/ML technology to predict the risk of mortality in patients with COPD [108, 109]. For instance, a total of 30 clinical, lung function, and chest imaging features obtained from 3,900 participants with moderate to severe COPD were analyzed to build a random forest model for mortality prediction. The novel model showed good prediction performance (C-index > 0.7), and the optimal risk predictors were the 6-min walk test, the FEV1 value, and the pulmonary artery-to-aorta ratio [109]. Hence, the novel ML model can be a useful tool to guide the early intervention of COPD to avoid further deterioration, but more external population cohorts are needed for validation.
Application of AI/ML in COPD treatment
AI/ML technologies can monitor, integrate, and analyze large-scale, heterogeneous clinical information from COPD patients; suggest optimal individualized treatments; and reduce over- or undertreatment caused by clinician errors. However, similar to asthma, we found only one study related to the application of AI/ML in COPD treatment. In that work, several ML models (e.g. sparse maximum-margin classifier, ensembles of boosted classifier, multitask neural network model) were developed based on 153 predictive factors derived from telemonitoring of physiological, symptom, and baseline data from 135 patients with moderate to severe COPD. The data included demography, severity, quality of life and hospital admissions, and the goal was to detect acute exacerbations and guide the corticosteroid therapy of COPD. Irrespective of acute exacerbations or corticosteroid use, the optimal ML model (multitask neural network) showed better AUC values than non-ML methods (0.74-0.77 vs 0.60-0.66), and its performance was not improved by adding weather data [110]. However, the evaluation of model performance relied on cross-validation rather than multiple independent cohorts, suggesting the need for further study.
Discussion and future directions
Given the structured data from pulmonary function testing and their important role in the diagnosis and management of COPD, AI/ML techniques were combined with such testing early in the field in order to help clinicians assess lung function [111]. Other studies have focused on the use of AI/ML methods to reduce the reliance on pulmonary function testing in clinical practice. Our literature review also showed that ML methods have been increasingly used in recent years for the identification of COPD phenotypes and for the prediction of acute exacerbation and death risk. However, similar to the limitations of AI/ML techniques in asthma, such as single-source populations and lack of external validation, the model still needs to be validated in more prospective studies using a large real-world dataset as well as clinical data from continuous monitoring.
Although there have been significant advances in understanding and managing COPD using AI/ML methods, there are still many unmet needs. For example, airflow limitation is the hallmark of COPD, and studies have found that even in patients with similar FEV1 levels, there are significant differences in the degree of respiratory symptoms, frequency of acute exacerbations, and activity endurance [112]. The underlying biological mechanism remains unknown, indicating the importance of phenotypic studies in COPD patients. The current four phenotypes of COPD proposed by the Global Obstructive Lung Disease Initiative are insufficient for individualized therapy; thus, the development of phenotype-specific therapeutic strategies should be extensively studied in the future [85]. We expect that the integration and analysis of different data using AI/ML technologies may provide new insights into the COPD phenotype.
AI/ML may also play a significant role in the treatment and management of patients with stable chronic airway diseases. Inhalation therapy is the basic therapeutic approach for stable COPD. However, stable COPD patients may misuse inhaled devices due to the diversity of inhaled drugs and devices and the lack of medical assistance. Therefore, the application of AI/ML methods for the early detection of errors and the improvement of patient compliance should be further investigated. AI/ML studies are also worth conducting in low-income individuals or regions, as they usually exhibit high morbidity and experience a heavy financial burden due to COPD; in addition, the relatively delayed generation of clinical data in their health systems leads to poor data integrity.
Conclusions
In recent years, the application of AI/ML techniques in the medical field has evolved rapidly, while several AI/ML tools have been extensively studied and applied to chronic airway diseases. This review summarizes the recent advances in the implementation of AI/ML techniques in the screening and diagnosis, classification and assessment, management and monitoring, as well as treatment of asthma and COPD. Our research supports that AI/ML techniques can be used effectively to analyze and integrate large, heterogeneous medical data, thus assisting physicians in making decisions and guiding clinical practice. In addition, these techniques can be applied to analyze different responses to treatment, providing therapeutic guidance for specific phenotypes required in precision medicine, and establish a management system for chronic airway disease patients during an infectious disease epidemic. However, the results about AI/ML tools should be interpreted and generalized with caution. The AI/ML techniques cannot yet replace clinicians in the diagnosis and treatment of chronic airway diseases, and further studies are needed to examine the effect of several parameters on ML model construction and to verify existing findings with larger samples and external data sources.
Abbreviations
ACO: asthma-COPD overlap; AECOPD: acute exacerbations of COPD; AI: artificial intelligence; ML: machine learning; AUC: area under the receiver operating characteristic curve; COPD: chronic obstructive pulmonary disease; ECLIPSE Study: “Evaluations of COPD longitudinally to identify predictive surrogate endpoints”; EHRs: electronic health records; FEV1: forced expiratory volume in 1 second.
Acknowledgements
This work was supported by grants from the Sichuan Science and Technology Project (2017SZ0068) to Hui Mao and from the Deyang Science and Technology Project (2018SZS056) to Yinhe Feng.
Competing Interests
The authors have declared that no competing interest exists.
References
1. Esteva A, Robicquet A, Ramsundar B. et al. A guide to deep learning in healthcare. Nat Med. 2019;25(1):24-29
2. Rajkomar A, Dean J, Kohane I. Machine Learning in Medicine. N Engl J Med. 2019;380(14):1347-1358
3. Erickson BJ, Korfiatis P, Akkus Z, Kline TL. Machine Learning for Medical Imaging. Radiographics. 2017;37(2):505-515
4. McBee MP, Awan OA, Colucci AT. et al. Deep Learning in Radiology. Acad Radiol. 2018;25(11):1472-1480
5. Ho D, Schierding W, Wake M, Saffery R, O', Sullivan J. Machine Learning SNP Based Prediction for Precision Medicine. Front Genet. 2019;10:267
6. van IJzendoorn D, Szuhai K, Briaire-de Bruijn IH, Kostine M, Kuijjer ML, Bovée J. Machine learning analysis of gene expression data reveals novel diagnostic and prognostic biomarkers and identifies therapeutic targets for soft tissue sarcomas. PLoS Comput Biol. 2019;15(2):e1006826
7. Feng Y, Teh HS, Cai Y. Deep Learning for Chest Radiology: A Review. Curr Radiol Rep. 2019;7(8):24
8. Angelini E, Dahan S, Shah A. Unravelling machine learning: insights in respiratory medicine. Eur Respir J. 2019;54(6):1901216
9. Shen Y, Huang S, Kang J. et al. Management of airway mucus hypersecretion in chronic airway inflammatory disease: Chinese expert consensus (English edition). Int J Chron Obstruct Pulmon Dis. 2018;13:399-407
10. Tan KS, Lim RL, Liu J. et al. Respiratory Viral Infections in Exacerbation of Chronic Airway Inflammatory Diseases: Novel Mechanisms and Insights From the Upper Airway Epithelium. Front Cell Dev Biol. 2020;8:99
11. Bao Z, Xiong J, Li W, Chen Z, Shen H, Ying S. Genomic instability in chronic airway inflammatory diseases. 2015; 38(2): 117-124.
12. Barnes PJ. Cellular and molecular mechanisms of asthma and COPD. Clin Sci (Lond). 2017;131(13):1541-1558
13. Exarchos KP, Beltsiou M, Votti CA, Kostikas K. Artificial intelligence techniques in asthma: a systematic review and critical appraisal of the existing literature. Eur Respir J. 2020;56(3):2000521
14. Buch VH, Ahmed I, Maruthappu M. Artificial intelligence in medicine: current trends and future possibilities. Br J Gen Pract. 2018;68(668):143-144
15. Mekov E, Miravitlles M, Petkov R. Artificial intelligence and machine learning in respiratory medicine. Expert Rev Respir Med. 2020;14(6):559-564
16. Sharafoddini A, Dubin JA, Lee J. Patient Similarity in Prediction Models Based on Health Data: A Scoping Review. JMIR Med Inform. 2017;5(1):e7
17. Bi Q, Goodman KE, Kaminsky J, Lessler J. What is Machine Learning? A Primer for the Epidemiologist. Am J Epidemiol. 2019;188(12):2222-2239
18. Artificial intelligence, medical imaging 2018. French Radiology Community white paper. Diagn Interv Imaging. 2018;99(11):727-742
19. Collins GS, Reitsma JB, Altman DG, Moons KG. Transparent reporting of a multivariable prediction model for individual prognosis or diagnosis (TRIPOD): the TRIPOD statement. BMJ. 2015;350:g7594
20. Lillywhite A, Wolbring G. Coverage of ethics within the artificial intelligence and machine learning academic literature: The case of disabled people. Assist Technol. 2019:1-7
21. Aaron SD, Boulet LP, Reddel HK, Gershon AS. Underdiagnosis and Overdiagnosis of Asthma. Am J Respir Crit Care Med. 2018;198(8):1012-1020
22. Aaron SD, Vandemheen KL, FitzGerald JM. et al. Reevaluation of Diagnosis in Adults with Physician-Diagnosed Asthma. JAMA. 2017;317(3):269-279
23. Wi CI, Sohn S, Rolfes MC. et al. Application of a Natural Language Processing Algorithm to Asthma Ascertainment. An Automated Chart Review. Am J Respir Crit Care Med. 2017;196(4):430-437
24. Wi CI, Sohn S, Ali M. et al. Natural Language Processing for Asthma Ascertainment in Different Practice Settings. J Allergy Clin Immunol Pract. 2018;6(1):126-131
25. Kaur H, Sohn S, Wi CI. et al. Automated chart review utilizing natural language processing algorithm for asthma predictive index. BMC Pulm Med. 2018;18(1):34
26. Alizadeh B, Safdari R, Zolnoori M, Bashiri A. Developing an Intelligent System for Diagnosis of Asthma Based on Artificial Neural Network. Acta Inform Med. 2015;23(4):220-223
27. Amaral J, Lopes AJ, Veiga J, Faria A, Melo PL. High-accuracy detection of airway obstruction in asthma using machine learning algorithms and forced oscillation measurements. Comput Methods Programs Biomed. 2017;144:113-125
28. Amaral J, Sancho AG, Faria A, Lopes AJ, Melo PL. Differential diagnosis of asthma and restrictive respiratory diseases by combining forced oscillation measurements, machine learning and neuro-fuzzy classifiers. Med Biol Eng Comput. 2020;58(10):2455-2473
29. Zhan J, Chen W, Cheng L, Wang Q, Han F, Cui Y. Diagnosis of Asthma Based on Routine Blood Biomarkers Using Machine Learning. Comput Intell Neurosci. 2020;2020:8841002
30. Sinha A, Desiraju K, Aggarwal K. et al. Exhaled breath condensate metabolome clusters for endotype discovery in asthma. J Transl Med. 2017;15(1):262
31. Pramono R, Bowyer S, Rodriguez-Villegas E. Automatic adventitious respiratory sound analysis: A systematic review. PLoS One. 2017;12(5):e0177926
32. Islam MA, Bandyopadhyaya I, Bhattacharyya P, Saha G. Multichannel lung sound analysis for asthma detection. Comput Methods Programs Biomed. 2018;159:111-123
33. Bou Chebl R, Madden B, Belsky J, Harmouche E, Yessayan L. Diagnostic value of end tidal capnography in patients with hyperglycemia in the emergency department. BMC Emerg Med. 2016;16:7
34. Singh OP, Ramaswamy P, Malarvili M. Automatic Quantitative Analysis of Human Respired Carbon Dioxide Waveform for Asthma and Non-Asthma Classification Using Support Vector Machine. IEEE Access. 2018:1-1
35. Tomita K, Nagao R, Touge H. et al. Deep learning facilitates the diagnosis of adult asthma. Allergol Int. 2019;68(4):456-461
36. Bacharier LB, Guilbert TW. Diagnosis and management of early asthma in preschool-aged children. J Allergy Clin Immunol. 2012;130(2):287-296
37. Sittka A, Vera J, Lai X, Schmeck BT. Asthma phenotyping, therapy, and prevention: what can we learn from systems biology. Pediatr Res. 2013;73(4 Pt 2):543-552
38. Howard R, Rattray M, Prosperi M, Custovic A. Distinguishing Asthma Phenotypes Using Machine Learning Approaches. Curr Allergy Asthma Rep. 2015;15(7):38
39. Korevaar DA, Westerhof GA, Wang J. et al. Diagnostic accuracy of minimally invasive markers for detection of airway eosinophilia in asthma: a systematic review and meta-analysis. Lancet Respir Med. 2015;3(4):290-300
40. Couto M, Stang J, Horta L. et al. Two distinct phenotypes of asthma in elite athletes identified by latent class analysis. J Asthma. 2015;52(9):897-904
41. Chen Q, Just AC, Miller RL. et al. Using latent class growth analysis to identify childhood wheeze phenotypes in an urban birth cohort. Ann Allergy Asthma Immunol. 2012;108(5):311-315
42. Weinmayr G, Keller F, Kleiner A. et al. Asthma phenotypes identified by latent class analysis in the ISAAC phase II Spain study. Clin Exp Allergy. 2013;43(2):223-232
43. Bochenek G, Kuschill-Dziurda J, Szafraniec K, Plutecka H, Szczeklik A, Nizankowska-Mogilnicka E. Certain subphenotypes of aspirin-exacerbated respiratory disease distinguished by latent class analysis. J Allergy Clin Immunol. 2014;133(1):98-103
44. Havstad S, Johnson CC, Kim H. et al. Atopic phenotypes identified with latent class analyses at age 2 years. J Allergy Clin Immunol. 2014;134(3):722-727
45. Ross MK, Yoon J, van der Schaar A, van der Schaar M. Discovering Pediatric Asthma Phenotypes on the Basis of Response to Controller Medication Using Machine Learning. Ann Am Thorac Soc. 2018;15(1):49-58
46. Wu W, Bang S, Bleecker ER. et al. Multiview Cluster Analysis Identifies Variable Corticosteroid Response Phenotypes in Severe Asthma. Am J Respir Crit Care Med. 2019;199(11):1358-1367
47. Prosperi MC, Marinho S, Simpson A, Custovic A, Buchan IE. Predicting phenotypes of asthma and eczema with machine learning. BMC Med Genomics. 2014;7(Suppl 1):S7
48. Krautenbacher N, Flach N, Böck A. et al. A strategy for high-dimensional multivariable analysis classifies childhood asthma phenotypes from genetic, immunological, and environmental factors. Allergy. 2019;74(7):1364-1373
49. Williams-DeVane CR, Reif DM, Hubal EC. et al. Decision tree-based method for integrating gene expression, demographic, and clinical data to determine disease endotypes. BMC Syst Biol. 2013;7:119
50. Siroux V, González JR, Bouzigon E. et al. Genetic heterogeneity of asthma phenotypes identified by a clustering approach. Eur Respir J. 2014;43(2):439-452
51. Mäkikyrö EM, Jaakkola MS, Jaakkola JJ. Subtypes of asthma based on asthma control and severity: a latent class analysis. Respir Res. 2017;18(1):24
52. Nabi FG, Sundaraj K, Lam CK, Palaniappan R. Characterization and classification of asthmatic wheeze sounds according to severity level using spectral integrated features. Comput Biol Med. 2019;104:52-61
53. Moustris KP, Douros K, Nastos PT. et al. Seven-days-ahead forecasting of childhood asthma admissions using artificial neural networks in Athens, Greece. Int J Environ Health Res. 2012;22(2):93-104
54. Messinger AI, Bui N, Wagner BD, Szefler SJ, Vu T, Deterding RR. Novel pediatric-automated respiratory score using physiologic data and machine learning in asthma. Pediatr Pulmonol. 2019;54(8):1149-1155
55. Xiang Y, Ji H, Zhou Y. et al. Asthma Exacerbation Prediction and Risk Factor Analysis Based on a Time-Sensitive, Attentive Neural Network: Retrospective Cohort Study. J Med Internet Res. 2020;22(7):e16981
56. Khatri KL, Tamil LS. Early Detection of Peak Demand Days of Chronic Respiratory Diseases Emergency Department Visits Using Artificial Neural Networks. IEEE J Biomed Health Inform. 2018;22(1):285-290
57. Grunwell JR, Gillespie S, Morris CR, Fitzpatrick AM. Latent Class Analysis of School-Age Children at Risk for Asthma Exacerbation. J Allergy Clin Immunol Pract. 2020;8(7):2275-2284.e2
58. Fitzpatrick AM, Bacharier LB, Jackson DJ. et al. Heterogeneity of Mild to Moderate Persistent Asthma in Children: Confirmation by Latent Class Analysis and Association with 1-Year Outcomes. J Allergy Clin Immunol Pract. 2020;8(8):2617-2627
59. Das LT, Abramson EL, Stone AE, Kondrich JE, Kern LM, Grinspan ZM. Predicting frequent emergency department visits among children with asthma using EHR data. Pediatr Pulmonol. 2017;52(7):880-890
60. Zhang O, Minku LL, Gonem S. Detecting asthma exacerbations using daily home monitoring and machine learning. J Asthma. 2020:1-10
61. Luo L, Liao C, Zhang F. et al. Applicability of internet search index for asthma admission forecast using machine learning. Int J Health Plann Manage. 2018:1-10
62. Ram S, Zhang W, Williams M, Pengetnze Y. Predicting asthma-related emergency department visits using big data. IEEE J Biomed Health Inform. 2015;19(4):1216-1223
63. Hosseini A, Buonocore CM, Hashemzadeh S. et al. Feasibility of a Secure Wireless Sensing Smartwatch Application for the Self-Management of Pediatric Asthma. Sensors (Basel). 2017;17(8):1780
64. Finkelstein J, Jeong IC. Machine learning approaches to personalize early prediction of asthma exacerbations. Ann N Y Acad Sci. 2017;1387(1):153-165
65. Huffaker MF, Carchia M, Harris BU. et al. Passive Nocturnal Physiologic Monitoring Enables Early Detection of Exacerbations in Children with Asthma. A Proof-of-Concept Study. Am J Respir Crit Care Med. 2018;198(3):320-328
66. Luo L, Yu X, Yong Z, Li C, Gu Y. Design comorbidity portfolios to improve treatment cost prediction of asthma using machine learning. IEEE J Biomed Health Inform. 2020 PP
67. Khasha R, Sepehri MM, Mahdaviani SA. An ensemble learning method for asthma control level detection with leveraging medical knowledge-based classifier and supervised learning. J Med Syst. 2019;43(6):158
68. Tsang K, Pinnock H, Wilson AM, Ahmar Shah S. Application of Machine Learning to Support Self-Management of Asthma with mHealth. Annu Int Conf IEEE Eng Med Biol Soc. 2020;2020:5673-5677
69. Hosseini SA, Jamshidnezhad A, Zilaee M, Fouladi Dehaghi B, Mohammadi A, Hosseini SM. Neural Network-Based Clinical Prediction System for Identifying the Clinical Effects of Saffron (Crocus sativus L) Supplement Therapy on Allergic Asthma: Model Evaluation Study. JMIR Med Inform. 2020;8(7):e17580
70. Mlodzinski E, Stone DJ, Celi LA. Machine Learning for Pulmonary and Critical Care Medicine: A Narrative Review. Pulm Ther. 2020;6(1):67-77
71. Andreeva E, Pokhaznikova M, Lebedev A, Moiseeva I, Kuznetsova O, Degryse J. Spirometry is not enough to diagnose COPD in epidemiological studies: a follow-up study. NPJ Prim Care Respir Med. 2017;27(5):62
72. Fulvio B, Pierachille S, Guido CA. et al. Chronic obstructive lung disease “expert system”: validation of a predictive tool for assisting diagnosis. Int J Chron Obstruct Pulmon Dis. 2018;13:1747-1753
73. Topalovic M, Das N, Burgel PR. et al. Artificial intelligence outperforms pulmonologists in the interpretation of pulmonary function tests. Eur Respir J. 2019;53(4):1801660
74. Matsumura K, Ito S. Novel biomarker genes which distinguish between smokers and chronic obstructive pulmonary disease patients with machine learning approach. BMC Pulm Med. 2020;20(1):29
75. Zheng H, Hu Y, Dong L. et al. Predictive diagnosis of chronic obstructive pulmonary disease using serum metabolic biomarkers and least-squares support vector machine. J Clin Lab Anal. 2020: e23641.
76. Melbye H, Garcia-Marcos L, Brand P, Everard M, Priftis K, Pasterkamp H. Wheezes, crackles and rhonchi: simplifying description of lung sounds increases the agreement on their classification: a study of 12 physicians' classification of lung sounds from video recordings. BMJ Open Respir Res. 2016;3(1):e000136
77. Sarkar M, Madabhavi I, Niranjan N, Dogra M. Auscultation of the respiratory system. Ann Thorac Med. 2015;10(3):158-168
78. Haider NS, Singh BK, Periyasamy R, Behera AK. Respiratory Sound Based Classification of Chronic Obstructive Pulmonary Disease: a Risk Stratification Approach in Machine Learning Paradigm. J Med Syst. 2019;43(8):255
79. Spathis D, Vlamos P. Diagnosing asthma and chronic obstructive pulmonary disease with machine learning. Health Informatics J. 2019;25(3):811-827
80. Gurbeta L, Badnjevic A, Maksimovic M, Omanovic-Miklicanin E, Sejdic E. A telehealth system for automated diagnosis of asthma and chronical obstructive pulmonary disease. J Am Med Inform Assoc. 2018;25(9):1213-1217
81. Rodrigo GJ, Neffen H, Plaza V. Asthma-chronic obstructive pulmonary disease overlap syndrome: a controversial concept. Curr Opin Allergy Clin Immunol. 2017;17(1):36-41
82. Al Sallakh MA, Rodgers SE, Lyons RA, Sheikh A, Davies GA. Identifying patients with asthma-chronic obstructive pulmonary disease overlap syndrome using latent class analysis of electronic health record data: a study protocol. NPJ Prim Care Respir Med. 2018;28(1):22
83. Nikolaou V, Massaro S, Fakhimi M, Stergioulas L, Price D. COPD phenotypes and machine learning cluster analysis: A systematic review and future research agenda. Respir Med. 2020;171:106093
84. Pikoula M, Quint JK, Nissen F, Hemingway H, Smeeth L, Denaxas S. Identifying clinically important COPD sub-types using data-driven approaches in primary care population based electronic health records. BMC Med Inform Decis Mak. 2019;19(1):86
85. Burgel PR, Paillasseur JL, Janssens W. et al. A simple algorithm for the identification of clinical COPD phenotypes. Eur Respir J. 2017;50(5):1701034
86. Yoon HY, Park SY, Lee CH. et al. Prediction of first acute exacerbation using COPD subtypes identified by cluster analysis. Int J Chron Obstruct Pulmon Dis. 2019;14:1389-1397
87. Kim WJ, Gupta V, Nishimura M. et al. Identification of chronic obstructive pulmonary disease subgroups in 13 Asian cities. Int J Tuberc Lung Dis. 2018;22(7):820-826
88. Castaldi PJ, Dy J, Ross J. et al. Cluster analysis in the COPDGene study identifies subtypes of smokers with distinct patterns of airway disease and emphysema. Thorax. 2014;69(5):415-422
89. Castaldi PJ, Boueiz A, Yun J. et al. Machine Learning Characterization of COPD Subtypes: Insights From the COPDGene Study. Chest. 2020;157(5):1147-1157
90. Bodduluri S, Nakhmani A, Reinhardt JM. et al. Deep neural network analyses of spirometry for structural phenotyping of chronic obstructive pulmonary disease. JCI Insight. 2020;5(13):e132781
91. Prieto-Centurion V, Rolle AJ, Au DH. et al. Multicenter study comparing case definitions used to identify patients with chronic obstructive pulmonary disease. Am J Respir Crit Care Med. 2014;190(9):989-995
92. Wu H, Wise RA, Medinger AE. Do Patients Hospitalized With COPD Have Airflow Obstruction. Chest. 2017;151(6):1263-1271
93. Akgün KM, Sigel K, Cheung KH. et al. Extracting lung function measurements to enhance phenotyping of chronic obstructive pulmonary disease (COPD) in an electronic health record using automated tools. PLoS One. 2020;15(1):e0227730
94. Gawlitza J, Sturm T, Spohrer K. et al. Predicting Pulmonary Function Testing from Quantified Computed Tomography Using Machine Learning Algorithms in Patients with COPD. Diagnostics (Basel). 2019;9(1):33
95. Westcott A, Capaldi D, McCormack DG, Ward AD, Fenster A, Parraga G. Chronic Obstructive Pulmonary Disease: Thoracic CT Texture Analysis and Machine Learning to Predict Pulmonary Ventilation. Radiology. 2019;293(3):676-684
96. González G, Ash SY, Vegas-Sánchez-Ferrero G. et al. Disease Staging and Prognosis in Smokers Using Deep Learning in Chest Computed Tomography. Am J Respir Crit Care Med. 2018;197(2):193-203
97. Peng J, Chen C, Zhou M, Xie X, Zhou Y, Luo CH. A Machine-learning Approach to Forecast Aggravation Risk in Patients with Acute Exacerbation of Chronic Obstructive Pulmonary Disease with Clinical Indicators. Sci Rep. 2020;10(1):3118
98. Goto T, Jo T, Matsui H, Fushimi K, Hayashi H, Yasunaga H. Machine Learning-Based Prediction Models for 30-Day Readmission after Hospitalization for Chronic Obstructive Pulmonary Disease. COPD. 2019;16(5-6):338-343
99. Min X, Yu B, Wang F. Predictive Modeling of the Hospital Readmission Risk from Patients' Claims Data Using Machine Learning: A Case Study on COPD. Sci Rep. 2019;9(1):2362
100. Cavailles A, Melloni B, Motola S. et al. Identification of Patient Profiles with High Risk of Hospital Re-Admissions for Acute COPD Exacerbations (AECOPD) in France Using a Machine Learning Model. Int J Chron Obstruct Pulmon Dis. 2020;15:949-962
101. Chen W, Sin DD, FitzGerald JM, Safari A, Adibi A, Sadatsafavi M. An Individualized Prediction Model for Long-term Lung Function Trajectory and Risk of COPD in the General Population. Chest. 2020;157(3):547-557
102. Ma X, Wu Y, Zhang L. et al. Comparison and development of machine learning tools for the prediction of chronic obstructive pulmonary disease in the Chinese population. J Transl Med. 2020;18(1):146
103. Lanclus M, Clukers J, Van Holsbeke C. et al. Machine Learning Algorithms Utilizing Functional Respiratory Imaging May Predict COPD Exacerbations. Acad Radiol. 2019;26(9):1191-1199
104. Wang C, Chen X, Du L, Zhan Q, Yang T, Fang Z. Comparison of machine learning algorithms for the identification of acute exacerbations in chronic obstructive pulmonary disease. Comput Methods Programs Biomed. 2020;188:105267
105. Hardinge M, Rutter H, Velardo C. et al. Using a mobile health application to support self-management in chronic obstructive pulmonary disease: a six-month cohort study. BMC Med Inform Decis Mak. 2015;15:46
106. Bonten MJ, Huijts SM, Bolkenbaas M. et al. Polysaccharide conjugate vaccine against pneumococcal pneumonia in adults. N Engl J Med. 2015;372(12):1114-1125
107. Luo L, Li J, Lian S. et al. Using machine learning approaches to predict high-cost chronic obstructive pulmonary disease patients in China. Health Informatics J. 2020;26(3):1577-1598
108. Morales DR, Flynn R, Zhang J, Trucco E, Quint JK, Zutis K. External validation of ADO, DOSE, COTE and CODEX at predicting death in primary care patients with COPD using standard and machine learning approaches. Respir Med. 2018;138:150-155
109. Moll M, Qiao D, Regan EA. et al. Machine Learning and Prediction of All-Cause Mortality in COPD. Chest. 2020;158(3):952-964
110. Orchard P, Agakova A, Pinnock H. et al. Improving Prediction of Risk of Hospital Admission in Chronic Obstructive Pulmonary Disease: Application of Machine Learning to Telemonitoring Data. J Med Internet Res. 2018;20(9):e263
111. Franssen FM, Alter P, Bar N. et al. Personalized medicine for patients with COPD: where are we. Int J Chron Obstruct Pulmon Dis. 2019;14:1465-1484
112. Agusti A, Calverley PM, Celli B. et al. Characterisation of COPD heterogeneity in the ECLIPSE cohort. Respir Res. 2010;11(1):122
Author contact
![]() Corresponding author: Hui Mao, Department of Respiratory and Critical Care Medicine, West China Hospital, Sichuan University, #37 Guoxue Alley, Chengdu 610041, China. Tel: (+86) 028-85423061, Fax: (+86) 028-2220098. E-mail: merrymhcom.
Corresponding author: Hui Mao, Department of Respiratory and Critical Care Medicine, West China Hospital, Sichuan University, #37 Guoxue Alley, Chengdu 610041, China. Tel: (+86) 028-85423061, Fax: (+86) 028-2220098. E-mail: merrymhcom.

 Global reach, higher impact
Global reach, higher impact