3.2
Impact Factor
ISSN: 1449-1907
Int J Med Sci 2021; 18(11):2347-2354. doi:10.7150/ijms.57990 This issue Cite
Review
The associations and roles of microRNA single-nucleotide polymorphisms in cervical cancer
Department of Immunogenetics, Institute of Medical Biology, Chinese Academy of Medical Sciences & Peking Union Medical College, Kunming 650118, Yunnan, China.
*These authors contributed equally to this work.
Received 2021-1-9; Accepted 2021-3-26; Published 2021-4-7
Abstract
Cervical cancer is one of the fourth most common gynecological malignancies and has been identified as the fourth leading cause of cancer death in women worldwide. MicroRNAs (miRNAs) are single-stranded sequences of noncoding RNAs that are approximately 22-24 nucleotides in length. They modulate posttranscriptional mRNA expression and play critical roles in cervical cancer. Single nucleotide polymorphisms (SNPs) in miRNA genes may alter miRNA expression and maturation and have been associated with various cancers. This review mainly focuses on the roles of SNPs in miRNA genes in the development of cervical cancer and summarizes the research progress of miRNA SNPs in cervical cancer and their molecular regulation mechanisms.
Keywords: cervical cancer, miRNAs, single nucleotide polymorphisms, regulation
Introduction
Cervical cancer
Cervical cancer is the fourth most diagnosed common cancer and the fourth death cause of cancer in women worldwide, which makes it a serious threat to women's life and health, especially in developing countries [1]. The occurrence and development of cervical cancer can be divided into two main stages: cervical intraepithelial neoplasia (CIN) and cervical cancer [2]. Studies have found that cervical cancer is a disease caused by a long-term interaction of multiple factors. Persistent infection with high-risk human papillomavirus (HPV) is a necessary condition for the occurrence of cervical cancer, whereas HPV infection alone is not sufficient to induce malignant transformation. Moreover, host genetic factors also play important roles in the occurrence and development of cervical cancer [3]. The persistent infection with high-risk HPV will cause HPV-DNA to integrate into the host genome, leading to increased expression of viral oncogenes E6 and E7 in cervical inflammation which is the cause of cervical cancer, at the same time, inflammation may also lead to abnormal expression of miRNA [4]. Shen et al. reported that miRNA could affect the replication of high risk HPV DNA, which may influence the life cycle of HPV and the mechanism of HPV-induced tumorigenesis [5]. Therefore, the miRNA may play a key role in high risk HPV tumorigenesis. Ellwanger et al. reported that the SNPs in miRNA might modulate immune response and viral restriction, cell cycle, proliferation and apoptosis, which may relate to HPV infection [6]. In addition, HPV's E6 and E7 oncoproteins are able to induce overexpression of DNA methyltransferase, which can cause abnormal methylation of miRNA genes and be associated with cervical cancer. Therefore, the interaction between the miRNAs and HPV genes may affect the occurrence and development of cervical cancer [4]. In recent years, studies on the role of host genetic factors in the development of cervical cancer, especially host miRNAs and their polymorphisms, have been research hot spots worldwide.
MiRNA
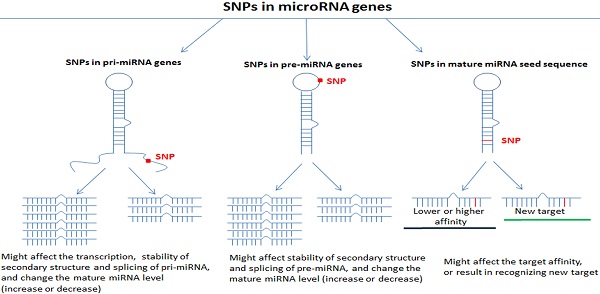
MiRNAs are a class of small, noncoding single-stranded RNA molecules that are encoded by endogenous genes; they are approximately 20-24 nucleotides in length and function as gene regulators in eukaryotes [7]. Mature miRNAs regulate the expression of target genes by binding to the 3'-UTR of the target gene through sequence complementation [8] and are involved in various cell biological processes, including cell proliferation, apoptosis, and migration [9]. MiRNAs encoding genes are transcribed to long primary transcripts (pri-miRNAs) by RNA polymerase in the cell nucleus. The pri-miRNAs are approximately 300 to 1000 nucleotides in length and are then cleaved to form miRNA precursors (pre-miRNAs) under the action of the Drosha enzyme [10]. Pre-miRNAs have stem-loop structures that are approximately 60-70 nucleotides in length, and they are transported from the nucleus to the cytoplasm by exportin-5 and processed into approximately 22-nt mature miRNA duplexes via cleavage by the Dicer enzyme [11]. Finally, mature and functional single-stranded miRNA molecules are processed and generated by helicase [12].
Single nucleotide polymorphisms (SNPs) in miRNAs
Single nucleotide polymorphisms are the most common type of human heritable variation and are caused by the mutation of a single nucleotide at the genome level in every 100-300 base pairs [13]. SNPs may occur in coding gene sequences or in noncoding regions, and approximately 90% of functional SNPs are located in noncoding regions, including promoter regions, enhancer regions and noncoding RNA sequences [14]. SNPs in miRNA genes, including pri-miRNAs, pre-miRNAs and mature miRNAs, may affect miRNA shearing, processing, and maturation processes; they play important roles in the expression level of mature miRNAs and the combination of miRNAs and their target genes and influence the development of diseases [15-17]. Many studies have reported that SNPs in miRNAs are related to different diseases, especially cancers [17-21]. SNPs in miRNA genes may affect the miRNA regulation of signaling pathways by affecting the expression of mature miRNAs or the recognition of target genes by miRNAs, thereby playing a role in the occurrence and development of diseases.
The roles of miRNAs and their SNPs in cervical cancer
A large number of miRNAs with abnormal expression in cervical cancer tissues and cells have been reported recently [22]. The abnormal expression of miRNAs in cervical cancer can affect its pathogenesis by affecting downstream signaling molecules and further affect the progression and prognosis of cervical cancer or may be related to the sensitivity of patients to radiotherapy and chemotherapy [23]. MiRNAs can affect the treatment effect and survival rate of patients and provide a reliable method for the diagnosis and effective treatment of cervical cancer [24].
These miRNAs with abnormal expression affect the expression of target genes in tumor-related signaling pathways and thus affect the regulation of signaling pathways and ultimately play a pivotal role in the occurrence and development of cervical cancer. For example, miRNAs may function as oncogenes or tumor suppressors to regulate the occurrence and development of cervical cancer through special signaling pathways, including the PI3K-AKT pathway [25], Notch pathway [26], E6-p53 pathway [27], E7-pRb pathway [28], Wnt/β-catenin pathway [29], NF-κB pathway [30], and Hedgehog pathway [31].
Studies have reported that SNPs in pri-miRNA and pre-miRNA genes may interfere with the maturation process or degradation of miRNAs by destroying the secondary structure, reducing the number of mature miRNAs and affecting the expression of mature miRNAs [32, 33]. The SNP site located in the mature miRNA may lose its combination with the mRNA target sequence or change its binding efficiency to the target sequence due to the mutation of the base site [34]. Therefore, SNPs in miRNA genes may affect the regulation of miRNAs on related signal pathways and the biological functions of cells in various ways by affecting the expression of mature miRNAs or the recognition of target genes by miRNAs [6, 35, 36]. Moreover, some miRNAs have targeting relationship with multiple genes, at the same time; a certain target gene could also bind to multiple miRNAs [37]. The regulation of complex miRNAs may have the ultimate biological effect, thereby playing a role in the occurrence and development of different kinds of diseases including cervical cancer (Table 1).
SNPs in microRNA genes involved in cervical cancer
| SNPs | MicroRNAs | Polymorphism | Consequence | Association in Cervical cancer | Populations [Case(n)/Control(n)] |
|---|---|---|---|---|---|
| rs4938723 | miR-34b/c | T>C | BTG4: Intron Variant; miR-34b/c: 2KB Upstream Variant | C allele associated with Increased risk | Chinese (328/568) [41] |
| rs11134527 | miR-218 | A>G | SLIT3: Intron Variant; miR-218-2: 2KB Upstream Variant | G allele associated with decreased risk [51, 52]; A allele associated with decreased risk [53] | Chinese (703/713) [51] Chinese (1584/1394) [52] Chinese (609/583) [53] |
| rs895819 | miR-27a | T>C | miR-27a: Non Coding Transcript Variant | T allele associated with decreased risk [55]; C allele associated with decreased risk [56] | Chinese (103/417) [55] Chinese (290/445) [56] |
| rs531564 | miR-124 | G>C | LINC0059: Non Coding Transcript Variant; miR-124-1: 500B Downstream Variant | G allele associated with decreased risk | Chinese (107/208) [60] Chinese (609/583) [53] |
| rs4636297 | miR-126 | C>T | EGFL7: Intron Variant miR-126: 500B Downstream Variant | T allele associated with increased risk | Chinese (547/567) [66] |
| rs1625579 | miR-137 | G>T | miR-137HG: Intron Variant | G allele associated with decreased risk | Chinese (290/445) [56] |
| rs1292037 | miR-21 | A>G | VMP1: 3 Prime UTR Variant; miR-21: 500B Downstream Variant | G allele associated with increased risk | Chinese (165) [70] |
| rs2292832 | miR-149 | T>C | miR-149: Non Coding Transcript Variant; GPC1: Intron Variant; LOC100130449: Intron Variant | C allele associated with increased risk | Chinese (954/1339) [75] |
| rs2910164 | miR-146a | G>C | miR-146a: Non Coding Transcript Variant; miR-3142HG: Non Coding Transcript Variant | C allele associated with increased risk | Chinese [82] |
| rs11614913 | miR-196a2 | T>C | miR-196a2: Non Coding Transcript Variant | T allele associated with decreased risk | Indian (150/150) [89] Chinese (547/567) [66] |
SNPs in pri-miRNAs and cervical cancer
MiR-34 is considered to be the first miRNA that is directly regulated by p53, and it has been reported to be dysregulated in many types of cancer [38, 39]. The miR-34 family consists of the 5p and 3p strands of miR-34a, miR-34b and miR-34c [40]. Yuan et al. reported that the SNP rs4938723 in the promoter of pri-miR-34b/c was significantly associated with cervical cancer, and the C allele was a risk factor for cervical cancer in a Chinese Han population. In addition, the CT genotype was significantly associated with the clinical staging and poor prognosis of patients [41]. Previous studies have shown that miR-34 family members can directly target p53, thereby inducing cell apoptosis, DNA repair, angiogenesis and other biological processes [27]. At the same time, p53 can regulate miR34 family members, thereby inhibiting the expression of sirtuin 1 to increase p53 activity and form a p53-miR-34 positive feedback loop [42]. Their study found that the CT and CC genotypes of rs4938723 combined with the CG and CC genotypes of TP53 Arg72Pro increased the risk of cervical cancer by 2.21 times. Studies have showed that the expression of miR-34a was downregulated in cervical cancer tissues [43] and miR-34a can bind to the 3'-UTR of Notch1 and Jagged1 and inhibit the invasiveness of cervical cancer cells by regulating the Notch pathway and its downstream matrix-degrading enzymes [26]. Therefore the two genotypes in rs4938723 and TP53 together may affect the expression of miR-34b/c and p53 and ultimately increase the risk of cervical cancer.
MiR-218 is encoded by the intron of the SLIT2 tumor suppressor gene [44], and its expression level is reduced in breast cancer [45], lung cancer [46], and gastric cancer [47]. Similar to that in other cancers, the expression level of miR-218 in cervical cancer cells and tissues is significantly lower than that in paracancerous tissues and normal tissues and can inhibit the proliferation, migration and invasion of cervical cancer cells [47]. Several studies have reported that the rs11134527 SNP in the intron of pri-miR-218 is related to hepatocellular carcinoma [48], gastric cancer [49] and breast cancer [50]. Research of Zhou and Shi et al. found that the SNP rs11134527 in miR-218 was associated with cervical cancer in a Chinese Han population, and the GG genotype can reduce the risk of cervical cancer [51, 52]. Predicting the folding of RNA by RNA-fold found that the mRNA structure of rs11134527 allele A to G has changed significantly. These findings further indicate that the SNP site may cause changes in the expression of miR-218 and affect the process of miRNA binding, and therefore is related to the susceptibility of cervical cancer [52]. Li et al. showed that the SNP rs11134527 was associated with the development of CIN in cervical cancer in a Chinese Han population, and the A allele of rs11134527 was associated with a lower risk of CIN [53]. Therefore, the SNP site located in pri-miR-218 mRNA may change its local secondary structure, resulting in changes in the expression of miR-218 and affecting the binding process of miRNA; therefore, this SNP may be related to susceptibility to cervical cancer.
SNPs in pre-miRNAs and cervical cancer
Previous study found that miR-27a was highly expressed in cervical cancer tissues and in HeLa and C33A cells [54]. Xiong et al.'s study found that the SNP rs895819 located in the promoter of pre-miR-27a has a significant association with the risk of cervical cancer in a southern Chinese Han population [55]. The T allele and CT and TT genotypes have a significant risk reduction compared with the C allele and CC genotype. Compared with the CC genotype, the TC and TT genotypes were significantly associated with a lower risk of cervical cancer in the recessive inheritance model. However, Subsequently Chen et al. reported that the SNP rs895819 was associated with cervical cancer in a dominant genetic model in a southern Chinese Han population [56]. Compared with the TT genotype, the TC and CC genotypes significantly reduce the risk of cervical cancer in a dominant inheritance model. The discrepancy between the studies of Xiong and Chen could be caused by different sample sizes and baseline characteristics [57]. Sun et al. reported that overexpression of miR-27a upregulates the expression of B4GALT3, promotes carcinogenic activity through the β1-integrin pathway, and may be a potential biomarker for cervical cancer [54]. Another similar study on miR-27a also showed that miR-27a was highly expressed in cervical cancer tissues, promoted the expression of the target gene inositol polyphosphate-1-phosphatase (INPP1), and promoted tumorigenic activity [58]. Much more research is needed to detect the potential related mechanisms and functions between rs895819 in miR-27a and cervical cancer in the future.
Mature miR-124 is processed by pre-miR-124-1, pre-miR-124-2 and pre-miR-124-3, all of which have reduced expression in a variety of tumor tissue types [59]. Xiong et al. reported that the G allele of the SNP rs531564 in the promoter of miR-124-1 was a protective factor in cervical cancer in a Chinese Han population [60]. Compared with individuals with the CC genotype, individuals with the CG and GG genotypes have a significantly lower risk of developing cervical cancer [60]. Subsequently, Li et al. showed that the C allele of rs531564 was the risk allele of cervical cancer in a Chinese Han population, and this SNP was related to the progression from CIN to cervical cancer [53]. Their research suggests that SNPs in miRNA genes may affect the expression or function of miRNAs [61]. Therefore, the SNP in miR-124 may be related to the expression of miR-124 and further related to cervical cancer. Study of Wan et al. found that miR-124 can inhibit the mimicry and cell movement of angiogenesis by targeting amotL1 [62]. Moreover, research of Shen et al. found that miR-124 negatively regulates the target lncRNA NEAT1 and affects the migration and invasion of HeLa cells, EMT and activation of the NF-κB pathway [30].
Several studies have found that miR-126 is usually underexpressed in human colorectal cancer [63], breast cancer [64] and cervical cancer [65]. Yan et al. reported that rs4636297 in miR-126, which was located 12 bp downstream of the pre-miR-126 sequence, was associated with CIN and cervical cancer in a Chinese Han population [66]. The results of the study by Yan et al. indicated that the T allele confers a higher risk of developing CIN and cervical cancer [66]. The association of the SNP rs4636297 with cervical cancer may be because the SNP is related to Drosha's recognition and cleavage of pri-miRNA [34]. Therefore, the SNP rs4636297 may affect the expression of miR-126, which affects the occurrence and development of cervical cancer. MiR-126 can inhibit the expression of MMP2 by targeting ZEB1, which inhibits the protein expression of p-JAK2 and p-STAT3 and inactivates the JAK2/STAT3 signaling pathway, which inhibits the proliferation, migration and invasion of cervical cancer cells [65]. Therefore, it will be valuable to carry out functional and associational researches to study the roles of rs4636297 in human cancers in the future for the reason that the location of this SNP might affect biogenesis.
MiR-137 is underexpressed in cervical cancer tissues and cell lines and can inhibit the development of tumors [67]. Chen et al. reported that the SNP rs1625579 in the intron of pre-miR-137 was associated with a significant reduction in the risk of cervical cancer in a Sourthern Chinese population. Compared with the TT genotype, the TG and GG genotypes had a better protective effect on cervical cancer patients [56]. They showed that the polymorphism of miR-137 rs1625579 may be positively correlated with the expression of miR-137, which in turn affects the occurrence of cervical cancer. More confirmation researches are required. Overexpression of miR-137 in cervical cancer cells can regulate the expression of downstream target genes (EZH2[67] or GEEM1[68]) to regulate signaling pathways such as TGF-β/Smad to inhibit cell proliferation and migration.
Lui et al. detected the expression of miRNAs in six types of cervical cancer cell lines and normal cervical samples, and they found that cancer samples can repeatedly show increased miR-21 expression [69]. Zhang et al. reported that there were significant differences in the alleles and genotypes of rs1292037, which is located 210 bps from the 3' end of pre-miR-21, in cervical cancer patients in the sensitive and resistant groups. The rs1292037 locus with the G allele in the miR-21 gene may increase the resistance of cervical cancer patients to cisplatin plus paclitaxel l in a Northern Chinese population [70]. Yao et al. confirmed that miR-21 was significantly overexpressed in human cervical squamous cell carcinoma tissues and cell lines and found that miR-21 regulates the proliferation, apoptosis and migration of HPV16-positive cervical squamous cells by regulating the expression of the target gene CCL20[71]. They also showed that the overexpression of miR-21 was related to lymph node metastasis and advanced disease [71]. Xu et al. reported that the overexpression of miR-21 inhibited the expression of the mRNA of the target gene PTEN in cervical cancer cell lines to promote the proliferation, migration and invasion of cervical cancer cells [72]. Du et al. detected the sensitivity of cervical cancer cells to paclitaxel and found that the inhibited expression of miR-21 can inhibit cell proliferation and colony formation by regulating the PTEN/AKT pathway and improve the PTX sensitivity of cervical cancer cells [25]. More research is needed to analyze the molecular regulation mechanism of rs1292037 on the transcription level and methylation level of miR-21 and its role in cervical cancer progression and chemoresistance, which may indicate its use in the treatment cervical cancer.
The expression of miR-149 is upregulated in cervical cancer tissues and cells [33]. A large number of studies have shown that the rs2292832 in the promoter of pre-miR-149 is related to a variety of cancers, including breast cancer [73] and lung cancer [74]. Wang et al. reported that for the polymorphic site rs2292832 of miR-149 in cervical cancer tissues and HeLa and SiHa cells, the TC and CC genotypes increased the risk of cervical cancer compared with the TT genotype [75]. We speculate that rs2292832 may affect the expression of miRNA by affecting the stability of the secondary structure of miR-149 and may further affect the regulation of miR-149 to target genes, thereby playing a role in the occurrence and development of cervical cancer.
The reason behind the association of SNPs with cervical cancer could be because SNPs located in pre-miRNAs may affect the expression of mature miRNAs and be involved in the binding of certain nuclear factors to miRNAs during miRNA processing.
SNPs in mature miRNAs and cervical cancer
MiR-146a is abnormally expressed in a variety of cancers [76-79]. Upregulating the expression of miR-146a may affect the proliferation of cancer cells [80]. The SNP rs2910164 is located in the 3p chain of mature miR-146a. Its polymorphic sites involve mismatches in the hairpin of pre-miR-146a, which leads to changes in its processing and reduces the expression of mature sequences [81], thereby affecting the expression of downstream signaling molecules. Hu et al. found that the expression of miR-146a was upregulated in HeLa cervical cancer cells and C33A cells, and the rs2910164 SNP increased the amount of mature miRNAs in cervical cancer cells. Similarly, Li et al. reported that miR-146a can act as an oncogene, affect the expression of the target gene TRAF6 and regulate the differentiation of Th17 cells through NF-κB signaling to regulate the growth and apoptosis of cervical cancer cells [82, 83]. The expression of miR-146a can downregulate the expression of receptor-related kinase 1 (IRAK1) and TNF receptor-related factor 6 (TRAF6), promote the expression of cyclin D1, and promote the occurrence of cervical cancer. It was found that after recombinant plasmid transfection, the expression of miR-146a in the C allele was higher than that in the G allele in cervical cancer cells, while the expression of miR-146a in the C allele in CRL-2615 cells was lower than that in the G allele. However, this SNP played the opposite role in immortalized nontumorigenic endocervical CRL-2615 cells [82]. Therefore, the role of this SNP site varied in different cervical cell lines.
Previous studies reported that the expression of miR-196a in human cervical cancer tissues and cell lines was significantly upregulated [84]. Many studies have found that the SNP rs11614913 located in mature miR-196a2 was associated with a variety of human cancers, including breast cancer [85], non-small cell lung cancer [86], ovarian cancer [87] and hepatocellular carcinoma [88]. Study of Nisha et al showed that the T allele in the SNP rs11614913 was associated with a reduced risk of cervical cancer development in an Indian population [89]. Similarly, the research results of Yan et al. in a Chinese Han population were consistent with those of Nisha et al. [66]. The SNP rs11614913, which is located in the mature sequence of miR-196a2, may affect the biological function of miRNA and the recognition of target mRNA, thereby affecting the occurrence and progression of cervical cancer [33]. The key role of miR-196a was in regulating cell cycle checkpoints and cervical cancer cell proliferation. Moreover, they confirmed the correlation between the proliferation of cervical cancer cells was mediated by miR-196a and the downregulation of the target gene FOXO1 p27Kip1 in the PI3K signaling pathway [84].
Therefore, the abnormal expression of miRNAs may directly or indirectly affect the expression of many target genes, and the expression levels of these target genes will also dynamically change under various cell and environmental stimulations. Combining these changes with SNP-related miRNA-level regulation can provide important internal mechanisms for disease development.
Conclusions and Future perspectives
The occurrence of tumors is the result of the abnormal regulation of signaling pathways related to cell proliferation, differentiation, apoptosis, and the cell cycle. Mutations in the genes that affect different cell signaling pathways could play important roles in the mechanisms of the occurrence and development of cervical cancer. Many studies have found that mutations in miRNA genes may affect the combination of mature miRNAs and downstream target genes by affecting miRNA expression, thereby affecting the regulation of signaling pathways and playing an important role in the occurrence and development of cervical cancer. This review summarizes the SNP sites of miRNAs related to cervical cancer, lays a foundation for in-depth research on the pathogenesis of cervical cancer and identifies new targets for the diagnosis and treatment of cervical cancer.
Acknowledgements
Funding
This work was supported by grants from the Special Funds for High-level Healthy Talents of Yunnan Province (L-201615 and H-2018014), Yunnan Provincial Science and Technology Department (2019HC0060). The funders had no role in study design, data collection and analysis, decision to publish or preparation of the manuscript.
Competing Interests
The authors have declared that no competing interest exists.
References
1. Sung H, Ferlay J, Siegel RL, Laversanne M, Soerjomataram I, Jemal A. et al. Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2021
2. Siegler E, Shiner M, Segev Y, Mackuli L, Lahat N, Lavie O. Prevalence and Genotype Distribution of HPV Types in Women at Risk for Cervical Neoplasia in Israel. The Israel Medical Association journal: IMAJ. 2017;19:635-9
3. de Freitas AC, Gurgel AP, Chagas BS, Coimbra EC, do Amaral CM. Susceptibility to cervical cancer: an overview. Gynecologic oncology. 2012;126:304-11
4. Sadri Nahand J, Moghoofei M, Salmaninejad A, Bahmanpour Z, Karimzadeh M, Nasiri M. et al. Pathogenic role of exosomes and microRNAs in HPV-mediated inflammation and cervical cancer: A review. International journal of cancer. 2020;146:305-20
5. Shen S, Zhang S, Liu P, Wang J, Du H. Potential role of microRNAs in the treatment and diagnosis of cervical cancer. Cancer genetics. 2020;248-249:25-30
6. Ellwanger JH, Zambra FMB, Guimarães RL, Chies JAB. MicroRNA-Related Polymorphisms in Infectious Diseases-Tiny Changes With a Huge Impact on Viral Infections and Potential Clinical Applications. Frontiers in immunology. 2018;9:1316
7. Bartel DP. Metazoan MicroRNAs. Cell. 2018;173:20-51
8. Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281-97
9. Ambros V. MicroRNA pathways in flies and worms: growth, death, fat, stress, and timing. Cell. 2003;113:673-6
10. Schanen BC, Li X. Transcriptional regulation of mammalian miRNA genes. Genomics. 2011;97:1-6
11. Loibl N, Arenz C, Seitz O. Monitoring Dicer-Mediated miRNA-21 Maturation and Ago2 Loading by a Dual-Colour FIT PNA Probe Set. Chembiochem: a European journal of chemical biology. 2020;21:2527-32
12. Lund E, Güttinger S, Calado A, Dahlberg JE, Kutay U. Nuclear export of microRNA precursors. Science (New York, NY). 2004;303:95-8
13. Abd El-Fattah AA, Sadik NAH, Shaker OG, Mohamed Kamal A. Single Nucleotide Polymorphism in SMAD7 and CHI3L1 and Colorectal Cancer Risk. Mediators of inflammation. 2018;2018:9853192
14. Tak YG, Farnham PJ. Making sense of GWAS: using epigenomics and genome engineering to understand the functional relevance of SNPs in non-coding regions of the human genome. Epigenetics & chromatin. 2015;8:57
15. Srivastava K, Srivastava A. Comprehensive review of genetic association studies and meta-analyses on miRNA polymorphisms and cancer risk. PloS one. 2012;7:e50966
16. Link A, Kupcinskas J, Wex T, Malfertheiner P. Macro-role of microRNA in gastric cancer. Digestive diseases (Basel, Switzerland). 2012;30:255-67
17. Mir R, Al Balawi IA, Duhier FMA. Involvement of microRNA-423 Gene Variability in Breast Cancer Progression in Saudi Arabia. Asian Pacific journal of cancer prevention: APJCP. 2018;19:2581-9
18. Moazeni-Roodi A, Hashemi M. Association between miR-124-1 rs531564 polymorphism and risk of cancer: An updated meta-analysis of case-control studies. EXCLI journal. 2018;17:608-19
19. Bodal VK, Sangwan S, Bal MS, Kaur M, Sharma S, Kaur B. Association between Microrna 146a and Microrna 196a2 Genes Polymorphism and Breast Cancer Risk in North Indian Women. Asian Pacific journal of cancer prevention: APJCP. 2017;18:2345-8
20. Latini A, Ciccacci C, Novelli G, Borgiani P. Polymorphisms in miRNA genes and their involvement in autoimmune diseases susceptibility. Immunologic research. 2017;65:811-27
21. Srivastava K, Tyagi K. Single nucleotide polymorphisms of microRNA in cardiovascular diseases. Clinica chimica acta; international journal of clinical chemistry. 2018;478:101-10
22. Gao C, Zhou C, Zhuang J, Liu L, Liu C, Li H. et al. MicroRNA expression in cervical cancer: Novel diagnostic and prognostic biomarkers. Journal of cellular biochemistry. 2018;119:7080-90
23. Magee P, Shi L, Garofalo M. Role of microRNAs in chemoresistance. Annals of translational medicine. 2015;3:332
24. Banno K, Iida M, Yanokura M, Kisu I, Iwata T, Tominaga E. et al. MicroRNA in cervical cancer: OncomiRs and tumor suppressor miRs in diagnosis and treatment. TheScientificWorldJournal. 2014;2014:178075
25. Du G, Cao D, Meng L. miR-21 inhibitor suppresses cell proliferation and colony formation through regulating the PTEN/AKT pathway and improves paclitaxel sensitivity in cervical cancer cells. Molecular medicine reports. 2017;15:2713-9
26. Pang RT, Leung CO, Ye TM, Liu W, Chiu PC, Lam KK. et al. MicroRNA-34a suppresses invasion through downregulation of Notch1 and Jagged1 in cervical carcinoma and choriocarcinoma cells. Carcinogenesis. 2010;31:1037-44
27. Zhang S, Qian J, Cao Q, Li P, Wang M, Wang J. et al. A potentially functional polymorphism in the promoter region of miR-34b/c is associated with renal cell cancer risk in a Chinese population. Mutagenesis. 2014;29:149-54
28. Zhao W, Liu Y, Zhang L, Ding L, Li Y, Zhang H. et al. MicroRNA-154-5p regulates the HPV16 E7-pRb pathway in Cervical Carcinogenesis by targeting CUL2. Journal of Cancer. 2020;11:5379-89
29. Song T, Hou X, Lin B. MicroRNA-758 inhibits cervical cancer cell proliferation and metastasis by targeting HMGB3 through the WNT/β-catenin signaling pathway. Oncology letters. 2019;18:1786-92
30. Shen X, Zhao W, Zhang Y, Liang B. Long Non-Coding RNA-NEAT1 Promotes Cell Migration and Invasion via Regulating miR-124/NF-κB Pathway in Cervical Cancer. OncoTargets and therapy. 2020;13:3265-76
31. Wen SY, Lin Y, Yu YQ, Cao SJ, Zhang R, Yang XM. et al. miR-506 acts as a tumor suppressor by directly targeting the hedgehog pathway transcription factor Gli3 in human cervical cancer. Oncogene. 2015;34:717-25
32. Hrovatin K, Kunej T. Classification of miRNA-related sequence variations. Epigenomics. 2018;10:463-81
33. Króliczewski J, Sobolewska A, Lejnowski D, Collawn JF, Bartoszewski R. microRNA single polynucleotide polymorphism influences on microRNA biogenesis and mRNA target specificity. Gene. 2018;640:66-72
34. Ha M, Kim VN. Regulation of microRNA biogenesis. Nature reviews Molecular cell biology. 2014;15:509-24
35. Cai Y, Yu X, Hu S, Yu J. A brief review on the mechanisms of miRNA regulation. Genomics, proteomics & bioinformatics. 2009;7:147-54
36. Duan S, Mi S, Zhang W, Dolan ME. Comprehensive analysis of the impact of SNPs and CNVs on human microRNAs and their regulatory genes. RNA biology. 2009;6:412-25
37. Drury RE, O'Connor D, Pollard AJ. The Clinical Application of MicroRNAs in Infectious Disease. Frontiers in immunology. 2017;8:1182
38. Hermeking H. MicroRNAs in the p53 network: micromanagement of tumour suppression. Nature reviews Cancer. 2012;12:613-26
39. Revathidevi S, Manikandan M, Rao AK, Vinothkumar V, Arunkumar G, Rajkumar KS. et al. Analysis of APOBEC3A/3B germline deletion polymorphism in breast, cervical and oral cancers from South India and its impact on miRNA regulation. Tumour biology: the journal of the International Society for Oncodevelopmental Biology and Medicine. 2016;37:11983-90
40. Córdova-Rivas S, Fraire-Soto I, Mercado-Casas Torres A, Servín-González LS, Granados-López AJ, López-Hernández Y. et al. 5p and 3p Strands of miR-34 Family Members Have Differential Effects in Cell Proliferation, Migration, and Invasion in Cervical Cancer Cells. International journal of molecular sciences. 2019 20
41. Yuan F, Sun R, Chen P, Liang Y, Ni S, Quan Y. et al. Combined analysis of pri-miR-34b/c rs4938723 and TP53 Arg72Pro with cervical cancer risk. Tumour biology: the journal of the International Society for Oncodevelopmental Biology and Medicine. 2016;37:6267-73
42. Okada N, Lin CP, Ribeiro MC, Biton A, Lai G, He X. et al. A positive feedback between p53 and miR-34 miRNAs mediates tumor suppression. Genes & development. 2014;28:438-50
43. Chen AH, Qin YE, Tang WF, Tao J, Song HM, Zuo M. MiR-34a and miR-206 act as novel prognostic and therapy biomarkers in cervical cancer. Cancer cell international. 2017;17:63
44. Griffiths-Jones S, Grocock RJ, van Dongen S, Bateman A, Enright AJ. miRBase: microRNA sequences, targets and gene nomenclature. Nucleic acids research. 2006;34:D140-4
45. Liu B, Tian Y, Li F, Zhao Z, Jiang X, Zhai C. et al. Tumor-suppressing roles of miR-214 and miR-218 in breast cancer. Oncology reports. 2016;35:3178-84
46. Yanaihara N, Caplen N, Bowman E, Seike M, Kumamoto K, Yi M. et al. Unique microRNA molecular profiles in lung cancer diagnosis and prognosis. Cancer cell. 2006;9:189-98
47. Tang S, Wang D, Zhang Q, Li L. miR-218 suppresses gastric cancer cell proliferation and invasion via regulation of angiopoietin-2. Experimental and therapeutic medicine. 2016;12:3837-42
48. Zhang LS, Liang WB, Gao LB, Li HY, Li LJ, Chen PY. et al. Association between pri-miR-218 polymorphism and risk of hepatocellular carcinoma in a Han Chinese population. DNA and cell biology. 2012;31:761-5
49. Wu Y, Jia Z, Cao D, Wang C, Wu X, You L. et al. Predictive Value of MiR-219-1, MiR-938, MiR-34b/c, and MiR-218 Polymorphisms for Gastric Cancer Susceptibility and Prognosis. Disease markers. 2017;2017:4731891
50. Danesh H, Hashemi M, Bizhani F, Hashemi SM, Bahari G. Association study of miR-100, miR-124-1, miR-218-2, miR-301b, miR-605, and miR-4293 polymorphisms and the risk of breast cancer in a sample of Iranian population. Gene. 2018;647:73-8
51. Zhou X, Chen X, Hu L, Han S, Qiang F, Wu Y. et al. Polymorphisms involved in the miR-218-LAMB3 pathway and susceptibility of cervical cancer, a case-control study in Chinese women. Gynecologic oncology. 2010;117:287-90
52. Shi TY, Chen XJ, Zhu ML, Wang MY, He J, Yu KD. et al. A pri-miR-218 variant and risk of cervical carcinoma in Chinese women. BMC cancer. 2013;13:19
53. Chuanyin L, Xiaona W, Zhiling Y, Yu Z, Shuyuan L, Jie Y. et al. The association between polymorphisms in microRNA genes and cervical cancer in a Chinese Han population. Oncotarget. 2017;8:87914-27
54. Sun Y, Yang X, Liu M, Tang H. B4GALT3 up-regulation by miR-27a contributes to the oncogenic activity in human cervical cancer cells. Cancer letters. 2016;375:284-92
55. Xiong XD, Luo XP, Cheng J, Liu X, Li EM, Zeng LQ. A genetic variant in pre-miR-27a is associated with a reduced cervical cancer risk in southern Chinese women. Gynecologic oncology. 2014;132:450-4
56. Chen G, Zhang M, Zhu J, Chen F, Yu D, Zhang A. et al. Common genetic variants in pre-microRNAs are associated with cervical cancer susceptibility in southern Chinese women. Journal of Cancer. 2020;11:2133-8
57. Weng Y, Wang B, Zheng L. Associated of rs895819 with risk of cervical cancer in Chinese women. Journal of Cancer. 2020;11:6286-7
58. Li P, Zhang Q, Tang H. INPP1 up-regulation by miR-27a contributes to the growth, migration and invasion of human cervical cancer. Journal of cellular and molecular medicine. 2019;23:7709-16
59. Zhang J, Huang X, Xiao J, Yang Y, Zhou Y, Wang X. et al. Pri-miR-124 rs531564 and pri-miR-34b/c rs4938723 polymorphisms are associated with decreased risk of esophageal squamous cell carcinoma in Chinese populations. PloS one. 2014;9:e100055
60. Xiong X, Cheng J, Liu X, Tang S, Luo X. [Correlation analysis between miR-124 rs531564 polymorphisms and susceptibility to cervical cancer]. Nan fang yi ke da xue xue bao = Journal of Southern Medical University. 2014;34:210-3
61. Qi L, Hu Y, Zhan Y, Wang J, Wang BB, Xia HF. et al. A SNP site in pri-miR-124 changes mature miR-124 expression but no contribution to Alzheimer's disease in a Mongolian population. Neurosci Lett. 2012;515:1-6
62. Wan HY, Li QQ, Zhang Y, Tian W, Li YN, Liu M. et al. MiR-124 represses vasculogenic mimicry and cell motility by targeting amotL1 in cervical cancer cells. Cancer letters. 2014;355:148-58
63. Ebrahimi F, Gopalan V, Wahab R, Lu CT, Smith RA, Lam AK. Deregulation of miR-126 expression in colorectal cancer pathogenesis and its clinical significance. Experimental cell research. 2015;339:333-41
64. Wang CZ, Yuan P, Li Y. MiR-126 regulated breast cancer cell invasion by targeting ADAM9. International journal of clinical and experimental pathology. 2015;8:6547-53
65. Xu J, Wang H, Wang H, Chen Q, Zhang L, Song C. et al. The inhibition of miR-126 in cell migration and invasion of cervical cancer through regulating ZEB1. Hereditas. 2019;156:11
66. Yan Z, Zhou Z, Li C, Yang X, Yang L, Dai S. et al. Polymorphisms in miRNA genes play roles in the initiation and development of cervical cancer. J Cancer. 2019;10:4747-53
67. Zhang H, Yan T, Liu Z, Wang J, Lu Y, Li D. et al. MicroRNA-137 is negatively associated with clinical outcome and regulates tumor development through EZH2 in cervical cancer. Journal of cellular biochemistry. 2018;119:938-47
68. Miao H, Wang N, Shi LX, Wang Z, Song WB. Overexpression of mircoRNA-137 inhibits cervical cancer cell invasion, migration and epithelial-mesenchymal transition by suppressing the TGF-β/smad pathway via binding to GREM1. Cancer cell international. 2019;19:147
69. Lui WO, Pourmand N, Patterson BK, Fire A. Patterns of known and novel small RNAs in human cervical cancer. Cancer research. 2007;67:6031-43
70. Zhang J, Li YH, Liu HL, Zhang Y, Zhang QS, Li SZ. Correlations of MicroRNA-21 Gene Polymorphisms With Chemosensitivity and Prognosis of Cervical Cancer. The American journal of the medical sciences. 2018;356:544-51
71. Yao T, Lin Z. MiR-21 is involved in cervical squamous cell tumorigenesis and regulates CCL20. Biochimica et biophysica acta. 2012;1822:248-60
72. Xu J, Zhang W, Lv Q, Zhu D. Overexpression of miR-21 promotes the proliferation and migration of cervical cancer cells via the inhibition of PTEN. Oncology reports. 2015;33:3108-16
73. Hu Z, Liang J, Wang Z, Tian T, Zhou X, Chen J. et al. Common genetic variants in pre-microRNAs were associated with increased risk of breast cancer in Chinese women. Human mutation. 2009;30:79-84
74. Vinci S, Gelmini S, Pratesi N, Conti S, Malentacchi F, Simi L. et al. Genetic variants in miR-146a, miR-149, miR-196a2, miR-499 and their influence on relative expression in lung cancers. Clinical chemistry and laboratory medicine. 2011;49:2073-80
75. Wang S, Zhu H, Ding B, Feng X, Zhao W, Cui M. et al. Genetic variants in microRNAs are associated with cervical cancer risk. Mutagenesis. 2019;34:127-33
76. Philippidou D, Schmitt M, Moser D, Margue C, Nazarov PV, Muller A. et al. Signatures of microRNAs and selected microRNA target genes in human melanoma. Cancer research. 2010;70:4163-73
77. Zavala V, Pérez-Moreno E, Tapia T, Camus M, Carvallo P. miR-146a and miR-638 in BRCA1-deficient triple negative breast cancer tumors, as potential biomarkers for improved overall survival. Cancer biomarkers: section A of Disease markers. 2016;16:99-107
78. Wang X, Tang S, Le SY, Lu R, Rader JS, Meyers C. et al. Aberrant expression of oncogenic and tumor-suppressive microRNAs in cervical cancer is required for cancer cell growth. PloS one. 2008;3:e2557
79. Shomali N, Mansoori B, Mohammadi A, Shirafkan N, Ghasabi M, Baradaran B. MiR-146a functions as a small silent player in gastric cancer. Biomedicine & pharmacotherapy = Biomedecine & pharmacotherapie. 2017;96:238-45
80. Chen G, Umelo IA, Lv S, Teugels E, Fostier K, Kronenberger P. et al. miR-146a inhibits cell growth, cell migration and induces apoptosis in non-small cell lung cancer cells. PloS one. 2013;8:e60317
81. Jazdzewski K, Murray EL, Franssila K, Jarzab B, Schoenberg DR, de la Chapelle A. Common SNP in pre-miR-146a decreases mature miR expression and predisposes to papillary thyroid carcinoma. Proc Natl Acad Sci U S A. 2008;105:7269-74
82. Hu Q, Song J, Ding B, Cui Y, Liang J, Han S. miR-146a promotes cervical cancer cell viability via targeting IRAK1 and TRAF6. Oncology reports. 2018;39:3015-24
83. Li T, Li M, Xu C, Xu X, Ding J, Cheng L. et al. miR-146a regulates the function of Th17 cell differentiation to modulate cervical cancer cell growth and apoptosis through NF-κB signaling by targeting TRAF6. Oncology reports. 2019;41:2897-908
84. Hou T, Ou J, Zhao X, Huang X, Huang Y, Zhang Y. MicroRNA-196a promotes cervical cancer proliferation through the regulation of FOXO1 and p27Kip1. British journal of cancer. 2014;110:1260-8
85. Catucci I, Yang R, Verderio P, Pizzamiglio S, Heesen L, Hemminki K. et al. Evaluation of SNPs in miR-146a, miR196a2 and miR-499 as low-penetrance alleles in German and Italian familial breast cancer cases. Human mutation. 2010;31:E1052-7
86. Hong YS, Kang HJ, Kwak JY, Park BL, You CH, Kim YM. et al. Association between microRNA196a2 rs11614913 genotypes and the risk of non-small cell lung cancer in Korean population. Journal of preventive medicine and public health = Yebang Uihakhoe chi. 2011;44:125-30
87. Song ZS, Wu Y, Zhao HG, Liu CX, Cai HY, Guo BZ. et al. Association between the rs11614913 variant of miRNA-196a-2 and the risk of epithelial ovarian cancer. Oncology letters. 2016;11:194-200
88. Qi P, Dou TH, Geng L, Zhou FG, Gu X, Wang H. et al. Association of a variant in MIR 196A2 with susceptibility to hepatocellular carcinoma in male Chinese patients with chronic hepatitis B virus infection. Human immunology. 2010;71:621-6
89. Thakur N, Singhal P, Mehrotra R, Bharadwaj M. Impacts of single nucleotide polymorphisms in three microRNAs (miR-146a, miR-196a2 and miR-499) on the susceptibility to cervical cancer among Indian women. Bioscience reports. 2019 39
Author contact
![]() Corresponding authors: Prof. Li Shi, Department of Immunogenetics, Institute of Medical Biology, Chinese Academy of Medical Sciences & Peking Union Medical College, Kunming 650118, Yunnan, China. E-mail: shili.imbcom; Dr. Yufeng Yao, Department of Immunogenetics, Institute of Medical Biology, Chinese Academy of Medical Sciences & Peking Union Medical College, Kunming 650118, Yunnan, China. E-mail: leoyyfcom or yufeng_yaocom.cn.
Corresponding authors: Prof. Li Shi, Department of Immunogenetics, Institute of Medical Biology, Chinese Academy of Medical Sciences & Peking Union Medical College, Kunming 650118, Yunnan, China. E-mail: shili.imbcom; Dr. Yufeng Yao, Department of Immunogenetics, Institute of Medical Biology, Chinese Academy of Medical Sciences & Peking Union Medical College, Kunming 650118, Yunnan, China. E-mail: leoyyfcom or yufeng_yaocom.cn.

 Global reach, higher impact
Global reach, higher impact