3.2
Impact Factor
ISSN: 1449-1907
Int J Med Sci 2021; 18(10):2093-2101. doi:10.7150/ijms.56551 This issue Cite
Research Paper
Long noncoding RNAs may impact podocytes and proximal tubule function through modulating miRNAs expression in Early Diabetic Kidney Disease of Type 2 Diabetes Mellitus patients
1. Dept. of Internal Medicine II - Division of Nephrology, “Victor Babes” University of Medicine and Pharmacy Timisoara, Romania, Eftimie Murgu Sq. no. 2, 300041 Timisoara, RO; County Emergency Hospital Timisoara, RO.
2. Centre for Molecular Research in Nephrology and Vascular Disease, Faculty of Medicine, “Victor Babes” University of Medicine and Pharmacy, Timisoara, Romania Eftimie Murgu Sq. no. 2, 300041 Timisoara, RO.
3. Department of Microbiology XIV- Division of Microbiology-Virusology, ”Victor Babes” University of Medicine and Pharmacy, Timisoara, Romania, Eftimie Murgu Sq. no. 2, 300041 Timisoara, RO.
4. Dept. of Internal Medicine II - Division of Diabetes and Metabolic Diseases, “Victor Babes” University of Medicine and Pharmacy Timisoara, Romania, Eftimie Murgu Sq. no. 2, 300041 Timisoara, RO; County Emergency Hospital Timisoara, RO.
5. Dept. of Internal Medicine II - Division of Endocrinology, “Victor Babes” University of Medicine and Pharmacy Timisoara, Romania, Eftimie Murgu Sq. no. 2, 300041 Timisoara, RO; County Emergency Hospital Timisoara, RO.
6. Dept. of Biochemistry and Pharmacology - Division of Pharmacology, “Victor Babes” University of Medicine and Pharmacy Timisoara, Romania, Eftimie Murgu Sq. no. 2, 300041 Timisoara, RO; County Emergency Hospital Timisoara, RO.
7. Dept. of Functional Sciences - Division of Public Health Medicine, Faculty of Medicine, “Victor Babes” University of Medicine and Pharmacy, Timisoara, Romania, Eftimie Murgu Sq. no. 2, 300041 Timisoara, RO; County Emergency Hospital Timisoara, RO.
8. Center for Translational Research and Systems Medicine, Faculty of Medicine, “Victor Babes” University of Medicine and Pharmacy, Timisoara, Romania, Eftimie Murgu Sq. no. 2, 300041 Timisoara, RO.
9. Dept. of Neurosciences - Division of Neurology, “Victor Babes” University of Medicine and Pharmacy Timisoara, Romania; Eftimie Murgu Sq. no. 2, 300041 Timisoara, RO; County Emergency Hospital Timisoara, RO.
10. Centre for Cognitive Research in Neuropsychiatric Pathology (Neuropsy-Cog), Faculty of Medicine, “Victor Babes” University of Medicine and Pharmacy, Timisoara, Romania, Eftimie Murgu Sq. no. 2, 300041 Timisoara, RO.
11. Dept. of Anatomy and Embryology- Division of Anatomy and Embryology; “Victor Babes” University of Medicine and Pharmacy Timisoara, Romania; Eftimie Murgu Sq. no. 2, 300041 Timisoara, RO.
12. Dept. of Surgery I- Division of Surgical Semiology I, “Victor Babes” University of Medicine and Pharmacy Timisoara, Romania; Eftimie Murgu Sq. no. 2, 300041 Timisoara, RO; Emergency Clinical Municipal Hospital Timisoara, RO.
13. Dept. of Surgery II- Division of Surgery I, “Victor Babes” University of Medicine and Pharmacy Timisoara, Romania; Eftimie Murgu Sq. no. 2, 300041 Timisoara, RO; County Emergency Hospital Timisoara, RO.
14. Dept. of Morphologic Microscopy - Division of Cellular and Molecular Biology; "Victor Babeș" University of Medicine and Pharmacy Timișoara, Romania; Eftimie Murgu Sq. no. 2, 300041 Timișoara, RO; County Emergency Hospital Timisoara, RO.
*Equal contributions. These authors drafted the article for intellectual content, and gave the final approval of the article to be published.
#Equal contributions to the laboratory assessments and article revision.
Received 2020-11-29; Accepted 2021-2-7; Published 2021-3-15
Abstract
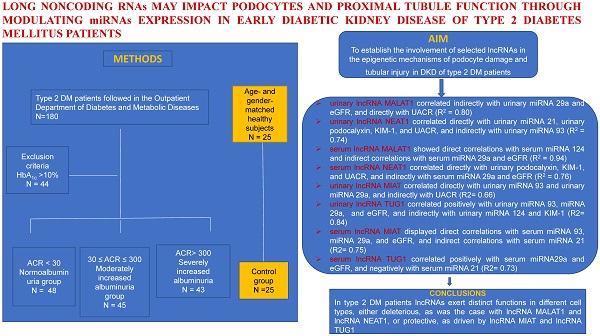
Aims: Long noncoding RNAs (lncRNAs) play key roles in the pathophysiology of DKD involving actions of microRNAs (miRNAs). The aims of the study were to establish the involvement of selected lncRNAs in the epigenetic mechanisms of podocyte damage and tubular injury in DKD of type 2 diabetes mellitus (DM) patients in relation to a particular miRNAs profile.
Methods: A total of 136 patients with type 2 DM and 25 healthy subjects were assessed in a cross-sectional study concerning urinary albumin: creatinine ratio (UACR), eGFR, biomarkers of podocyte damage (synaptopodin, podocalyxin) and of proximal tubule (PT) dysfunction (Kidney injury molecule-1-KIM-1, N-acetyl-D-glucosaminidase-NAG), urinary lncRNA metastasis-associated lung adenocarcinoma transcript 1 (MALAT1), nuclear-enriched abundant transcript 1 (NEAT1), myocardial infarction-associated transcript (MIAT), taurine-upregulated gene 1 (TUG1), urinary miRNA21, 124, 93, 29a.
Results: Multivariable regression analysis showed that urinary lncMALAT1 correlated directly with urinary synaptopodin, podocalyxin, KIM-1, NAG, miRNA21, 124, UACR, and negatively with eGFR, miRNA93, 29a (p<0.0001; R2=0.727); urinary lncNEAT1 correlated directly with synaptopodin, KIM-1, NAG, miRNA21, 124, and negatively with eGFR, miRNA93, 29a (p<0.0001; R2=0.702); urinary lncMIAT correlated directly with miRNA93 and 29a, eGFR (p<0.0001; R2=0.671) and negatively with synaptopodin, KIM-1, NAG, UACR, miRNA21, 124 (p<0.0001; R2=0.654); urinary lncTUG1 correlated directly with eGFR, miRNA93, 29a, and negatively with synaptopodin, podocalyxin, NAG, miRNA21, 124 (p<0.0001; R2=0.748).
Conclusions: In patients with type 2 DM lncRNAs exert either deleterious or protective functions within glomeruli and PT. LncRNAs may contribute to DKD through modulating miRNAs expression and activities. This observation holds true independently of albuminuria and DKD stage.
Keywords: lncRNA, miRNA, podocyte, proximal tubule, diabetes mellitus
Introduction
Diabetic kidney disease (DKD), as a major microvascular complication of both type 1 and type 2 diabetes mellitus (DM), accounts for over 40% of patients which reach end-stage renal disease and are referred to renal replacement therapies [1].
The tubulocentric concept with regard to DKD has emphasized the pivotal role of the proximal tubule (PT) and of the tubulointerstitial compartment in the development of DKD [2]. The glomerular theory raises a similar interest, with a special focus on the contribution of the podocyte injury in the course of DKD.
Particular molecular signatures and epigenetic profiles have emerged to support the complexity of DKD, pointing to the pathogenic importance of microRNAs (miRNAs) and of long noncoding RNAs (lncRNAs) in the development of DKD.
MiRNAs are endogenous small noncoding RNAs of about 20-25 nucleotides that silence the expression of target genes either through translational repression and/or mRNA degradation [3]. MiRNAs play important roles in modulating gene expression and promoting the initiation and progression of various diseases, including DKD [4, 5].
MiRNAs with negative or protective roles, either at renal level or both at renal and vascular level, have been thoroughly studied in the course of DKD [4, 5, 6, 7, 8].
MiRNAs 21 and 124 have been proved to enhance podocyte injury and PT lesions [6, 7, 8, 9, 10], while other miRNAs proved to be protective at the same levels, such as miRNA 192, 125a, 126, 146a [6, 7, 8].
New key miRNAs have been proposed as protective and prompted further research, such as miRNA 93 and miRNA 29. Downregulation of miRNA 93 produces important podocyte and endothelial injury [11]. A significant decrease of the miRNA 29 family expression leads to tubular lesions and severe renal fibrosis [12].
LncRNAs are a group of transcription materials with over 200 nucleotides, which lack a complete open reading frame and have no protein-coding functions [13]. LncRNAs regulate gene expression, including epigenetic transcription and posttranscriptional modifications [14].
Dysregulated lncRNAs have been involved in key biological processes by mediating podocyte damage [15] and high glucose-induced PT injury [16].
Recently, a particular interest has been shown towards the intricate connection between miRNAs and lncRNAs. Cytoplasmic lncRNAs may act as molecular sponge and bind to miRNAs, thus indirectly enhancing or decreasing protein expression [17], through inhibiting their mRNA targets [18].
Recent studies in DKD revealed interactions of podocytes and PTs with lncRNAs posesing deleterious roles, among which metastasis - associated lung adenocarcinoma transcript 1 (MALAT1) and nuclear enriched abundant transcript 1 (NEAT1), and with renal protective lncRNAs, such as taurine - upregulated gene 1 (TUG1) and myocardial infarction - associated transcript (MIAT) [14].
The aim of the study was to establish the involvement of selected lncRNAs in the epigenetic mechanisms of podocyte damage and tubular injury in DKD of type 2 DM patients. The molecular mechanisms of lncRNAs intervention were evaluated in relation to a particular miRNAs profile.
Materials and Methods
Subjects
A total of consecutive 180 patients with type 2 DM attending the Outpatient Department of Diabetes and Metabolic Diseases (from January 2019 through December 2019), aged between 20 and 69 years, were screened for the study according to person visits and chart reviews. The inclusion criteria were DM duration higher than 5 years, and therapy with oral antidiabetic drugs (metformin, gliclazide), angiotensin-converting enzyme inhibitors or angiotensin receptor blockers, and statins. The exclusion criterion was decompensated DM (HbA1c >10%). A total of 136 patients [48 patients with normoalbuminuria (group 1), 45 patients with moderately increased albuminuria (group 2), and 43 patients with severely increased albuminuria (group 3)] and 25 age- and gender-matched healthy control subjects attending a general practitioner's office for routine check-up, without known history of renal diseases and in whom diabetes or pre-diabetes were excluded by a value of HbA1c ≤5.6% (group 4) were enrolled in this case series study. The remaining 48 patients out of the 73 subjects screened were excluded due to HbA1c levels.
The County Emergency Hospital Ethics Committee (Board of Human Studies) approved the protocol (approval number 8/10.01.2019), and every patient provided written informed consent before enrolment.
Laboratory assessments
The serum and urinary specimens of patients and controls utilized for the assessment of UACR, cystatin C, KIM-1, NAG, synaptopodin, and podocalyxin were frozen at -80 °C and thawed before assay. Urinary biomarkers were evaluated in the same first morning urine sample [8, 19, 20]. The biomarkers of podocyte injury (synaptopodin, podocalyxin) and of PT dysfunction (KIM-1, NAG), were reported as per urinary creatinine ratio. Urinary tract infections were ruled out by negative urine cultures in all patients. CKD was defined according to the KDIGO Guideline for the Evaluation and Management of Chronic Kidney Disease [21].
miRNA and lncRNA assessment
Total RNA was isolated from the blood and urine samples using the mirVana™ miRNA Isolation Kit (Life Technologies, 5791 Van Allen Way, Carlsbad, California 92008., Cat. no. AM1560) and microRNA Purification Kit (Norgen Biotek Corporation, 3430 Schmon Parkway, Thorold, ON, Canada Cat. no. 29000). The quantity and quality of total RNA was verified using NanoDrop ND-1000 spectrophotometer (Analytik Jena). Total RNA was stored at -80 ºC, and reverse transcription was performed using the TaqMan MicroRNA Reverse Transcription Kit (Applied Biosystems, Foster City, CA). cDNA were amplified using the TaqMan® Universal PCR Master Mix and specific primers (TaqMan Assay) for miRNAs (miRNA21, miRNA124, miRNA93, miRNA29a) and lncRNAs (lncRNA MALAT1, lncRNA NEAT1, lncRNA MIAT, lncRNA TUG1) were utilized. The PCR reaction was performed on a 7900HT real-time PCR System. All samples were run in triplicate, and relative expression level of specific gene was calculated using the SDS software v.2.4., and U6 small nuclear RNA and GAPDH were utilized as the endogenous controls. The relative expression rate was measured by the 2-ΔΔCT method (ΔΔCT=ΔCT patients-ΔCT healthy controls) [22].
Statistical analysis
Clinical and biological data are presented as medians and interquartile range (IQR), as for variables with skewed distribution. Depending on the distribution of the values, differences between subgroups were analyzed with the Mann-Whitney U test for comparison of 2 groups and the Kruskal-Wallis test for comparison of 4 groups. Univariable and multivariable linear regression analyses were conducted in order to assess the significance of the relation of serum and urinary lncRNAs with serum and urinary miRNAs, as well as with other continuous variables. The statistical methods were utilised as required by a case series study which is an exploratory study. Case series have a descriptive study design. In our case series analyses were performed in order to investigate the association between lncRNAs and a particular miRNA profile. Statistical significance was set at p<0.05 and the analyses applied were conducted with Stata 15.1 (Statacorp, Texas, USA).
Results
Demographic, clinical, and biological data of the study subjects are presented in Table 1 as medians and IQR. In Table 1, p values were corrected for multiple comparisons.
LncRNA MALAT1 and lncRNA NEAT1 are upregulated in early DKD and inhibit miRNA 93 and miRNA 29a function, while enhancing miRNA 21 and miRNA 124 activities
In univariable linear regression analysis, we found a direct relationship between urinary and serum lncRNA MALAT1, and urinary and serum miRNA 21, miRNA 124, urinary podocalyxin, urinary synaptopodin, urinary KIM-1, urinary NAG, and UACR (p<0.001). This lncRNA showed negative correlations with urinary and serum miRNA 93 and miRNA 29a, as well as with eGFR (p<0.001). Urinary and serum lncRNA NEAT1 followed the same trend as lncRNA MALAT1 (Table 2 and 3).
Multivariable regression analysis yielded models which showed that urinary lncRNA MALAT1 correlated indirectly with urinary miRNA 29a and eGFR, and directly with UACR (R2 = 0.80). Also, we found a direct relationship between urinary lncRNA NEAT1 and urinary miRNA 21, urinary podocalyxin, KIM-1, and UACR, and an indirect relationship with urinary miRNA 93 (R2 = 0.74) (Table 4).
Serum lncRNA MALAT1 showed direct correlations with serum miRNA 124 and indirect correlations with serum miRNA 29a and eGFR (R2 = 0.94). Serum lncRNA NEAT1 correlated directly with urinary podocalyxin, KIM-1, and UACR, and indirectly with serum miRNA 29a and eGFR (R2 = 0.76) (Table 5).
These data point to the deleterious involvement of lncRNA MALAT1 and lncRNA NEAT1 upon the glomeruli and the PT in early DKD, as documented by the direct correlation of these lncRNAs with podocyte damage and PT dysfunction biomarkers. Their negative effects are also supported by the indirect correlations with eGFR and the two miRNAs, miRNA 93 and miRNA 29a, respectively, known for their protective effects in the course of DKD.
Clinical and biological data of the patients studied
| Parameter | Group 1 (healthy controls) | Group 2 (normoalbuminuria) | Group 3 (moderately increased albuminuria) | Group 4 (severely increased albuminuria) |
|---|---|---|---|---|
| Number of subjects | 25 | 48 | 45 | 43 |
| Age (years) | 61 (55, 69) | 59 (53.5;63) | 60 (55;67) | 60 (57;66) |
| DM duration (years) | 8 (7;10) ♣ | 8 (6;13) | 10 (8;13) | |
| BMI | 26.67 (23.88;29.76) # | 32.02 (29.05;35.06) ⁋ | 32 (28;38.09) | 34 (30;37) |
| SBP (mmHg) | 120 (120;130) # | 130 (125;140) † | 130 (125;140) ♦ | 150 (145;170) |
| DBP (mmHg) | 70 (70;80) # | 75 (70;80) † | 75 (70;80) ♦ | 85 (75;90) |
| Hb (g/dl) | 13.6 (12.5;14.5) # | 12.65 (12;13.65) † | 13.4 (12.2;14) ♦ | 11.25 (10.5;12.9) |
| Serum creatinine (mg/dl) | 0.92 (0.78;1.01) # | 0.78 (0.69;0.87) † | 0.74 (0.69;0.81) ♦ | 1.65 (1.47;1.94) |
| eGFR (ml/min/1.73m2) | 99.35 (91.46; 105.27) # | 39.60 (36.44;99.77) | 48.89 (46.30;52.70) ♦ | 43.40 (36.49;46.55) |
| Glycaemia (fasting) (mg/dl) | 99 (90;100) # | 150.5 (117;203) ⸿, ‡ | 165 (125;264) | 176 (140;238) |
| HbA1c (%) | 5.8 (5.7;6) # | 6.95 (6.5;7.4) *, † | 7.8 (6.94;9.51) | 8.43 (7.69;9.67) |
| Cholesterol (mg/dl) | 160 (125; 181) # | 203 (180;243) ∆ | 202 (172;288) | 223 (187;276) |
| Triglycerides (mg/dl) | 132 (107;148) ¤ | 161 (129;191.5) | 158 (117;205) | 167 (122;236) |
| hsCRP (mg/dl) | 0.85 (0.8;2.78) # | 3.25 (2.31;4.75) ●, ∏ | 10 (8.55;16.87) | 18.81 (2.6;27.68) |
| UACR (mg/g) | 10.82 (10.12;14.32) # | 18.38 (16.63;20.01) ●, † | 83.33 (44.28;117.38) ♦ | 541.45 (486.34;981.91) |
| Cystatin C (mg/L) | 0.66 (0.57;0.67) # | 3 (0.72;3.24) ⁑, ◊ | 2.1 (1.94;2.23) ♦ | 1.46 (1.31;1.53) |
| Nephrin/creat (mg/g) | 0.08 (0.04;0.09) # | 0.12 (0.11;0.15) ●, † | 0.78 (0.53;1.1) ♦ | 6.25 (4.6;7.2) |
| Podocalyxin/creat (mg/g) | 32.5 (28.6;38.4) # | 65.6 (58.9;70.35) ●, † | 121.6 (114.2;155.7) ♦ | 870.3 (384.9;950.1) |
| Synaptopodin/creat (mg/g) | 9.33 (6.88;9.64) # | 18.6 (15.38;21.34) ●, † | 26.58 (24.89;27.65) ♦ | 130.5 (72.4;137.78) |
| KIM-1/creat (ng/g) | 43.82 (26;47.5) # | 74.18 (63.6;87.28) ●, † | 120.56 (114.73;133.78) ♦ | 684.83 (570.4;831.65) |
| NAG/creat (UI/g) | 2.22 (1.69;2.41) # | 3.17 (1.88;5.28) ●, † | 10.5 (9.92;11.96) ♦ | 16.74 (16.05;18) |
| Alpha -1 microglobulin | 3.16 (2.49; 3.24) # | 4.16 (3.76;5.16) ●, † | 7.73 (6.71;9.08) ♦ | 56.27 (31.18;65) |
| u-miRNA-29a | 2.95 (2.75;3.08) # | 1.80 (1.7;2.03) ●, † | 0.76 (0.6;0.91) ♦ | 0.47 (0.41;0.6) |
| p-miRNA-29a | 3.64 (3.52;3.75) # | 2.64 (2.53;2.73) ●, † | 1.54 (1.39;1.71) ♦ | 0.91 (0.78;0.97) |
| u-miRNA-21p | 1.02 (0.92;1.02) # | 1.19 (1.14;1.26) ●, † | 1.82 (1.67;1.90) ♦ | 2.67 (2.27;2.75) |
| p-miRNA-21p | 1.09 (1.06;1.17) # | 1.26 (1.16;1.44) ●, † | 2.34 (2.25;2.54) ♦ | 2.94 (2.85;3.04) |
| u-miRNA-124 | 0.54 (0.44;0.73) # | 1.5 (1.25;1.71) ●, † | 2.74 (2.57;2.85) ♦ | 3.23 (3.18;3.37) |
| p-miRNA-124 | 0.81 (0.77;0.96) # | 1.95 (1.67;2.44) ●, † | 3.38 (3.07;3.59) ♦ | 3.86 (3.79;4.01) |
| u-miRNA-93 | 2.21 (1.9;3.25) # | 2.89 (2.20;3.08) ●, † | 1.7 (1.38;1.92) ♦ | 1.14 (0.89;1.25) |
| p-miRNA-93 | 3.45 (3.24;4.49) # | 2.98 (2.4;3.93) ●, † | 2.42 (2.26;2.65) ♦ | 1.44 (1.21;1.57) |
| u-lnc-MALAT-1 | 1.13 (1.1;1.28) # | 2.3 (1.89;2.85) ●, † | 3.53 (3.08;3.88) ♦ | 3.88 (3.66;4.23) |
| p-lnc-MALAT-1 | 1.61 (1.54;1.64) # | 2.51 (2.13;2.83) ●, † | 4.04 (3.64;4.46) ♦ | 4.53 (4.39;4.72) |
| u-lnc-NEAT | 0.78 (0.68;0.82) # | 0.90 (0.68;1.03) ●, † | 1.24 (1.10;1.57) ♦ | 2.27 (1.92;2.53) |
| p-lnc-NEAT | 1.06 (1;1.13) # | 1.38 (1.19;1.54) ●, † | 1.67 (1.53;2.01) ♦ | 2.82 (2.19;3.1) |
| u-lnc-MIAT | 2.53 (2.4;2.91) # | 1.77 (1.37;2.07) ▀, † | 1.4 (1.26;1.84) ♦ | 1.03 (0.62;1.16) |
| p-lnc-MIAT | 2.77 (2.61;3.12) # | 2.67 (1.73;2.86) ●, † | 1.83 (1.43;1.95) ♦ | 1.28 (0.95;1.38) |
| u-lnc-TUG | 3.13 (2.84;3.21) # | 2.65 (2.18;3.31) ●, † | 1.99 (1.7;2.24) ♦ | 1.32 (1.05;1.41) |
| p-lnc-TUG | 3.38 (3.09;3.57) # | 3.10 (2.67;3.77) ●, † | 2.46 (2.14;2.62) ♦ | 2.07 (1.75;2.26) |
Clinical and biological data are presented as medians and IQR, as for variables with skewed distribution.
#: Significance between group 1 vs group 2 vs group 3 vs group 4; p < 0.001; ¤: Significance between group 1 vs group 2 vs group 3 vs group 4; p = 0.001; ● Significance between group 2 vs group 3; p < 0.001; ⸿: Significance between group 2 vs group 3; p = 0.044; *Significance between group 2 vs group 3; p = 0.001; ⁑ Significance between group 2 vs group 3; p = 0.015; ▀ Significance between group 2 vs group 3; p = 0.030; † Significance between group 2 vs group 4; p < 0.001; ♣ Significance between group 2 vs group 4; p = 0.009; ⁋ Significance between group 2 vs group 4; p = 0.047; ‡ Significance between group 2 vs group 4; p = 0.014; ∆: Significance between group 2 vs group 4; p = 0.046; ∏ Significance between group 2 vs group 4; p = 0.006; ◊ Significance between group 2 vs group 4; p = 0.016; ♦ Significance between group 3 vs group 4; p < 0.001.
BMI: body mass index; SBP: systolic blood pressure; DBP: diastolic blood pressure; DM: diabetes mellitus; eGFR: estimated glomerular filtration rate; hsCRP: high-sensitive C reactive protein; UACR: urinary albumin: creatinine ratio; KIM-1/creat: urinary kidney injury molecule-1:creatinine ratio; NAG/creat: urinary N-beta-D-acetyl-glucosaminidase/creat; Nephrin/creat: Nephrin/creatinine ratio; Podocalyxin/creat: Podocalyxin/creatinine ratio; Synaptopodin/creat: Synaptopodin/creatinine ratio; Hb: serum haemoglobin; HbA1C: glycated haemoglobin; p-miRNA: plasmatic microRNA; u-miRNA: urinary microRNA; u-lncRNA: urinary long noncoding RNA; p-lncRNA: plasmatic long noncoding RNA; MALAT-1: metastasis associated lung adenocarcinoma transcript 1 gene; MIAT: myocardial infarction associated transcript gene; NEAT: nuclear enrich abundant transcript gene; TUG: taurine upregulated gene.
Univariable regression analysis for urinary lnc-RNA
| Parameter | Variable | R2 | Coef β | P |
|---|---|---|---|---|
| u-lnc- MALAT-1 | u-miRNA-29a | 0.78 | -1.003 | <0.001 |
| p-miRNA-29a | 0.78 | -0.911 | <0.001 | |
| u-miRNA-21p | 0.62 | 1.295 | <0.001 | |
| p-miRNA-21p | 0.64 | 1.063 | <0.001 | |
| u-miRNA-124 | 0.73 | 0.885 | <0.001 | |
| p-miRNA-124 | 0.81 | 0.825 | <0.001 | |
| u-miRNA-93 | 0.34 | -0.678 | <0.001 | |
| p-miRNA-93 | 0.44 | -0.658 | <0.001 | |
| Podocalyxin/creat | 0.35 | 0.001 | <0.001 | |
| Synaptopodin/creat | 0.36 | 0.013 | <0.001 | |
| KIM-1/creat | 0.35 | 0.002 | <0.001 | |
| NAG/creat | 0.46 | 0.102 | <0.001 | |
| UACR | 0.34 | 0.001 | <0.001 | |
| eGFR | 0.25 | -0.018 | <0.001 | |
| u-lnc-NEAT | u-miRNA-29a | 0.47 | -0.473 | <0.001 |
| p-miRNA-29a | 0.59 | -0.476 | <0.001 | |
| u-miRNA-21p | 0.62 | 0.784 | <0.001 | |
| p-miRNA-21p | 0.57 | 0.602 | <0.001 | |
| u-miRNA-124 | 0.54 | 0.458 | <0.001 | |
| p-miRNA-124 | 0.52 | 0.400 | <0.001 | |
| u-miRNA-93 | 0.42 | -0.450 | <0.001 | |
| p-miRNA-93 | 0.40 | -0.379 | <0.001 | |
| Podocalyxin/creat | 0.62 | 0.001 | <0.001 | |
| Synaptopodin/creat | 0.59 | 0.010 | <0.001 | |
| KIM-1/creat | 0.61 | 0.001 | <0.001 | |
| NAG/creat | 0.40 | 0.058 | <0.001 | |
| UACR | 0.42 | 0.001 | <0.001 | |
| eGFR | 0.21 | -0.010 | <0.001 | |
| u-lnc-MIAT | u-miRNA-29a | 0.58 | 0.618 | <0.001 |
| p-miRNA-29a | 0.59 | 0.562 | <0.001 | |
| u-miRNA-21p | 0.47 | -0.806 | <0.001 | |
| p-miRNA-21p | 0.47 | -0.644 | <0.001 | |
| u-miRNA-124 | 0.55 | -0.549 | <0.001 | |
| p-miRNA-124 | 0.52 | -0.470 | <0.001 | |
| u-miRNA-93 | 0.19 | 0.361 | <0.001 | |
| p-miRNA-93 | 0.30 | 0.388 | <0.001 | |
| Podocalyxin/creat | 0.27 | -0.001 | <0.001 | |
| Synaptopodin/creat | 0.33 | -0.009 | <0.001 | |
| KIM-1/creat | 0.30 | -0.001 | <0.001 | |
| NAG/creat | 0.33 | -0.062 | <0.001 | |
| UACR | 0.38 | -0.001 | <0.001 | |
| eGFR | 0.17 | 0.011 | <0.001 | |
| u-lnc-TUG | u-miRNA-29a | 0.64 | 0.703 | <0.001 |
| p-miRNA-29a | 0.70 | 0.671 | <0.001 | |
| u-miRNA-21p | 0.65 | -1.030 | <0.001 | |
| p-miRNA-21p | 0.72 | -0.870 | <0.001 | |
| u-miRNA-124 | 0.70 | -0.671 | <0.001 | |
| p-miRNA-124 | 0.57 | -0.539 | <0.001 | |
| u-miRNA-93 | 0.55 | 0.659 | <0.001 | |
| p-miRNA-93 | 0.43 | 0.508 | <0.001 | |
| Podocalyxin/creat | 0.45 | -0.001 | <0.001 | |
| Synaptopodin/creat | 0.48 | -0.011 | <0.001 | |
| KIM-1/creat | 0.47 | -0.001 | <0.001 | |
| NAG/creat | 0.51 | -0.083 | <0.001 | |
| UACR | 0.48 | -0.001 | <0.001 | |
| eGFR | 0.11 | 0.009 | <0.001 |
UACR: urinary albumin:creatinine ratio; eGFR: estimated glomerular filtration rate; KIM-1/creat: urinary kidney injury molecule-1:creatinine ratio; NAG/creat: urinary N-beta-D-acetyl-glucosaminidase/creat; Podocalyxin/creat: Podocalyxin/creatinine ratio; Synaptopodin/creat: Synaptopodin/creatinine ratio; p-miRNA: plasmatic microRNA; u-miRNA: urinary microRNA; u-lncRNA: urinary long noncoding RNA; MALAT-1: metastasis associated lung adenocarcinoma transcript 1 gene; MIAT: myocardial infarction associated transcript gene; NEAT: nuclear enrich abundant transcript gene; TUG: taurine upregulated gene.
Univariable regression analysis for serum lnc-RNA
| Parameter | Variable | R2 | Coef β | P |
|---|---|---|---|---|
| p-lnc-MALAT-1 | u-miRNA-29a | 0.86 | -1.191 | <0.001 |
| p-miRNA-29a | 0.88 | -1.094 | <0.001 | |
| u-miRNA-21p | 0.71 | 1.568 | <0.001 | |
| p-miRNA-21p | 0.76 | 1.300 | <0.001 | |
| u-miRNA-124 | 0.83 | 1.067 | <0.001 | |
| p-miRNA-124 | 0.91 | 0.990 | <0.001 | |
| u-miRNA-93 | 0.47 | -0.895 | <0.001 | |
| p-miRNA-93 | 0.48 | -0.776 | <0.001 | |
| Podocalyxin/creat | 0.40 | 0.002 | <0.001 | |
| Synaptopodin/creat | 0.43 | 0.016 | <0.001 | |
| KIM-1/creat | 0.40 | 0.002 | <0.001 | |
| NAG/creat | 0.55 | 0.126 | <0.001 | |
| UACR | 0.40 | 0.002 | <0.001 | |
| eGFR | 0.35 | -0.025 | <0.001 | |
| p-lnc-NEAT | u-miRNA-29a | 0.51 | -0.527 | <0.001 |
| p-miRNA-29a | 0.62 | -0.528 | <0.001 | |
| u-miRNA-21p | 0.63 | 0.852 | <0.001 | |
| p-miRNA-21p | 0.56 | 0.648 | <0.001 | |
| u-miRNA-124 | 0.57 | 0.509 | <0.001 | |
| p-miRNA-124 | 0.56 | 0.447 | <0.001 | |
| u-miRNA-93 | 0.40 | -0.477 | <0.001 | |
| p-miRNA-93 | 0.41 | -0.416 | <0.001 | |
| Podocalyxin/creat | 0.63 | 0.001 | <0.001 | |
| Synaptopodin/creat | 0.62 | 0.011 | <0.001 | |
| KIM-1/creat | 0.60 | 0.001 | <0.001 | |
| NAG/creat | 0.42 | 0.063 | <0.001 | |
| UACR | 0.41 | 0.001 | <0.001 | |
| eGFR | 0.29 | -0.013 | <0.001 | |
| p-lnc-MIAT | u-miRNA-29a | 0.64 | 0.716 | <0.001 |
| p-miRNA-29a | 0.65 | 0.653 | <0.001 | |
| u-miRNA-21p | 0.55 | -0.959 | <0.001 | |
| p-miRNA-21p | 0.54 | -0.764 | <0.001 | |
| u-miRNA-124 | 0.60 | -0.630 | <0.001 | |
| p-miRNA-124 | 0.51 | -0.516 | <0.001 | |
| u-miRNA-93 | 0.26 | 0.458 | <0.001 | |
| p-miRNA-93 | 0.35 | 0.464 | <0.001 | |
| Podocalyxin/creat | 0.31 | -0.001 | <0.001 | |
| Synaptopodin/creat | 0.35 | -0.010 | <0.001 | |
| KIM-1/creat | 0.32 | -0.001 | <0.001 | |
| NAG/creat | 0.42 | -0.077 | <0.001 | |
| UACR | 0.35 | -0.001 | <0.001 | |
| eGFR | 0.09 | 0.009 | <0.001 | |
| p-lnc-TUG | u-miRNA-29a | 0.55 | 0.567 | <0.001 |
| p-miRNA-29a | 0.58 | 0.530 | <0.001 | |
| u-miRNA-21p | 0.53 | -0.804 | <0.001 | |
| p-miRNA-21p | 0.63 | -0.706 | <0.001 | |
| u-miRNA-124 | 0.59 | -0.536 | <0.001 | |
| p-miRNA-124 | 0.46 | -0.420 | <0.001 | |
| u-miRNA-93 | 0.48 | 0.537 | <0.001 | |
| p-miRNA-93 | 0.34 | 0.390 | <0.001 | |
| Podocalyxin/creat | 0.31 | -0.001 | <0.001 | |
| Synaptopodin/creat | 0.32 | -0.008 | <0.001 | |
| KIM-1/creat | 0.31 | -0.001 | <0.001 | |
| NAG/creat | 0.42 | -0.065 | <0.001 | |
| UACR | 0.35 | -0.001 | <0.001 | |
| eGFR | 0.05 | 0.006 | <0.001 |
UACR: urinary albumin:creatinine ratio; eGFR: estimated glomerular filtration rate; KIM-1/creat: urinary kidney injury molecule-1:creatinine ratio; NAG/creat: urinary N-beta-D-acetyl-glucosaminidase/creat; Podocalyxin/creat: Podocalyxin/creatinine ratio; Synaptopodin/creat: Synaptopodin/creatinine ratio; p-miRNA: plasmatic microRNA; u-miRNA: urinary microRNA; p-lncRNA: plasmatic long noncoding RNA; MALAT-1: metastasis associated lung adenocarcinoma transcript 1 gene; MIAT: myocardial infarction associated transcript gene; NEAT: nuclear enrich abundant transcript gene; TUG: taurine upregulated gene.
Multivariable regression analysis for urinary lnc-RNA
| Parameter | Variable | Coef β | P | 95% CI | Prob>F | R2 |
|---|---|---|---|---|---|---|
| u-lnc-MALAT-1 | Constant | 3.963 | 0.0001 | 3.750 to 4.175 | 0.00001 | 0.80 |
| u-miRNA-29a | -1.027 | 0.0001 | -1.149 to -0.906 | |||
| UACR | 0.0003 | 0.011 | 0.00007 to 0.0005 | |||
| eGFR | 0.004 | 0.007 | 0.001 to 0.008 | |||
| u-lnc-NEAT | Constant | 0.670 | 0.0001 | 0.327 to 1.013 | 0.00001 | 0.74 |
| u-miRNA-21p | 0.352 | 0.0001 | 0.189 to 0.516 | |||
| u-miRNA-93 | -0.103 | 0.011 | -0.182 to -0.024 | |||
| Podocalyxin/creat | 0.0006 | 0.0001 | 0.0003 to 0.0009 | |||
| KIM-1/creat | 0.0006 | 0.0001 | 0.0002 to 0.001 | |||
| UACR | -0.0003 | 0.003 | -0.0006 to -0.0001 | |||
| u-lnc-MIAT | Constant | 1.422 | 0.0001 | 1.191 to 1.653 | 0.00001 | 0.66 |
| u-miRNA-29a | 0.599 | 0.0001 | 0.496 to 0.702 | |||
| u-miRNA-93 | -0.217 | 0.0001 | -0.324 to -0.111 | |||
| UACR | -0.0007 | 0.0001 | -0.0009 to -0.0004 | |||
| u-lnc-TUG | Constant | 2.528 | 0.0001 | 1.828 to 3.229 | 0.00001 | 0.84 |
| u-miRNA-29a | 0.364 | 0.0001 | 0.184 to 0.544 | |||
| u-miRNA-124 | -0.274 | 0.005 | -0.465 to -0.083 | |||
| u-miRNA-93 | 0.272 | 0.0001 | 0.186 to 0.358 | |||
| KIM-1/creat | -0.0005 | 0.0001 | -0.0007 to -0.0003 | |||
| eGFR | -0.011 | 0.0001 | -0.013 to -0.008 |
UACR- urinary albumin:creatinine ratio; eGFR-estimated glomerular filtration rate; KIM-1/creat-urinary kidney injury molecule-1:creatinine ratio; Podocalyxin/creat - Podocalyxin/creatinine ratio; u-miRNA- urinary microRNA; u-lncRNA - urinary long noncoding RNA; MALAT-1 - metastasis associated lung adenocarcinoma transcript 1 gene; MIAT - myocardial infarction associated transcript gene; NEAT - nuclear enrich abundant transcript gene; TUG - taurine upregulated gene.
Multivariable regression analysis for serum lnc-RNA
| Parameter | Variable | Coef β | P | 95% CI | Prob>F | R2 |
|---|---|---|---|---|---|---|
| p-lnc-MALAT-1 | Constant | 1.183 | 0.002 | 0.447 to 1.919 | 0.00001 | 0.94 |
| p-miRNA-29a | -0.294 | 0.0001 | -0.445 to -0.144 | |||
| p-miRNA-124 | 0.864 | 0.0001 | 0.719 to 1.009 | |||
| eGFR | 0.007 | 0.0001 | 0.004 to 0.009 | |||
| p-lnc NEAT | Constant | 2.102 | 0.0001 | 1.888 to 2.316 | 0.00001 | 0.76 |
| p-miRNA-29a | -0.237 | 0.0001 | -0.332 to -0.142 | |||
| Podocalyxin/creat | 0.0008 | 0.0001 | 0.0005 to 0.001 | |||
| KIM-1 | 0.0006 | 0.001 | 0.0002 to 0.0009 | |||
| UACR | -0.0002 | 0.034 | -0.0005 to -0.00002 | |||
| eGFR | -0.002 | 0.048 | -0.005 to -0.00002 | |||
| p-lnc-MIAT | Constant | -0.089 | 0.815 | -0.842 to -0.663 | 0.00001 | 0.75 |
| p-miRNA-29a | 0.995 | 0.0001 | 0.812 to 1.177 | |||
| p-miRNA-21p | 0.264 | 0.015 | 0.052 to 0.476 | |||
| p-miRNA-93 | 0.108 | 0.021 | 0.016 to 0.200 | |||
| eGFR | -0.012 | 0.0001 | -0.016 to -0.009 | |||
| p-lnc-TUG | Constant | 3.026 | 0.0001 | 2.390 to 3.661 | 0.00001 | 0.73 |
| p-miRNA-29a | 0.461 | 0.0001 | 0.299 to 0.624 | |||
| p-miRNA-21p | -0.352 | 0.0001 | -0.536 to -0.167 | |||
| eGFR | -0.010 | 0.0001 | -0.012 to -0.007 |
UACR- urinary albumin:creatinine ratio; eGFR-estimated glomerular filtration rate; KIM-1/creat-urinary kidney injury molecule-1:creatinine ratio; Podocalyxin/creat - Podocalyxin/creatinine ratio; p-miRNA- plasmatic microRNA; p-lncRNA - plasmatic long noncoding RNA; MALAT-1 - metastasis associated lung adenocarcinoma transcript 1 gene; MIAT - myocardial infarction associated transcript gene; NEAT - nuclear enrich abundant transcript gene; TUG - taurine upregulated gene.
LncRNA MIAT and lncRNA TUG1 are downregulated and enhance miRNA 93 and miRNA 29a expression in DKD patients, and suppress miRNA 21 and miRNA 124 activities
In univariable linear regression analysis, we found a direct relationship between urinary and serum lncRNA MIAT, and urinary and serum miRNA 93 and miRNA 29a, as well as eGFR, and an indirect relationship with urinary and serum miRNA 21, miRNA 124, UACR, urinary podocalyxin, urinary synaptopodin, KIM-1, and NAG. Urinary and serum lncRNA-TUG1 displayed similar correlations with lncRNA MIAT (Table 2 and 3).
Multivariable regression analysis produced models in which urinary lncRNA MIAT correlated directly with urinary miRNA 93 and urinary miRNA 29a, and indirectly with UACR (R2= 0.66). Urinary lncRNA TUG1 correlated positively with urinary miRNA 93, miRNA 29a and eGFR, and indirectly with urinary miRNA 124 and KIM-1 (R2= 0.84) (Table 4).
Serum lncRNA MIAT displayed direct correlations with serum miRNA 93, miRNA 29a, and eGFR, and indirect correlations with serum miRNA 21 (R2= 0.75). Serum lncRNA TUG1 correlated positively with serum miRNA29a and eGFR, and negatively with serum miRNA 21 (R2= 0.73) (Table 5).
The results presented show that lncRNA MIAT and lncRNA TUG1 enhance the expression and potentiate the protective effects of miRNA 93 and miRNA 29a. Moreover, these lncRNAs display protective activities upon the podocytes and the PT, as supported by their indirect correlations with the podocyte injury and PT dysfunction biomarkers.
Discussion
Interactions among multiple molecules and signaling pathways contribute to the pathogenesis and progression of DKD. LncRNAs play key roles in the pathophysiology of DKD, involving actions of miRNAs. The present study aimed at establishing the involvement of selected lncRNAs in the epigenetic mechanisms of DKD.
We found that in patients with type 2 DM lncRNAs exert distinct functions in different cell types, either deleterious or protective, at both glomerular and tubular level. These properties were displayed through modulating miRNAs expression and activities.
LncRNA MALAT1 and lncRNA NEAT1 are dysregulated in DKD and involved in podocyte and PT injury via miRNAs modulation
LncRNA MALAT1 is highly expressed in the kidney [23] and significantly upregulated in early DKD [24]. This lncRNA is an initiator of inflammation and oxidative stress through upregulation of several inflammatory mediators [14, 25]. LncRNA MALAT1 could be attributed severe podocyte damage via its interplay with β-catenin, a key mediator of podocyte injury and renal fibrosis [15]. LncRNA MALAT1 is upregulated in PT cells [16, 26] and intervenes in PT dysfunction by regulating tubular cell viability [27, 23, 24].
In our study, lncRNA MALAT1 was upregulated and exerted negative effects upon the podocytes and the PT, as supported by the association with the levels of podocyte damage and PT dysfunction biomarkers.
LncRNAs regulate miRNAs through two major modes of regulation, either by enhancing or by inhibiting their function [28]. Profibrotic miRNAs, such as miRNA 21 [9] and miRNA 124, may exert a role in podocyte damage under mechanical stress [10]. In our study, serum and urinary miRNA 21 was upregulated and was associated with podocyte and PT injury biomarkers, a fact which points to its potential involvement within the glomerulus and the tubulo-interstitial compartment. These data are in keeping with other studies in which miRNA 21 has been ascribed pro-fibrotic and pro-inflammatory properties [6, 7, 8, 29]. The same trend was followed by miRNA 124 which may impair podocyte structural and functional integrity, and may induce tubulointerstitial fibrosis [6, 7, 8, 10]. LncRNA MALAT1 enhanced the activity of both miRNA 21 and miRNA 124 across all the groups of patients with type 2 DM studied, even in the early stage of DKD.
MiRNA 93 is downregulated in the podocytes and tubular cells of DKD patients [11, 30] and plays a key role in preserving podocyte integrity [30] and in preventing renal interstitial fibrosis in DKD [31]. Although miRNA 29 family displays intriguing roles in DKD, its expression being both increased and decreased [32], miRNA 29a is considered a renoprotective miRNA involved in inflammation-mediated podocyte and tubular injury under diabetic conditions [12, 33, 34, 35].
In our patients, lncRNA MALAT1 decreased the expression of miRNA 93 and miRNA 29a. This observation was concurrent with the increased levels of podocyte injury and of PT dysfunction biomarkers, even in normoalbuminuric patients.
LncRNA NEAT1 is known to intervene in fibrogenesis by extracellular matrix accumulation and epithelial-mesenchymal transition in DKD [36, 37]. In our study, lncRNA NEAT1 increased the expression of both miRNA 21 and miRNA 124, and decreased the activity of miRNA 93 and miRNA 29a, irrespective of UACR and level of renal function. This data is in keeping with the results derived from experimental [36] and clinical studies [37], which show that these effects of lncRNA NEAT1 are exerted through mediating miRNAs expression and activities.
The deleterious properties of lncRNA NEAT1 were extended at both glomerular and tubular level, as supported by its association with podocyte damage and PT dysfunction biomarkers. A recent study by Li N et al demonstrated that lncRNA NEAT1 facilitated proliferation, fibrosis, and epithelial-mesenchymal transition through sponging miRNA 23c in diabetic nephropathy. In their study the authors approached the PT dysfunction biomarkers, KIM-1 and NGAL, by assessing the expression of their mRNA in serum, which proved to be enhanced in parallel with lncRNA NEAT1 activities [38].
LncRNA MIAT and lncRNA TUG1 exert renoprotective functions in DKD through interactions with specific miRNAs
Among other lncRNAs which act as regulators of diabetic tubular injury in DKD [39, 40, 41], the expression of lncRNA MIAT was reduced and negatively correlated with serum creatinine and blood urea nitrogen levels [24, 42]. It is assumed that lncRNAs could be potential regulators of high glucose-induced renal tubular injury. In the study by Zhou L et al, lncRNA MIAT was decreased in the proximal tubules homogenate of STZ-induced diabetic kidneys from Wistar rats. LncRNA MIAT expression correlated negatively with the biomarkers of kidney function. The authors showed that lncRNA MIAT may regulate PT cell viability via stabilizing nuclear factor erythroid 2-related factor 2 expression, which is the key molecule of cellular defense against high blood glucose - induced oxidative stress [16].
In our study, lncRNA MIAT was downregulated and showed a strong association with decreased levels of podocyte injury and PT dysfunction biomarkers. We hypothesized that lncRNA MIAT could exert protective effects at both glomerular and tubular level. Furthermore, due to the statistically significant association of lncRNA MIAT with the enhanced expression of miRNA 93 and miRNA 29a, and with the decreased activity of miRNA 21 and miRNA 124, we speculated that lncRNA MIAT could modulate the activity of these miRNAs. As a direct consequence of lncRNA MIAT intervention, the protective miRNAs 93 and 29a could prevail, thus promoting the mechanism of defense against podocyte and PT injury from the early stages of DKD.
LncRNA TUG1 plays a key role in the initiation and progression of DKD [14]. LncRNA TUG1 is significantly repressed in the podocytes and produces metabolic alterations in these cells. This lncRNA regulates the peroxisome proliferator-activated receptor γ co-activator α (PGC-1α), thus rescuing this important member of the nuclear receptor superfamily [14].
LncRNA TUG1 participated in the regulation of mitochondrial function in podocytes in a murine model of DKD [14], phenomenon regulated by PGC-1α, known for its role in mitochondrial bioenergetics and respiration [43]. The modulation of mitochondrial metabolism by lncRNA TUG1 was documented by Long J et al in a mouse model through recruitment of PGC-1α to its own promoter [44]. By interacting with miRNAs, lncRNA TUG 1 may interfere with extracellular matrix accumulation and pro-inflammatory cytokines secretion in DKD [14].
In our study, lncRNA TUG1 paralleled the activities of lncRNA MIAT, thus pointing to its protective properties in the course of DKD, even in normoalbuminuric patients. Our results are in keeping with those provided by the experimental study by Wang F et al, in which the authors reported that lncRNA TUG1 can improve renal fibrosis in diabetic nephropathy by modulating miRNA 21 targeting tissue inhibitor of metalloproteinase 3 (TIMP 3). The intervention of lncRNA TUG1 via miRNA 21 modulation regulates TIMP3, known as a potent inhibitor of metalloproteinase 3 [45].
There are several limitations of our study. First, the small sample size of the patients studied may reduce the statistical power of the study. Second, cell-specific and tissue-specific expression and activities of miRNAs and lncRNAs could interfere interpretation of data with regard to their interactions. Finally, the study follows the design of a case series study, which requires validation in a longitudinal study, in order to prove a statistically significant relation of causality between the miRNAs and the lncRNAs studied.
The strength of our study derives from the documentation in a human study of an important association of lncRNAs which poses well defined roles in the pathogenesis of DKD, such as lncRNA MALAT1, NEAT1, MIAT, and TUG1, with miRNAs which have concurrent negative (miRNA 21, 124), or positive (miRNA 93, 29a) effects, respectively, even in the early stages of DKD.
In conclusion, in patients with type 2 DM, lncRNAs exert distinct functions in different cell types, either deleterious, as was the case with lncRNA MALAT1 and lncRNA NEAT1, or protective, as driven by lncRNA MIAT and lncRNA TUG1.
These properties were documented at both glomerular and PT level due to their strong association with podocyte damage and PT dysfunction biomarkers. LncRNAs may contribute to initiation and DKD progression through modulating miRNAs expression and activities.
The results of our study forward several lncRNAs and miRNAs as candidate biomarkers in the early diagnosis of DKD. The study is based on translational research from epigenetic mechanisms to clinical practice, which may enable a precision medicine approach to therapeutic strategies in the early stages of renal involvement in the course of DM. The interactions between these molecular pathways could provide a theoretical basis which may allow for a tailored therapy of DKD in its early stages.
Acknowledgements
Funding source
This research received funding from an Internal Grant of “Victor Babes” University of Medicine and Pharmacy Timisoara (grant number PIII-C5-PCFI-2017/2018).
Ethics Approval
This study was performed in line with the principles of the Declaration of Helsinki. The County Emergency Hospital Ethics Committee (Board of Human Studies) approved the protocol (approval number 8/10.01.2019), and every patient provided written informed consent before enrolment.
Consent to participate
All patients provided written informed consent before enrolment.
Availability of data and material
The datasets generated during and/or analysed during the current study are available from the corresponding author on reasonable request.
Author contributions
All authors contributed to the study conception and design. Material preparation, data collection and analysis were performed by Ligia Petrica, Elena Hogea, Florica Gadalean, Adrian Vlad, Mihaela Vlad, Victor Dumitrascu, Silvia Velciov, Cristina Gluhovschi, Flaviu Bob, Sorin Ursoniu, Dragos Catalin Jianu, Petru Matusz, Agneta-Maria Pusztai, Andrei Motoc, O.M.Cretu, Dana Radu, Oana Milas, Alina Golea-Secara, Anca Simulescu, Maria Mogos-Stefan, Mihaela Patruica, Lavinia Balint, Silvia Ienciu, Daliborca Vlad and Roxana Popescu. The first draft of the manuscript was written by PETRICA LIGIA and ELENA HOGEA and all authors commented on previous versions of the manuscript. All authors read and approved the final manuscript.
Competing Interests
The authors have declared that no competing interest exists.
References
1. United States Renal Data System. 2019 USRDS annual data report: Epidemiology of kidney disease in the United States. National Institutes of Health, National Institute of Diabetes and Digestive and Kidney Diseases, Bethesda, MD, 2019. https://doi.org/10.1053/j.ajkd.2019.09.003
2. Vallon V, Thomson SC. Renal function in diabetic disease models: the tubular system in the pathophysiology of the diabetic kidney. Annu Rev Physiol. 2012;74:351-375 DOI: 10.1146/annurev-physiol-020911-153333
3. Kato M. Noncoding RNAs as therapeutic targets in early-stage diabetic kidney disease. Kidney Res Clin Pract. 2018;37:197-209
4. Bhatt K, Lanting L, Jia Y, Yadav S, Reddy M, Magilnick N. et al. Anti-inflammatory role of microRNA-146a in the pathogenesis of diabetic nephropathy. J Am Soc Nephrol. 2016;27:2277-2288 DOI: 10.1681/ASN.2015010111
5. Trionfini P, Benigni A. MicroRNAs as master regulators of glomerular function in health and disease. J Am Soc Nephrol. 2017;28:1686-1696 DOI: 10.1681/ASN.2016101117
6. Petrica L, Pusztai A-M, Vlad M, Vlad A, Gadalean F, Dumitrascu V. et al. MiRNA Expression is Associated with Clinical Variables Related to Vascular Remodeling in the Kidney and the Brain in Type 2 Diabetes Mellitus Patients. Endocr Res. 2020;45:119-130
7. Petrica L, Milas O, Vlad M, Vlad A, Gadalean F, Dumitrascu V. et al. Interleukins and miRNAs intervene in the early stages of diabetic kidney disease in Type 2 diabetes mellitus patients. Biomark Med. 2019;13(18):1577-1588 DOI: 10.2217/bmm-2019-0124
8. Milas O, Gadalean F, Vlad A, Dumitrascu V, Gluhovschi C, Gluhovschi G. et al. Deregulated profiles of urinary micro-RNAs may explain podocyte injury and proximal tubule dysfunction in normoalbuminuric patients with type 2 diabetes mellitus. J Investig Med. 2018;66:747-754 DOI: 10.1136/jim-2017-000556
9. Wang J, Gao Y, Ma M, Li M, Zou D, Yang J. et al. Effect of miR-21 on renal fibrosis by regulating MMP-9 and TIMP1 in kk-ay diabetic nephropathy mice. Cell Biochem Biophys. 2013;67:537-546 DOI: 10.1007/s12013-013-9539-2
10. Li D, Lu Z, Jia J, Zheng Z, Lin S. Changes in microRNAs associated with podocytic adhesion damage under mechanical stress. J Renin Angiotensin Aldosterone Syst. 2013;14:97-102 DOI: 10.1177/1470320312460071
11. Badal SS, Wang Y, Long J, Corcoran DL, Chang BH, Truong LD. et al. MiR-93 regulates Msk2-mediated chromatin remodelling in diabetic nephropathy. Nat Commun. 2016;17:12076. DOI: 10.1038/ncomms12076
12. Wang B, Komers R, Carew R, Winbanks CE, Xu B, Herman-Edelstein M. et al. Suppression of microRNA-29 expression by TGF-β1 promotes collagen expression and renal fibrosis. J Am Soc Nephrol. 2012;23:252-265 DOI: 10.1681/ASN.2011010055
13. Kapranov P, Cheng J, Dike S, Nix DA, Duttagupta R, Willingham AT. et al. RNA maps reveal new RNA classes and a possible function for pervasive transcription. Science. 2007;316:1484-1488 DOI: 10.1126/science.1138341
14. Li Y, Xu K, Xu K, Chen S, Cao Y, Zhan H. Roles of identified long noncoding RNA in diabetic nephropathy. J Diabetes Res. 2019;2019:5383010 DOI 10.1155/2019/5383010
15. Hu M, Wang R, Li X, Fan M, Lin J, Zhen J. et al. LncRNA MALAT1 is dysregulated in diabetic nephropathy and involved in high glucose-induced podocyte injury via its interplay with β-catenin. J Cell Mol Med. 2017;21:2732-2747 DOI: 10.1111/jcmm.13189
16. Zhou L, Xu DY, Sha WG, Shen L, Lu GY, Yin X. Long non-coding MIAT mediates high glucose-induced renal tubular epithelial injury. Biochem Biophys Res Commun. 2015;468:726-732 DOI: 10.1016/j.bbrc.2015.11.023
17. Kashi K, Henderson L, Bonetti A, Carninci P. Discovery and functional analysis of lncRNAs: Methodologies to investigate an uncharacterized transcriptome. Biochim Biophys Act. 2016;1(859):3-15 DOI: 10.1016/j.bbagrm.2015.10.010
18. Paraskevopoulou MD, Hatzigeorgiou AG. Analyzing MiRNA-LncRNA interactions. Methods in Molecular Biology. 2016;1402:271-286 DOI: 10.1007/978-1-4939-3378-5_21
19. Petrica L, Vlad A, Gluhovschi G, Gadalean F, Dumitrascu V, Vlad D. et al. Glycated peptides are associated with the endothelial variability in the kidney and cerebral vessels in type 2 diabetes mellitus patients. J Diabetes Complications. 2015;29:230-237
20. Petrica Ligia, Sorin Ursoniu, Florica Gadalean, Adrian Vlad, Gheorghe Gluhovschi, Victor Dumitrascu. et al. Urinary podocyte-associated mRNA levels correlate with proximal tubule dysfunction in early diabetic nephropathy of type 2 diabetes mellitus. Diabetology and Metabolic Syndrome. 2017;9:31-43 DOI: 10.1016/j.jdiacomp.2014.11.014
21. KDIGO Clinical Practice Guideline for the Evaluation and Management of Chronic Kidney Disease. Kidney Int. 2013; 3(Suppl): 1-150.
22. Schmittgen TD, Livak KJ. Analyzing real-time PCR data by the comparative CT method. Nature Protocols. 2008;3:1101-1108 DOI: 10.1038/nprot.2008.73
23. Zhou X, Liu S, Cai G, Kong L, Zhang T, Ren Y. et al. Long noncoding RNA MALAT1 promotes tumor growth and metastasis by inducing epithelial-mesenchymal transition in oral squamous cell carcinoma. Scientific Reports. 2015;5:15972 DOI: 10.1038/srep15972
24. Yan B, Tao ZF, Li XM, Zhang H, Yao J, Jiang Q. Aberrant expression of long noncoding RNAs in early diabetic retinopathy. Investigative Ophthalmology and Visual Science. 2014;55:941-951 DOI: 10.1167/iovs.13-13221
25. Puthanveetil P, Chen S, Feng B, Gautam A, Chakrabarti S. Long Non-coding RNA MALAT1 regulates hyperglycaemia induced inflammatory process in the endothelial cells. J Cell Mol Me. 2015;19(6):1418-1425 DOI: 10.1111/jcmm.12576
26. Liu JY, Yao J, Li XM, Song YC, Wang XQ, Li YJ. et al. Pathogenic role of lncRNA-MALAT1 in endothelial cell dysfunction in diabetes mellitus. Cell Death Dis. 2014;5:e1506 DOI: 10.1038/cddis.2014.466
27. Li X, Zeng L, Cao C, Lu C, Lian W, Han J. et al. Long noncoding RNA MALAT1 regulates renal tubular epithelial pyroptosis by modulated miR-23c targeting of ELAVL1 in diabetic nephropathy. Exp Cell Res. 2017;350:327-335 DOI: 10.1016/j.yexcr.2016.12.006
28. Thomson DW, Dinger ME. Endogenous microRNA sponges: evidence and controversy. Nat Rev Genet. 2016;17:272-283 DOI: 10.1038/nrg.2016.20
29. Zhong X, Chung AC, Chen HY, Dong Y, Meng XM, Li R. et al. MiR-21 is a key therapeutic target for renal injury in a mouse model of type 2 diabetes. Diabetologia. 2013;56:663-674 https://doi.org/10.1007/s00125-012-2804-x
30. Long J, Wang Y, Wang W, Chang B, Danesh F. Identification of microRNA-93 as a novel regulator of vascular endothelial growth factor in hyperglycemic conditions. J Biol Chem. 2010Jul23;285:23457-23465 DOI: 10.1074/jbc.M110.136168
31. Yang J, Shen Y, Yang X, Long Y, Chen S, Lin X. et al. Silencing of long noncoding RNA XIST protects against renal interstitial fibrosis in diabetic nephropathy via microRNA-93-5p-mediated inhibition of CDKN1A. Am J Physiol Renal Physiol. 2019;317:F1350-F1358 DOI: 10.1152/ajprenal.00254.2019
32. Qin W, Chung A, Huang X, Meng XM, Hui D, Yu CM. et al. TGF-β/Smad3 signaling promotes renal fibrosis by inhibiting miR-29. J Am Soc Nephrol. 2011;22:1462-147 DOI: 10.1681/ASN.2010121308
33. Sun SF, Tang PHK, Feng M, Xiao J, Huang XR, Li P. et al. Novel lncRNA Erbb4-IR promotes diabetic kidney injury in db/db mice by targeting miR-29b. Diabetes. 2018;67:731-744 DOI: 10.2337/db17-0816
34. Chen HY, Zhong X, Huang XR, Meng XM, You Y, Chung ACK. et al. MicroRNA-29b inhibits diabetic nephropathy in db/db mice. Mol Ther. 2014;22:842-853 DOI: 10.1038/mt.2013.235
35. Lin CL, Lee PH, Hsu YC, Lei CC, Ko JY, Chuang, P C. et al. MicroRNA-29a promotion of nephrin acetylation ameliorates hyperglycemia-induced podocyte dysfunction. J Am Soc Nephrol. 2014;25:1698-1709 DOI: 10.1681/ASN.2013050527
36. Wang X, Xu Y, Zhu YC, Wang YK, Li J, Li XY. et al. LncRNA NEAT1 promotes extracellular matrix accumulation and epithelial-to-mesenchymal transition by targeting miR-27b-3p and ZEB1 in diabetic nephropathy. J Cell Physiol. 2019;234:12926-12933 DOI: 10.1002/jcp.27959
37. Huang S, Xu Y, Ge X, Xu B, Peng W, Jiang X. et al. Long noncoding RNA NEAT1 accelerates the proliferation and fibrosis in diabetic nephropathy through activating Akt/mTOR signaling pathway. J Cell Physiol. 2019;234:11200-11207 DOI: 10.1002/jcp.27770
38. Li N, Jia T, Li YR. LncRNA NEAT1 accelerates the occurrence and development of diabetic nephropathy by sponging miR-23c. Eur Rev Med Pharmacol Sci. 2020;24(3):1325-1337 doi: 10.26355/eurrev_202002_20190
39. Leti F, DiStefano JK. Long Noncoding RNAs as diagnostic and therapeutic targets in type 2 diabetes and related complications. Genes (Basel). 2017;8:207 DOI: 10.3390/genes8080207
40. He X, Ou C, Xiao Y, Han Q, Li H, Zhou S. LncRNAs: key players and novel insights into diabetes mellitus. Oncotarget. 2017;8:71325-71341 DOI: 10.18632/oncotarget.19921
41. Raut SK, Khullar M. The big entity of new RNA world: Long non-coding RNAs in microvascular complications of diabetes. Front Endocrinol (Lausanne). 2018;9:300. DOI: 10.3389/fendo.2018.00300
42. Sun C, Huang L, Li Z, Leng K, Xu Y, Jiang X. et al. Long non-coding RNA MIAT in development and disease: a new player in an old game. J Biomed Sci. 2018;25:23 DOI: 10.1186/s12929-018-0427-3
43. Hock MB, Kralli A. Transcriptional control of mithochondrial biogenesis and function. Annu Rev Pathol. 2009;71:177-2023 DOI: 10.1146/annurev.physiol.010908.163119
44. Long J, Badal SS, Ye Z, Wang Y, Ayanga BA, Galvan DL. et al. Long noncoding RNA Tug1 regulates mitochondrial bioenergetics in diabetic nephropathy. J Clin Invest. 2016;126:4205-4218 DOI: 10.1172/JCI87927
45. Wang F, Gao X, Zhang R, Zhao P, Sun Y, Li C. LncRNA TUG1 ameliorates diabetic nephropathy by inhibiting miR-21 to promote TIMP3-expression. Int J Clin Exp Pathol. 2019;12(3):717-729 PMID: 319338791
Author contact
![]() Corresponding author: Florica Gadalean; Tel.: +407 4134 6890; Fax: +402 5637 6908; fnnicoricicom. Roxana Popescu, popescurxncom
Corresponding author: Florica Gadalean; Tel.: +407 4134 6890; Fax: +402 5637 6908; fnnicoricicom. Roxana Popescu, popescurxncom

 Global reach, higher impact
Global reach, higher impact