3.2
Impact Factor
ISSN: 1449-1907
Int J Med Sci 2021; 18(11):2339-2346. doi:10.7150/ijms.57886 This issue Cite
Research Paper
Impact of pentraxin 3 genetic variants on uterine cervical cancer clinicopathologic characteristics
1. Institute of Medicine, Chung Shan Medical University, Taichung, Taiwan.
2. Department of Obstetrics and Gynecology, Chi-Mei Foundation Medical Center, Tainan, Taiwan.
3. Department of Medical Imaging and Radiological Sciences, Chung Shan Medical University, Taichung, Taiwan.
4. Department of Radiation Oncology, Chung Shan Medical University Hospital, Taichung, Taiwan.
5. Department of Medicine, National Taiwan University, Taipei, Taiwan.
6. School of Medicine, Chung Shan Medical University, Taichung, Taiwan.
7. Department of Obstetrics and Gynecology, Changhua Christian Hospital, Changhua, Taiwan.
8. Department of Obstetrics and Gynecology, Chiayi Chang Gung Memorial Hospital Chiayi, Taiwan.
9. Department of Nursing, Chang Gung University of Science and Technology, Chiayi Campus, Chiayi, Taiwan.
10. Department of Medical Research, Chung Shan Medical University Hospital, Taichung, Taiwan.
11. Department of Obstetrics and Gynecology, Chung Shan Medical University Hospital, Taichung, Taiwan.
#Equal contribution as first authors.
Received 2021-1-6; Accepted 2021-3-24; Published 2021-4-7
Abstract
The aims of this study were to investigate the relationships among pentraxin 3 (PTX3) genetic variants and development and clinicopathological characteristics of uterine cervical cancer, and patient survival in Taiwanese women. The study enrolled 125 patients with invasive cancer and 98 patients with precancerous lesions of uterine cervix, and 325 control women. PTX3 genetic variants rs2120243, rs3816527, rs2305619 and rs1840680 were selected and their genotypic distributions were determined by real-time polymerase chain reaction. Our results indicated that patients with genotype CC in PTX3 rs2120243 and genotype GG in rs1840680 had more chance to have adenocarcinoma but not squamous cell carcinoma, as compared to those with CA/AA and those with GA/AA, respectively. No other clinicopatholgical characteristics were associated with PTX3 genetic variants. In addition, PTX3 genetic variants were not associated with 5 years survival of cervical cancer patients. In conclusions, PTX3 genetic variants are not associated with carcinogenesis and clinicopathological variables of uterine cervix and patient survival in Taiwanese women. The only independent predictor for the 5 years survival is pelvic lymph node metastasis.
Keywords: pentraxin 3, genetic variants, uterine cervical cancer, pelvic lymph node metastasis
Introduction
Pentraxin 3 (PTX3) is the first member of the pentraxin superfamily of multifunctional and multimeric proteins, and originally found as a long-chain pentameric protein of 40-50 kilodaltons [1-3]. It could be produced and released by a variety of cells such as neutrophils, macrophages and fibroblasts [4,5]. PTX3 is recognized to be an acute-phase reactive protein because its levels in plasma are low under normal conditions, but elevated rapidly during sepsis and other inflammatory situation and infection [6,7]. In addition to the regulation in inflammation, PTX3 also exhibits biological reactions in cancer and tissue repair [8].
When the shared sequence of a gene exhibits the difference in the single nucleotides between the members of a species or paired chromosomes in more than 1% of certain population, single-nucleotide polymorphisms (SNPs) will appear [9]. SNPs may influence the gene expression by exerting an impact on the promoter area, exon and 3'-untranslated region, and then genetic susceptibility is involved and affects the development of diseases and cancers such as oral cancer and oral submucous fibrosis [9-12]. There are 22 SNPs spaning the PTX3 gene on chromosome 3. Two of them are situated at intronic regions of the gene (rs2305619 at intron 1, and rs1840680 at intron 2), and one (rs3816527 at exon 2) presenting as a non-synonymous variant, which leads to a single amino acid alanine to aspartic acid change. They may be concerned with the susceptibility to various infections and cancer-related infection [13-15].
Uterine cervical cancer is the fourth most common cancer among women globally, accounting for approximately 580,000 new diagnoses in 2018 [16]. Approximately 90% of deaths from cervical cancer occur in low- and middle-income countries. The worldwide annual incidence of uterine cervical cancer was reported to be 14.0 per 100,000 populations [17]. Uterine cervical cancer ranked the eighth most common malignancy in female individuals based on 2014 Annual Cancer Registry Report in Taiwan. Cervical carcinogenesis is a continuous multistep process. Cervical intraepithelial neoplasias (CINs) can remain persistent progression into invasive cancer.18 Cervical intraepithelial neoplasia 1 (CIN 1) is called low-grade squamous intraepithelial lesion in cytology (LSIL; also referred to low-grade CIN or low-grade dysplasia in histology), in which mitotic and immature cells occupy only the lower third of the epithelium; whereas CIN 2 and CIN 3 are denominated (collectively referred to as high-grade squamous intraepithelial lesion, HSIL in cytology; also known as high-grade CIN or high-grade dysplasia in histology) when mitotic and immature cells respectively exist in the middle and upper thirds, according to the Bethesda system [19,20]. Approximately 90% of CIN 1 was estimated to regress to normal. In contrast, high-grade CIN progress to invasive cancer in considerable rate [21], and is therefore regarded as precancerous lesions.
Pentraxin 3 single nucleotide polymorphisms (rs2305619 and rs1840680) and plasma levels have been reported to be correlated with hepatocellular carcinoma [22]. The implication of four PTX genetic variants (rs1840680, rs2305619, rs3816527, and rs2120243) has also been investigated and revealed that rs3816527 in smokers is associated with the development of late-stage cancer and increased lymph node metastasis [23]. So far, there have been no studies linking PTX3 genetic variants to cervical cancer tin Taiwanese women. We inferred that different PTX3 SNPs exert different effects on genes expression and their protein, and therefore exhibit different susceptibilities to the development of cervical neoplasias. Then, we conducted this study to explore the relationships between PTX3 genetic variants and the development of cervical cancer and patient survival.
Materials and methods
Subjects
The design of this retrospective study was to enroll 125 patients with invasive cancer and 98 women with precancerous lesions (high-grade CIN) from the Department of Obstetrics and Gynecology affiliated to Chung Shan Medical University Hospital in Taichung, Taiwan, from February 1994 to February 2015. At the same time, 325 women without previous cervical neoplasias who received routine examinations in the outpatient department of the hospital were transferred to the control group. The control women were included as having no cervical neoplasias based on the normal cytologic report from cervical Papanicolaou smear and the report was further verified by normal colposcopic results in this general examination. All subjects are Taiwanese women living in central Taiwan. The marital status and education level were compatible in patients and controls. Colposcopy-directed cervical biopsy was done and pathological report was obtained to diagnose the patients with invasive cervical cancer and precancerous lesions. Patients with invasive cervical cancer and precancerous lesions are regarded as patients with cervical neoplasia. These patients underwent the standard treatment protocols, which has been revised in accordance with guidelines provided by National Comprehensive Cancer Network. The Institutional Review Board of the Affiliated Hospital of Chung Shan Medical University approved this study (CSMUH number: CS18208) with informed consents.
Extract deoxyribonucleic acid (DNA) from the blood samples of all female individuals and select PTX3 genetic variants
Laboratory staff used venipuncture techniques to draw blood samples from all female individuals. The samples were collected into Vacutainer tubes mingled with ethylenediaminetetraacetic acid, and immediately stored at 4 °C. DNA was extracted from white blood cells according to previous study [24]. The obtained DNA was then dissolved in pH 7.8 TE buffer. After that, it was quantified by the measurement of OD260. The OD260/OD280 ratio was checked and the range of 1.8-2.0 met our criteria and defined as pure to prevent its cross reactivity from the current homologous RNA in the samples. The final products were then stored at -20 °C and used as templates for the polymerase chain reaction (PCR).
Based on the data of the international HapMap project and the previous investigation of Yeh et al., four genetic variants of PTX3 were selected [22,23]. PTX3 genetic variant rs2120243 is located at transcription factor binding site on chromosome 3. rs3816527 is at exon 2 on chromosome 3 and is a non-synonymous variant in which amino acid is changed from alanine to aspartic acid. rs2305619 and rs1840680 are intron variants on chromosome 3. PTX3 SNPs rs2120243, rs3816527, rs2305619 and rs1840680 were detected by ABI StepOne Real-Time PCR System (Applied Biosystems, Foster City, CA, USA), and determined with SDS vers. 3.0 software, as our previous study [24].
Statistical analysis
The genotypic distribution of PTX3 genetic variants rs2120243 reached Hardy-Weinberg equilibrium in the control women [χ2 value: 2.07, p=0.356, degree of freedom (d.f.)=2]. The genotypic frequencies of other PTX3 genetic variants rs3816527, rs2305619 and rs1840680 all satisfied to the equilibrium (χ2 value: 0.39, p=0.821, d.f.=2; χ2 value: 1.00, p=0.607, d.f.=2; χ2 value: 1.95, p=0.377, d.f.=2; respectively).
Analysis of variance (ANOVA) was performed to compare the age distribution of the studied subjects and Bonferroni test for post hoc analysis. Chi-square test was applied to relate genotypic distributions of PTX3 genetic variants to the incidence of cervical neoplasias. Because the age of patients suffering from cervical precancerous lesions is earlier than that of cervical invasive cancer, age must be adjusted. The p values were determined by chi-square test, logistic or multinomial logistic regression models even for age adjustment. The adjusted odds ratios (AORs) with their 95% confidence intervals (CIs) were used to define the associations among genotypic distributions of PTX3 genetic variants and the incidence of cervical neoplasias (including precancerous lesions and invasive cancer) using the logistic and multinomial logistic regression models after controlling for age. Chi-square test was applied to associate PTX3 genetic variants with clinicopathological factors. The Kaplan-Meier curve model (univariate analysis over time) was used to determine the significances of PTX3 genetic variants and clinicopathological parameters for patient survival, which were significantly related to 5 years survival of patients with invasive cervical cancer. The log-rank test was applied to discriminate the differences among them. The influences of PTX3 genetic variants and these clinicopathological characteristics on 5 years survival of these patients were dertimined using the Cox proportional hazard model for multivariate analysis in relation to survival time. The hazard ratios (HRs) were therefore calculated. The SPSS, version 18.0 and WinPepi Software, version 10.0 were performed for statistical analysis. P <0.05 was known as statistically significant difference.
Results
The age distribution of cervical neoplasm patients was significantly different from that of control women (50.9 ± 13.8 vs. 44.0 ± 10.0, p<0.001). The results by the ANOVA using Bonferroni test for post hoc analysis revealed that the age distribution between cervical cancer patients and control women was statistically different (56.2 ± 12.4 vs. 44.0 ± 10.0, p<0.001), and the age distribution between patients with cervical cancer and those with precancerous lesion was also different (56.2 ± 12.4 vs. 44.2 ± 12.5, p<0.001). However, the age distribution between patients with precancerous and control women was not statistically different (44.2 ± 12.5 vs. 44.9 ± 10.0, p= 1.000).
There was no significant difference for the genetic distribution C/C, C/A and A/A in PTX3 genetic variant rs2120243 (p=0.516) between patients with cervical neoplasias and control women. The distributions of other PTX3 genetic variants, rs3816527, rs2305619 and rs1840680 showed no significant difference between patients with cervical neoplasias and control women (p=0.480, 0.956 and 0.320, respectively; Table 1). After controlling for age, there were still no significant difference in PTX3 genetic variants between these patients and control women (p=0.511, 0.749, 0.956 and 0.342, respectively; Table 1).
After patients with cervical neoplasias group was classified into subgroups of patients with invasive cancer and those with precancerous lesions, there was still no significant difference for the genetic distribution C/C, C/A and A/A in PTX3 genetic variant rs2120243 among invasive cancer, precancerous and control subgroups (p=0.746; Table 2). It also showed no significant difference in the genotypic distributions of other PTX3 genetic variants, rs3816527, rs2305619 and rs1840680 (p=0.228, 0.997 and 0.506, respectively; Table 2). The risk of invasive cancer and precancerous lesion of uterine cervix did not rise for these PTX3 genetic variants (Table 2).
Compared with CA/AA, patients with cervical invasive carcinoma with genotype CC in PTX3 rs2120243 had a higher chance of developing adenocarcinoma, but no squamous cell carcinoma (OR for adenocarcinoma: 0.21, 95% CI: 0.05-0.78, CA/AA vs CC; Table 3). There were no relationships of PTX3 rs2120243 with other clinicopathological parameters. Patients with genotype GG in PTX3 rs1840680 also had a higher chance to have adenocarcinoma as compared to those with GA/AA (OR for adenocarcinoma: 0.23, 95% CI: 0.06-0.88, GA/AA vs GG). There were still no relationships of PTX3 rs1840680 with other clinicopathological parameters. Moreover, there were no significant relationships of other PTX3 genetic variants, rs3816527 and rs2305619, with clinicopathological variables.
Genetic variant distributions of pentraxin 3 in women with cervical neoplasias and control women in Taiwan
| Genetic variants | Controls (n =325) | Cervical neoplasiasa (n=223) | ORs (95% CIs) | p values | AORs (95% CIs)b | Adjusted p valuesb |
|---|---|---|---|---|---|---|
| rs2120243 | 0.516 | 0.511 | ||||
| C/Cc | 123 | 95 | 1.00 | 1.00 | ||
| C/A | 140 | 92 | 0.85 (0.59-1.24) | 0.399 | 0.82 (0.55-1.21) | 0.315 |
| A/A | 28 | 25 | 1.16 (0.63-2.11) | 0.637 | 1.08 (0.57-2.02) | 0.819 |
| C/Cc | 123 | 95 | 1.00 | 0.570 | 1.00 | 0.435 |
| C/A & A/A | 168 | 117 | 0.90 (0.63-1.29) | 0.86 (0.59-1.25) | ||
| C/C & C/Ac | 263 | 187 | 1.00 | 0.434 | 1.00 | 0.562 |
| A/A | 28 | 25 | 1.26 (0.71-2.22) | 1.19 (0.66-2.17) | ||
| rs3816527 | 0.480 | 0.749 | ||||
| A/Ac | 186 | 143 | 1.00 | 1.00 | ||
| A/C | 101 | 67 | 0.86 (0.59-1.26) | 0.444 | 0.88 (0.59-1.31) | 0.526 |
| C/C | 11 | 12 | 1.42 (0.61-3.31) | 0.418 | 1.16 (0.48-2.82) | 0.747 |
| A/Ac | 186 | 143 | 1.00 | 0.640 | 1.00 | 0.627 |
| A/C & C/C | 112 | 79 | 0.92 (0.64-1.32) | 0.91 (0.62-1.33) | ||
| A/A & A/Cc | 287 | 210 | 1.00 | 0.347 | 1.00 | 0.674 |
| C/C | 11 | 12 | 1.49 (0.65-3.44) | 1.21 (0.50-2.91) | ||
| rs2305619 | 0.956 | 0.956 | ||||
| G/Gc | 130 | 95 | 1.00 | 1.00 | ||
| G/A | 149 | 103 | 0.95 (0.66-1.36) | 0.765 | 0.95 (0.65-1.39) | 0.797 |
| A/A | 34 | 24 | 0.97 (0.54-1.74) | 0.908 | 0.93 (0.51-1.71) | 0.819 |
| G/Gc | 130 | 95 | 1.00 | 0.771 | 1.00 | 0.771 |
| G/A & A/A | 183 | 127 | 0.95 (0.67-1.35) | 0.95 (0.66-1.36) | ||
| G/G & G/Ac | 279 | 198 | 1.00 | 0.985 | 1.00 | 0.879 |
| A/A | 34 | 24 | 1.00 (0.57-1.73) | 0.96 (0.54-1.70) | ||
| rs1840680 | 0.320 | 0.342 | ||||
| G/Gc | 124 | 102 | 1.00 | 1.00 | ||
| G/A | 142 | 91 | 0.78 (0.54-1.13) | 0.188 | 0.76 (0.52-1.12) | 0.170 |
| A/A | 29 | 26 | 1.09 (0.60-1.97) | 0.775 | 1.02 (0.55-1.90) | 0.949 |
| G/Gc | 124 | 102 | 1.00 | 0.305 | 1.00 | 0.254 |
| G/A & A/A | 171 | 117 | 0.83 (0.59-1.18) | 0.81 (0.56-1.17) | ||
| G/G & G/Ac | 266 | 193 | 1.00 | 0.459 | 1.00 | 0.602 |
| A/A | 29 | 26 | 1.24 (0.71-2.17) | 1.17 (0.65-2.11) |
Statistical analysis: logistic regression model or chi-square test.
aCervical neoplasias comprised precancerous lesions and invasive cancer of the uterine cervix.
bThe adjusted p values as well as adjusted odds ratios (AORs) and their 95% confident intervals (95% CIs) were determined through logistic regression model after controlling age.
cUsed as a reference for comparison to calculate the odds ratios of other genotypes.
Genetic variant distributions of pentraxin 3 in women with uterine cervical invasive cancer or precancerous lesion and control women in Taiwan
| Genetic variants | Controls (n =325) | Pre-cancerous lesions (n =98) | Invasive cancer (n =125) | p values | AORs (95% CIs)a | Ad. p values | AORs (95% CIs)b | Ad. p values |
|---|---|---|---|---|---|---|---|---|
| rs2120243 | ||||||||
| C/Cc | 123 | 39 | 56 | 0.746 | 1.00 | 1.00 | ||
| C/A | 140 | 42 | 50 | 0.94 (0.57-1.55) | 0.817 | 0.69 (0.41-1.14) | 0.146 | |
| A/A | 28 | 12 | 13 | 1.34 (0.62-2.88) | 0.459 | 0.87 (0.39-1.94) | 0.728 | |
| C/Cc | 123 | 39 | 56 | 0.644 | 1.00 | 1.00 | ||
| C/A & A/A | 168 | 54 | 63 | 1.01 (0.63-1.62) | 0.972 | 0.72 (0.44-1.16) | 0.176 | |
| C/C & C/Ac | 263 | 81 | 106 | 0.660 | 1.00 | 1.00 | ||
| A/A | 28 | 12 | 13 | 1.38 (0.67-2.84) | 0.383 | 1.05 (0.49-2.25) | 0.904 | |
| rs3816527 | ||||||||
| A/Ac | 186 | 59 | 84 | 0.228 | 1.00 | 1.00 | ||
| A/C | 101 | 35 | 32 | 1.09 (0.67-1.77) | 0.717 | 0.67 (0.39-1.13) | 0.133 | |
| C/C | 11 | 3 | 9 | 0.85 (0.23-3.15) | 0.807 | 1.36 (0.48-3.80) | 0.563 | |
| A/Ac | 186 | 59 | 84 | 0.556 | 1.00 | 1.00 | ||
| A/C & C/C | 112 | 38 | 41 | 1.07 (0.67-1.71) | 0.781 | 0.75 (0.46-1.23) | 0.249 | |
| A/A & A/Cc | 287 | 94 | 116 | 0.245 | 1.00 | 1.00 | ||
| C/C | 11 | 3 | 9 | 0.82 (0.22-3.01) | 0.767 | 1.54 (0.56-4.25) | 0.407 | |
| rs2305619 | ||||||||
| G/Gc | 130 | 41 | 54 | 0.997 | 1.00 | 1.00 | ||
| G/A | 149 | 46 | 57 | 0.98 (0.61-1.59) | 0.935 | 0.90 (0.55-1.46) | 0.659 | |
| A/A | 34 | 11 | 13 | 1.02 (0.48-2.20) | 0.953 | 0.86 (0.39-1.88) | 0.697 | |
| G/Gc | 130 | 41 | 54 | 0.928 | 1.00 | 1.00 | ||
| G/A & A/A | 183 | 57 | 70 | 0.99 (0.62-1.57) | 0.960 | 0.89 (0.56-1.42) | 0.616 | |
| G/G & G/Ac | 279 | 87 | 111 | 0.984 | 1.00 | 1.00 | ||
| A/A | 34 | 11 | 13 | 1.03 (0.50-2.13) | 0.927 | 0.91 (0.43-1.91) | 0.794 | |
| rs1840680 | ||||||||
| G/Gc | 124 | 41 | 61 | 0.506 | 1.00 | 1.00 | ||
| G/A | 142 | 43 | 48 | 0.91 (0.56-1.49) | 0.720 | 0.61 (0.37-1.02) | 0.058 | |
| A/A | 29 | 12 | 14 | 1.24 (0.58-2.65) | 0.580 | 0.84 (0.39-1.84) | 0.669 | |
| G/Gc | 124 | 41 | 61 | 0.352 | 1.00 | 1.00 | ||
| G/A & A/A | 171 | 55 | 62 | 0.97 (0.61-1.55) | 0.896 | 0.66 (0.41-1.05) | 0.081 | |
| G/G & G/Ac | 266 | 84 | 109 | 0.734 | 1.00 | 1.00 | ||
| A/A | 29 | 12 | 14 | 1.30 (0.63-2.66) | 0.475 | 1.07 (0.51-2.24) | 0.865 |
aAdjusted p values and adjusted odds ratios with their 95% CIs were assessed using multinomial logistic regression models after controlling for age between patients with uterine cervical precancerous lesions and control women.
bAdjusted p values and adjusted odds ratios with their 95% CIs were assessed using multinomial logistic regression models after controlling for age between patients with uterine cervical invasive cancer and control women.
cUsed as a reference for comparison to calculate the odds ratios of other genotypes.
AORs, adjusted odds ratios; 95% CIs, 95% confidence intervals; Ad. p, adjusted p.
Associations between genotypic distributions of pentraxin 3 and clinicopathological characteristics of the patients with cervical invasive cancer
| Characteristicsa | rs2120243 | rs1840680 | ||||||
|---|---|---|---|---|---|---|---|---|
| CCb | CA/AA | ORs (95% CIs) | p value | GGb | GA/AA | ORs (95% CIs) | p value | |
| Clinical stage | 0.405 | 0.854 | ||||||
| stage Ib | 31 | 39 | 1.00 | 35 | 36 | 1.00 | ||
| ≥ stage II | 25 | 23 | 0.73 (0.35-1.53) | 26 | 25 | 0.94 (0.46-1.92) | ||
| Pathologic type | 0.012 | 0.021 | ||||||
| squamous cell carcinomab | 45 | 60 | 1.00 | 50 | 59 | 1.00 | ||
| adenocarcinoma | 11 | 3 | 0.21 (0.05-0.78) | 11 | 3 | 0.23 (0.06-0.88) | ||
| Cell grading | 0.433 | 0.290 | ||||||
| well (grade 1)b | 10 | 8 | 1.00 | 11 | 7 | 1.00 | ||
| moderate & poor (grades 2/3) | 46 | 55 | 1.50 (0.55-4.10) | 50 | 55 | 1.73 (0.62-4.80) | ||
| Stromal invasion depth | 0.530 | 0.783 | ||||||
| ≤ 10 mmb | 27 | 33 | 1.00 | 31 | 29 | 1.00 | ||
| > 10 mm | 26 | 25 | 0.79 (0.37-1.66) | 27 | 28 | 1.11 (0.53-2.31) | ||
| Tumor diameter | 0.689 | 0.935 | ||||||
| ≤ 4cmb | 31 | 36 | 1.00 | 34 | 33 | 1.00 | ||
| > 4 cm | 25 | 25 | 0.86 (0.41-1.79) | 27 | 27 | 1.03 (0.50-2.11) | ||
| Parametrium | 0.968 | 0.526 | ||||||
| no invasionb | 36 | 39 | 1.00 | 40 | 36 | 1.00 | ||
| invasion | 20 | 22 | 1.02 (0.48-2.16) | 21 | 24 | 1.27 (0.61-2.66) | ||
| Vagina | 0.190 | 0.207 | ||||||
| no invasionb | 33 | 43 | 1.00 | 36 | 42 | 1.00 | ||
| invasion | 23 | 18 | 0.60 (0.28-1.29) | 25 | 18 | 0.62 (0.29-1.31) | ||
| Pelvic lymph node | 0.181 | 0.441 | ||||||
| no metastasisb | 39 | 49 | 1.00 | 43 | 46 | 1.00 | ||
| metastasis | 17 | 12 | 0.56 (0.24-1.32) | 18 | 14 | 0.73 (0.32-1.64) | ||
Statistical analyses: chi-square;
aSome clinicopathological data could not be obtained from the patients with cervical invasive cancer because of incomplete medical charts or records.
bAs a reference. ORs, odds ratios; 95% CIs, 95% confidence intervals.
In univariate analysis, PTX3 genetic variants rs2120243 and rs1840680 had nothing to do with 5 years survival in cervical cancer patients (HR: 0.62, 95% CI: 0.24-1.59, p=0.318 for rs2120243; HR: 0.76, 95% CI: 0.31-1.89, p=0.556 for rs1840680; Table 4). In contrast, HRs with poor 5 years survival could be found in cervical patients with more advanced stages (≥ stage II), deep stromal invasion, large tumor diameter, positive parametrium invasion and positive pelvic lymph node metastasis (Table 4).
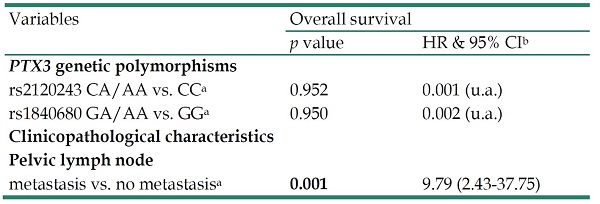
In multivariate analysis, PTX3 genetic variants rs2120243 and rs1840680 were also not associated with 5 years survival. However, HR with significantly poor 5 years survival was only found in cervical patients with positive pelvic lymph node metastasis (HR: 9.79, 95% CI: 2.43-37.75, p=0.001; Table 5).
Univariate analysis of pentraxin 3 (PTX3) genetic variants rs2120243 and rs1840680 and clinicopathological characteristics for 5 years survival in cervical cancer patients
| Variablesa | 5 years survival | |||
|---|---|---|---|---|
| + | ─ | p value | ORs (95% CIs) | |
| rs2120243 | 0.318 | |||
| CCb | 45 | 11 | 1.00 | |
| CA/AA | 53 | 7 | 0.62 (0.24-1.59) | |
| rs1840680 | 0.556 | |||
| GGb | 50 | 77 | 1.00 | |
| GA/AA | 51 | 8 | 0.76 (0.31-1.89) | |
| Clinical stage | 0.016 | |||
| stage Ib | 62 | 6 | 1.00 | |
| ≥ stage II | 39 | 14 | 3.23 (1.24-8.40) | |
| Pathologic type | 0.253 | |||
| squamous cell carcinomab | 92 | 16 | 1.00 | |
| adenocarcinoma | 10 | 4 | 1.90 (0.63-5.67) | |
| Cell grading | 0.915 | |||
| well (grade 1)b | 15 | 3 | 1.00 | |
| moderate & poor (grades 2/3) | 87 | 17 | 1.07 (0.31-3.65) | |
| Stromal invasion depth | 0.011 | |||
| ≤10 mmb | 53 | 4 | 1.00 | |
| > 10 mm | 42 | 15 | 4.18 (1.39-12.61) | |
| Tumor diameter | 0.010 | |||
| ≤ 4cmb | 59 | 5 | 1.00 | |
| >4 cm | 41 | 15 | 3.77 (1.37-10.37) | |
| Parametrium | 0.015 | |||
| no invasionb | 67 | 7 | 1.00 | |
| invasion | 33 | 13 | 3.148 (1.25-7.88) | |
| Vagina | 0.129 | |||
| no invasionb | 66 | 10 | 1.00 | |
| invasion | 34 | 10 | 1.96 (0.82-4.75) | |
| Pelvic lymph node | <0.001 | |||
| no metastasisb | 82 | 5 | 1.00 | |
| metastasis | 18 | 15 | 9.33 (3.39-25.71) | |
Statistical analyses: Kaplan-Meier curve model;
aSome clinicopathological data could not be obtained from the patients with cervical invasive cancer because of incomplete records of medical chart.
bAs a reference.
Survival; +, survival, -, mortality.
Discussion
It was reported that PTX3 is related to various cancers such as prostate cancer and lung cancer [25-28]. In addition, it has been found that PTX3 contributes to carcinogenesis and metastasis of human cervical cancer cells and regarded as a possible biomarker for cervical cancer [29]. PTX3 has been identified as a signature of 11 proteins for invasive cervical cancer, both individually and multivariate signatures [30]. PTX3 genetic variant rs2120243 is located at transcription factor binding site on chromosome 3 [23]. Variation at this site can change transcriptional regulation, which may influence the circulating plasma PTX3 levels [31]. rs3816527 is at exon 2 on chromosome 3 and is a non-synonymous variant in which amino acid is changed from alanine to aspartic acid. This could potentially have an interference with the N-terminal-mediated binding of PTX3 to its ligands and affect its function [32]. rs2305619 and rs1840680 are intron variants on chromosome 3 [23]. Previous studies have found that PTX3 SNPs rs2305619 and rs1840680 have functional significance and revealed that the mutant allele A of PTX3 rs2305619 and rs1840680 is related to higher plasma levels of PTX3 protein [31,33]. PTX genetic variants may have an impact on the expression of PTX3 gene or its protein levels. The implication of four PTX3 genetic variants rs1840680, rs2305619, rs3816527, and rs2120243 has been investigated by few authors [22,23]. To date, no study investigates the involvement of PTX3 genetic variants in the development of cervical cancer and patient prognosis in Taiwanese women. Thus, we designed this study to investigate the significance of PTX3 genetic variants in the occurrence of cervical cancer.
Multivariate analysis for the associations of pentraxin 3 (PTX3) genetic variants rs2120243 and rs1840680 as well as various clinicopatholgical characteristics with 5 years survival of cervical cancer patients
| Variables | Overall survival | |
|---|---|---|
| p value | HR & 95% CIb | |
| PTX3 genetic polymorphisms | ||
| rs2120243 CA/AA vs. CCa | 0.952 | 0.001 (u.a.) |
| rs1840680 GA/AA vs. GGa | 0.950 | 0.002 (u.a.) |
| Clinicopathological characteristics | ||
| Pelvic lymph node | ||
| metastasis vs. no metastasisa | 0.001 | 9.79 (2.43-37.75) |
Statistical analyses: Cox proportional hazard model;
aAs a comparison reference;
bHR, hazard ratio and 95% CI, 95% confidence interval for PTX3 genetic variants rs2120243 and rs1840680 and clinicopathological variables, compared to their respective controls.
u.a.: unavailable.
In this study, four PTX3 gene variants rs2120243, rs3816527, rs2305619 and rs1840680 in Taiwanese women with cervical neoplasias and control women did not find significant differences in genotypic distribution. Even after subdividing patients with cervical neoplasias into invasive and precancerous subgroups and adjusting for age, no significant genotypic distribution of these PTX3 variants was found. As far as we know, few studies have revealed a significant association between PTX3 genetic variants and cancer development. However, it was showed that PTX3 genetic variants rs2305619 and rs3816527 were associated with invasive mold infections in acute leukemia patients undergoing intense chemotherapy [13]. Fisher et al. reported that PTX3 genetic variants rs7252229 and rs3816527 may be associated with invasive aspergillosis following hematopoietic cell transplantation [34]. PTX3 SNP rs3816527 has been reported to be associated with invasive pulmonary aspergillosis in non-neutropenic patients [35]. Zhang et al. suggested that PTX3 genetic variant rs3845978 was related to ankylosing spondylitis, but rs2305619 was not [36]. In contrast, Barbati et al. found that PTX3 polymorphisms, rs2305619, rs3816527, rs1840680, were not linked to the risk of acute myocardial infarction [33].
We further investigated the relationships between PTX3 genetic variants and clinopathological parameters of cervical cancer. Compared with CA/AA and GA/AA, patients with genotype CC in PTX3 rs2120243 and genotype GG in rs1840680 were more likely to develop adenocarcinoma. Otherwise, PTX3 genetic variants had nothing to do with clinicopathological characteristics. However, a high PTX3 expression has been proved to be related to pancreatic cancer metastasis [27]. In addition, several studies have demonstrated the role of PTX3 in epithelial cancer metastasis because of epithelial-mesenchymal transition [37-39]. It was suggested that oral cancer patients with PTX3 genetic variant rs3816527 in smoking were linked to the development of late-stage cancer and the increase of lymph node metastasis [23]. Carmo et al. demonstrated a significant association between PTX3 polymorphisms and hepatocellular occurrence [22]. PTX3 rs2305619 polymorphism has been reported to be associated with Child-Pugh scores B and C in hepatocellular carcinoma individuals. Brunel et al. found that the 6-month cumulative incidence of invasive mold infections for patients, carrying two copies of the minor allele of PTX3 SNPs rs2305619 and/or rs3816527, who had acute myeloid leukemia, acute lymphoblastic leukemia, or refractory anemia with excess blasts-2 and received intense chemotherapy, was significantly different from those without these single SNPs (21% vs 10%; log-rank test p=0.04) in the Lausanne University Hospital [13]. However, based on National Center for Biotechnology Information dbSNP database, the distribution of PTX3 genetic variants rs2120243, rs3816527, rs2305619 and rs1840680 in Taiwanese are not similar to other population of the world, the conflicting findings in cancer development and progression may occur.
In the univariate analysis of the effects of PTX3 genetic variants and clinicopathological characteristics on patient survival, although cervical cancer patients with the genotype CC in rs2120243 and the genotype GG in PTX3 rs1840680 had a higher chance of developing adenocarcinoma, no PTX3 variants were found to be predictive of 5 years survival. Hazard ratios with poor 5 years survival could be found in cervical patients with more advanced stages (≥ stage II), deep stromal invasion, large tumor diameter, positive parametrium invasion and positive pelvic lymph node metastasis. Moreover, after controlling the clinicopathological parameters through multivariate analysis, we also found that PTX3 genetic variants had no significant influence on 5 years survival in cervical cancer patients. Pelvic lymph node metastasis was suggested to be the only independent predictor of 5 years survival in cervical cancer patients through multivariate analysis in Taiwanese women among those significant predictor variables in univariate analysis. This suggestion is reasonable because it has been reported that pelvic lymph node metastasis is the most important prognostic indicator for the survival of cervical cancer patients [40,41].
This study has two main limitations. First, we only studied the population of central Taiwan and did not enroll residents from other areas. Therefore, the recruited sample size was relatively small. Thus, statistical analysis of the impact of PTX3 genetic variants on patient survival may not be enough to achieve a significant correlation. This may limit the external validity of this investigation. Second, women in the control group were recruited from the outpatient clinic of our hospital for routine examination. Due to their conservative attitude, there is no routine detection for human papillomavirus infection. The possible influence of this factor has not been analyzed.
Acknowledgements
We would like to thank the Human Biobank of Chung Shan Medical University Hospital for providing the biological specimen and related clinical data for our research. This study was supported by research grants from Chung Shan Medical University and Chi-Mei Foundation Medical Center (CSMU-CMMC-108-02). This study was also supported by Chung Shan Medical University Hospital (CSH-2020-D-007).
Competing Interests
The authors have declared that no competing interest exists.
References
1. Peri G, Introna M, Corradi D. et al. PTX3, A prototypical long pentraxin, is an early indicator of acute myocardial infarction in humans. Circulation. 2000;102(6):636-641
2. Mantovani A, Garlanda C, Doni A, Bottazzi B. Pentraxins in innate immunity: from C-reactive protein to the long pentraxin PTX3. J Clin Immunol. 2008;28(1):1-13
3. Lee GW, Lee TH, Vilcek J. TSG-14, a tumor necrosis factor- and IL-1-inducible protein, is a novel member of the pentaxin family of acute phase proteins. J Immunol. 1993;150(5):1804-1812
4. Garlanda C, Bottazzi B, Bastone A, Mantovani A. Pentraxins at the crossroads between innate immunity, inflammation, matrix deposition, and female fertility. Annu Rev Immunol. 2005;23:337-366
5. Bottazzi B, Garlanda C, Salvatori G, Jeannin P, Manfredi A, Mantovani A. Pentraxins as a key component of innate immunity. Curr Opin Immunol. 2006;18(1):10-15
6. Azzurri A, Sow OY, Amedei A. et al. IFN-gamma-inducible protein 10 and pentraxin 3 plasma levels are tools for monitoring inflammation and disease activity in Mycobacterium tuberculosis infection. Microbes Infect. 2005;7(1):1-8
7. Mauri T, Bellani G, Patroniti N. et al. Persisting high levels of plasma pentraxin 3 over the first days after severe sepsis and septic shock onset are associated with mortality. Intensive Care Med. 2010;36(4):621-629
8. Garlanda C, Bottazzi B, Magrini E, Inforzato A, Mantovani A. PTX3, a Humoral Pattern Recognition Molecule, in Innate Immunity, Tissue Repair, and Cancer. Physiol Rev. 2018;98(2):623-639
9. Shastry BS. SNPs: impact on gene function and phenotype. Methods Mol Biol. 2009;578:3-22
10. Hua KT, Liu YF, Hsu CL. et al. 3'UTR polymorphisms of carbonic anhydrase IX determine the miR-34a targeting efficiency and prognosis of hepatocellular carcinoma. Scientific reports. 2017;7(1):4466
11. Su CW, Chien MH, Lin CW. et al. Associations of genetic variations of the endothelial nitric oxide synthase gene and environmental carcinogens with oral cancer susceptibility and development. Nitric oxide: biology and chemistry. 2018;79:1-7
12. Cheng RH, Wang YP, Chang JY, Pan YH, Chang MC, Jeng JH. Genetic Susceptibility and Protein Expression of Extracellular Matrix Turnover-Related Genes in Oral Submucous Fibrosis. Int J Mol Sci. 2020 21(21)
13. Brunel AS, Wojtowicz A, Lamoth F. et al. Pentraxin-3 polymorphisms and invasive mold infections in acute leukemia patients receiving intensive chemotherapy. Haematologica. 2018;103(11):e527-e530
14. Cunha C, Aversa F, Lacerda JF. et al. Genetic PTX3 deficiency and aspergillosis in stem-cell transplantation. N Engl J Med. 2014;370(5):421-432
15. Chiarini M, Sabelli C, Melotti P. et al. PTX3 genetic variations affect the risk of Pseudomonas aeruginosa airway colonization in cystic fibrosis patients. Genes Immun. 2010;11(8):665-670
16. Cubie HA, Campbell C. Cervical cancer screening - The challenges of complete pathways of care in low-income countries: Focus on Malawi. Womens Health (Lond). 2020;16:1745506520914804
17. Portnoy A, Clark S, Ozawa S, Jit M. The impact of vaccination on gender equity: conceptual framework and human papillomavirus (HPV) vaccine case study. Int J Equity Health. 2020;19(1):10
18. Nasiell K, Roger V, Nasiell M. Behavior of mild cervical dysplasia during long-term follow-up. Obstet Gynecol. 1986;67(5):665-669
19. Petignat P, Roy M. Diagnosis and management of cervical cancer. BMJ. 2007;335(7623):765-768
20. Khan MJ, Castle PE, Lorincz AT. et al. The elevated 10-year risk of cervical precancer and cancer in women with human papillomavirus (HPV) type 16 or 18 and the possible utility of type-specific HPV testing in clinical practice. J Natl Cancer Inst. 2005;97(14):1072-1079
21. Martin CM, O'Leary JJ. Histology of cervical intraepithelial neoplasia and the role of biomarkers. Best Pract Res Clin Obstet Gynaecol. 2011;25(5):605-615
22. Carmo RF, Aroucha D, Vasconcelos LR, Pereira LM, Moura P, Cavalcanti MS. Genetic variation in PTX3 and plasma levels associated with hepatocellular carcinoma in patients with HCV. J Viral Hepat. 2016;23(2):116-122
23. Yeh CM, Lin CW, Chuang CY. et al. Functional Genetic Variant of Long Pentraxin 3 Gene Is Associated With Clinical Aspects of Oral Cancer in Male Patients. Front Oncol. 2019;9:581
24. Sun YH, Chou YH, Ou CC. et al. Investigation of metastasis-associated in colon cancer-1 genetic variants in the development and clinicopathologcial characteristics of uterine cervical cancer in Taiwanese women. Int J Med Sci. 2020;17(4):490-497
25. Locatelli M, Ferrero S, Martinelli Boneschi F. et al. The long pentraxin PTX3 as a correlate of cancer-related inflammation and prognosis of malignancy in gliomas. J Neuroimmunol. 2013;260(1-2):99-106
26. Stallone G, Cormio L, Netti GS. et al. Pentraxin 3: a novel biomarker for predicting progression from prostatic inflammation to prostate cancer. Cancer Res. 2014;74(16):4230-4238
27. Kondo S, Ueno H, Hosoi H. et al. Clinical impact of pentraxin family expression on prognosis of pancreatic carcinoma. Br J Cancer. 2013;109(3):739-746
28. Hu FQ, Qiao T, Xie X, Hu R, Xiao HB. Knockdown of the inflammatory factor pentraxin-3 suppresses growth and invasion of lung adenocarcinoma through the AKT and NF-kappa B pathways. J Biol Regul Homeost Agents. 2014;28(4):649-657
29. Ying TH, Lee CH, Chiou HL. et al. Knockdown of Pentraxin 3 suppresses tumorigenicity and metastasis of human cervical cancer cells. Sci Rep. 2016;6:29385
30. Berggrund M, Enroth S, Lundberg M. et al. Identification of Candidate Plasma Protein Biomarkers for Cervical Cancer Using the Multiplex Proximity Extension Assay. Mol Cell Proteomics. 2019;18(4):735-743
31. Diamond JM, Meyer NJ, Feng R. et al. Variation in PTX3 is associated with primary graft dysfunction after lung transplantation. Am J Respir Crit Care Med. 2012;186(6):546-552
32. Inforzato A, Rivieccio V, Morreale AP. et al. Structural characterization of PTX3 disulfide bond network and its multimeric status in cumulus matrix organization. J Biol Chem. 2008;283(15):10147-10161
33. Barbati E, Specchia C, Villella M. et al. Influence of pentraxin 3 (PTX3) genetic variants on myocardial infarction risk and PTX3 plasma levels. PLoS One. 2012;7(12):e53030
34. Fisher CE, Hohl TM, Fan W. et al. Validation of single nucleotide polymorphisms in invasive aspergillosis following hematopoietic cell transplantation. Blood. 2017;129(19):2693-2701
35. Tang T, Dai Y, Zeng Q. et al. Pentraxin-3 polymorphisms and pulmonary fungal disease in non-neutropenic patients. Ann Transl Med. 2020;8(18):1142
36. Zhang X, Ding W. Association of Genetic Variants in Pentraxin 3 Gene with Ankylosing Spondylitis. Med Sci Monit. 2016;22:2911-2916
37. Scimeca M, Antonacci C, Toschi N. et al. Breast Osteoblast-like Cells: A Reliable Early Marker for Bone Metastases From Breast Cancer. Clin Breast Cancer. 2018;18(4):e659-e669
38. Ronca R, Di Salle E, Giacomini A. et al. Long pentraxin-3 inhibits epithelial-mesenchymal transition in melanoma cells. Mol Cancer Ther. 2013;12(12):2760-2771
39. Scimeca M, Antonacci C, Colombo D, Bonfiglio R, Buonomo OC, Bonanno E. Emerging prognostic markers related to mesenchymal characteristics of poorly differentiated breast cancers. Tumour Biol. 2016;37(4):5427-5435
40. Kamura T, Tsukamoto N, Tsuruchi N. et al. Multivariate analysis of the histopathologic prognostic factors of cervical cancer in patients undergoing radical hysterectomy. Cancer. 1992;69(1):181-186
41. Choi KH, Kim JY, Lee DS. et al. Clinical impact of boost irradiation to pelvic lymph node in uterine cervical cancer treated with definitive chemoradiotherapy. Medicine (Baltimore). 2018;97(16):e0517
Author contact
![]() Corresponding author: Po-Hui Wang, MD, PhD, Institute of Medicine, Chung Shan Medical University, 110, Section 1, Chien-Kuo North Road, Taichung, 40201, Taiwan. Tel.: 886-4-24739595 ext. 21721; Fax: 884-4-24738493; E-mail: wang082160com.
Corresponding author: Po-Hui Wang, MD, PhD, Institute of Medicine, Chung Shan Medical University, 110, Section 1, Chien-Kuo North Road, Taichung, 40201, Taiwan. Tel.: 886-4-24739595 ext. 21721; Fax: 884-4-24738493; E-mail: wang082160com.

 Global reach, higher impact
Global reach, higher impact