Impact Factor
ISSN: 1449-1907
Int J Med Sci 2021; 18(8):1739-1745. doi:10.7150/ijms.51235 This issue Cite
Research Paper
Machining learning predicts the need for escalated care and mortality in COVID-19 patients from clinical variables
1. Department of Family, Population and Preventive Medicine, Renaissance School of Medicine at Stony Brook University, Stony Brook, New York, United States of America.
2. Department of Radiology, Renaissance School of Medicine at Stony Brook University, Stony Brook, New York, United States of America.
3. Department of Radiology, Montefiore Medical Center and Albert Einstein College of Medicine, Bronx, New York, United States of America.
Received 2020-7-28; Accepted 2021-1-21; Published 2021-2-18
Abstract
Objective: This study aimed to develop a machine learning algorithm to identify key clinical measures to triage patients more effectively to general admission versus intensive care unit (ICU) admission and to predict mortality in COVID-19 pandemic.
Materials and methods: This retrospective study consisted of 1874 persons-under-investigation for COVID-19 between February 7, 2020, and May 27, 2020 at Stony Brook University Hospital, New York. Two primary outcomes were ICU admission and mortality compared to COVID-19 positive patients in general hospital admission. Demographic, vitals, symptoms, imaging findings, comorbidities, and laboratory tests at presentation were collected. Predictions of mortality and ICU admission were made using machine learning with 80% training and 20% testing. Performance was evaluated using receiver operating characteristic (ROC) area under the curve (AUC).
Results: A total of 635 patients were included in the analysis (age 60±11, 40.2% female). The top 6 mortality predictors were age, procalcitonin, C-creative protein, lactate dehydrogenase, D-dimer and lymphocytes. The top 6 ICU admission predictors are procalcitonin, lactate dehydrogenase, C-creative protein, pulse oxygen saturation, temperature and ferritin. The best machine learning algorithms predicted mortality with 89% AUC and ICU admission with 79% AUC.
Conclusion: This study identifies key independent clinical parameters that predict ICU admission and mortality associated with COVID-19 infection. The predictive model is practical, readily enhanced and retrained using additional data. This approach has immediate translation and may prove useful for frontline physicians in clinical decision making under time-sensitive and resource-constrained environment.
Keywords: coronavirus 2 (SARS-CoV-2), pneumonia, artificial intelligence, lung infection
Introduction
Coronavirus Disease 2019 (COVID-19) [1-3], first reported in Wuhan, China in December 2019, was declared a pandemic on March 11, 2020 by the World Health Organization [4]. As of June 26, 2020, COVID-19 has already infected 10 million and killed over 500,000 individuals worldwide [5]. The actual numbers are likely to be much higher due to testing kit shortages and potential under-reporting. The sudden outbreak and rapid spread of COVID-19 have strained hospital resources, such as personal protective equipment, intensive care unit beds and mechanical ventilators. There will likely be a second wave and recurrence [6]. There are currently no established prognostic biomarkers to accurately predict which patients are at imminent risk of death or require immediate escalated care, making resource allocation difficult. This challenge is further magnified by the large number of clinical lab markers being affected by COVID-19 infection (see reviews [7-9]), the incompletely understood disease course, and the heterogeneous presentations. For example, some patients have mild or asymptomatic infections, while others exhibit severe symptoms. Some patients exhibit a mild disease course, while others deteriorate rapidly with multi-organ failure. Together, these challenges underscore the need to effectively triage and manage the care of COVID-19 patients, particularly in resource-constrained environments. There is currently no consensus as to which clinical variables are predictive of mortality and the needs for escalated care.
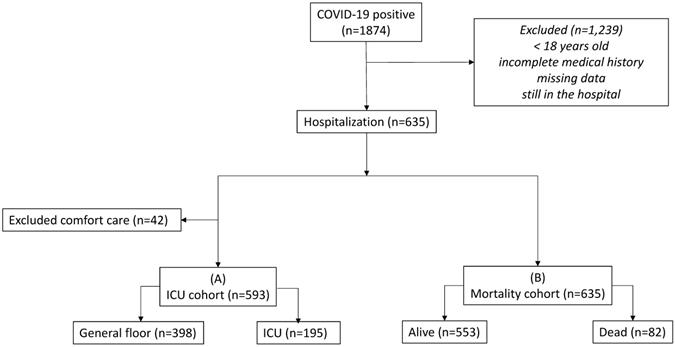
Flowchart of patient selection.
A few studies have attempted to develop models to predict mortality and disease severity based on a large array of clinical variables associated with COVID-19 infection [10-14]. Most of these prediction studies investigated patients from China, used logistic regression, and had small sample size and small number of clinical variables. Machine learning (ML) is increasingly being used in medicine, because of its ability to analyze large number of variables [15-17]. ML uses computer algorithms to learn relationships amongst different data elements to inform outcomes without the need to explicitly specify the exact relationship, in contrast to conventional analysis methods. Many studies have shown that machine learning methods outperform humans in many tasks in medicine [18]. With increasing computing power and growing relevance of big data in medicine, ML is expected to play an important role in clinical practice.
The goal of this study was to develop and compare different machine learning algorithms to predict the likelihood of ICU admission and mortality in COVID-19 patients. We identified the top few variables amongst the large array of clinical variables that were most predictive of the likelihood of ICU admission and mortality.
Methods
Patient population and data description
The retrospective study was approved by the Human Subjects Committee with an exemption for informed consent and HIPAA waiver. The data were collected from Electronic Medical Record and REDCAP database of the COVID-19 Patient under Investigation (PUI) registry of Stony Brook Hospital from March 9, 2020 to May 27, 2020. A subset of this dataset has been analyzed for a different purpose previously [19]. The inclusion criteria were patients tested positive for severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) and admitted to the hospital. Exclusion criteria were patients younger than 21 years old, patients still in the hospital, and missing data.
Figure 1 shows the flowchart. There were 1874 patients tested positive for COVID-19. After applying the inclusion and exclusion criteria, 635 COVID-19 positive patients were used in our analysis. In the alive versus dead comparison, there were 553 alive and 82 dead. In the general floor versus ICU comparison, patients treated for comfort care were excluded (n=42) because these patients would have been sent to ICU if they were full code. There were 195 admitted to ICU and 398 to general floor. This ICU group included direct ICU admission and upgraded from general floor to ICU.
The input variables included demographic information (age, gender, ethnicity and race), chronic comorbidities (smoking, diabetes, hypertension, asthma, chronic obstructive pulmonary disease (COPD), coronary artery disease, heart failure, cancer, immunosuppression and chronic kidney disease), vital signs (heart rate, respiratory rate, pulse oxygen saturation (SpO2), systolic blood pressure and temperature), and laboratory tests at admission (alanine aminotransferase (ALT), brain natriuretic peptide, C-creative protein (CRP), D-dimer, ferritin, lactate dehydrogenase (LDH), leukocytes, lymphocytes, procalcitonin and troponin). Demographic information and chronic comorbidities were collected at admission to the Emergency Department.
Machine learning model building
Two separate models were constructed for evaluation using different machine learning algorithms: 1) death vs discharged alive; 2) ICU vs general admission. Categorical predictors (e.g. ethnicity and race) were coded as dummy variables and continuous predictor (e.g. age, vital signs and laboratory tests) were standardized before machine learning. Multiple imputation with predictive mean matching method was used to impute missing values in vital signs and laboratory tests using the Multivariate Imputation by Chained Equations in R (statistical analysis software 4.0) [20]. No imputation was performed for a predictor with missing more than 15%. Brain natriuretic peptide and troponin had more than 30% missing and were excluded from the analyses. As a result, 24 predictors were included as input features for machine learning models.
Logistic regression and four machine learning algorithms were considered for prediction models: random forest, Xgboost, kernel support vector machine (SVM) and neural network (packages “randomForest”, “xgboost”, “caret”, and “h2o” in R Statistical Analysis software, v4.0). Breiman's algorithm was performed for the random forest model. The number of trees to grow was set at a large number of 500. The minimum node size is set as 1 for the dichotomous classification and the maximum node size is not limited. In Xgboost models, gbtree was used for gradient boosting and the logistic regression objective function was used for classification. The learning rate parameter eta was set at 0.2 since the typical range for eta is from 0.01 to 0.3 as a lower value requires more computation and more iterations. The maximum number of iterations for gradient descent converge was set 100. Linear, polynomial and radial kernel were explored for SVM and the optimal kernel method was selected based on the prediction performance. As a result, the linear kernel achieved the highest AUC under ROC curves among the three kernel methods and were used for both mortality and ICU models. For neural network model, rectifier activation function for deep learning was used for classification. Two layers and ten nodes for each layer were set for the initiative model and the number of data iteration was set at 100 times.
Feature importance was determined using different methods in different machine learning algorithms. Random forest ranks predictors by mean decrease in Gini index [21]. The corrected permutation approach was performed in the following two steps. First, the outcome was permuted 50 times and a nonparametric null distribution of feature importance was obtained, and second, the random forests with selected significant features were obtained. In Xgboost, mean decrease in Gini index and gain in the improvement in accuracy is used to evaluate the contribution of each predictor and neural network uses the Gedeon method for feature importance [22]. To select top important predictors, 1000 rounds of permutation tests was performed. In each round, the original dataset was randomly split into training and testing sets with a ratio of 80%:20%. Predictors were then ranked by their importance in each of the four machine learning models. Percentages of times ranked on top 5 over 1000 permutation tests for each predictor were then calculated and used to determine the final rank of feature importance. Prediction performance was evaluated by area under the curve (AUC) of the receiver operating characteristic (ROC) curve for the test data set, sensitivity, and specificity. The average prediction performance was obtained with 1000 runs.
Results
There were 635 COVID-19 positive hospitalized patients in our analysis. Descriptive statistics of demographics, chronic comorbidities, vital signs, and laboratory tests are presented. Table 1 shows the comparison between the discharged alive versus dead group. Age, race, Hispanic were significantly different (p<0.05) between groups. Coronary artery disease, COPD, CKD, hypertension, heart failure, and smoking were significantly different between group (p<0.05), except asthma, carcinoma, diabetes and immunosuppression (p>0.05). CRP, D-dimer, LDH, leukocytes, lymphocytes, procalcitonin, SpO2, respiration rates and temperature were statistically different between groups (p<0.05), but not ALT, ferritin, heart rate, and SBP (p>0.05).
Table 2 shows the comparison ICU vs general floor admission group. Patients treated for comfort care were excluded from this group (n=42) because these patients would have been sent to ICU if they were full code. Coronary artery disease, COPD, carcinoma, CKD, hypertension, heart failure, and smoking were significantly different between group (p<0.05), but not asthma, diabetes and immunosuppression (p>0.05). CRP, D-dimer, ferritin, LDH, leukocytes, lymphocytes, procalcitonin, SpO2, respiration rates, and temperature were statistically different between groups (p<0.05), but not ALT, heart rate, and SBP (p>0.05).
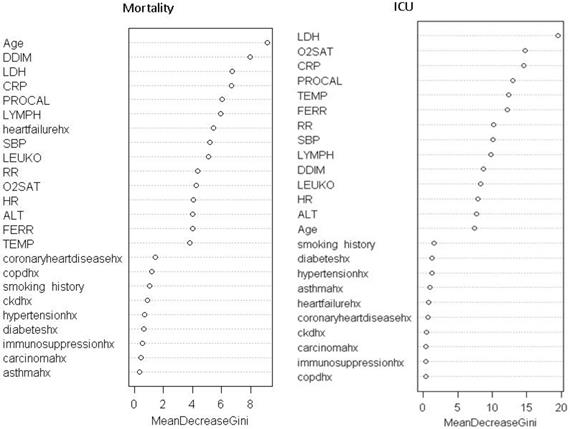
Feature rank of random forest models.
Demographic, comorbidity, laboratory findings by survived and non-survived group
| Survived (n=553) | Non-Survived (n=82) | P value | |
|---|---|---|---|
| Demographics | |||
| Age, mean (std), y | 57.71 (16.76) | 73.62 (13.53) | <0.0001 |
| Male | 327 (59.1%) | 53 (64.6%) | 0.343 |
| Race | 0.002 | ||
| Caucasian | 251 (45.4%) | 53 (64.6%) | |
| African American | 38 (6.87%) | 7 (8.5%) | |
| Other | 264 (47.7%) | 22 (26.8%) | |
| Hispanic | 169 (30.6%) | 8 (9.8%) | <0.0001 |
| Comorbidity | |||
| Asthma | 38 (6.9%) | 3 (3.7%) | 0.269 |
| Coronary artery disease | 59 (10.7%) | 25 (30.5%) | <0.0001 |
| Chronic obstructive pulmonary disease | 31 (5.6%) | 15 (18.3%) | <0.0001 |
| Carcinoma | 32 (5.8%) | 7 (8.5%) | 0.333 |
| Chronic kidney disease | 37 (6.7%) | 14 (17.1%) | 0.001 |
| Diabetes | 148 (26.8%) | 25 (30.5%) | 0.48 |
| Hypertension | 243 (43.9%) | 52 (63.4%) | 0.001 |
| Heart failure | 12 (2.2%) | 22 (26.8%) | <0.0001 |
| Immunosuppression | 31 (5.6%) | 8 (9.8%) | 0.144 |
| Smoking | 118 (21.3%) | 36 (43.9%) | <0.0001 |
| Laboratory test and vital sign | |||
| Alanine aminotransferase, U/L | 45.05 (48.22) | 44.81 (54.07) | 0.967 |
| C-reactive protein, mg/L | 10.22 (8.74) | 15.69 (10.53) | <0.0001 |
| D-dimer, nmol/L | 835.89 (3519.6) | 3037.9 (7692.1) | <0.0001 |
| Ferritin, μg/L | 1130.7 (1425.3) | 1350.0 (1877.6) | 0.255 |
| Lactate dehydrogenase, U/L | 381.24 (170.30) | 489.38 (242.88) | <0.0001 |
| Leukocytes×109 /liter | 7.86 (4.21) | 9.22 (5.41) | 0.009 |
| Lymphocytes % | 15.35 (9.21) | 11.72 (10.21) | 0.001 |
| Procalcitonin, ng/mL | 0.80 (4.75) | 4.18 (22.28) | 0.002 |
| Heart Rate, bpm | 101.58 (66.72) | 98.94 (21.39) | 0.722 |
| Pulse oxygen saturation, % | 92.88 (7.01) | 90.32 (8.63) | 0.003 |
| Respiratory rate, rate/min | 21.95 (6.76) | 24.67 (8.29) | 0.001 |
| SBP, mmHg | 127.61 (24.43) | 129.30 (29.83) | 0.57 |
| Temperature, °C | 37.68 (0.90) | 37.33 (0.78) | 0.002 |
P values are based on Chi-square test or two-sample t-test.
Predictive performance
Predictive performance of each machine learning algorithms is shown in Table 3. The AUC of the mortality model ranged from 84% to 89%. The AUC of the ICU model ranged from 72% to 78%. Specificity was generally better than sensitivity. Random forest and Xgboost achieved better prediction AUC both in mortality and ICU models than the support vector machine and neural network. Random forest has the highest AUC for mortality (89%) and Xgboost has high AUC for ICU (79%).
Feature importance
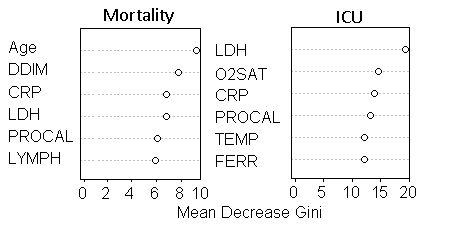
To evaluate the contribution of each predictor, predictors were ranked by their importance through 1000 the permutation tests. Table 4 shows the feature ranking of all clinical variables based on individual AUCs and the permutation tests using logistic regression and machine learning algorithms. For all predictive models of mortality, the top common predictors were consistent across model and they were age, procalcitonin, C-creative protein, lactate dehydrogenase, and D-dimer. For all predictive models of ICU admission, the top common predictors were comparatively less consistent across models and they were procalcitonin, lactate dehydrogenase, C-creative protein, pulse oxygen saturation, ferritin and temperature. The common top predictors of mortality and ICU admission were procalcitonin, LDH, CRP, and SpO2.
Demographic, comorbidity, laboratory findings by general floor and ICU group
| General floor (n=398) | ICU (n=195) | P value | |
|---|---|---|---|
| Demographics | |||
| Age, mean (std), y | 57.68 (17.57) | 59.70 (14.82) | 0.168 |
| Male | 222 (55.8%) | 136 (67.9%) | 0.001 |
| Race | 0.080 | ||
| Caucasian | 190 (47.7%) | 81 (41.5%) | |
| African American | 32 (8%) | 10 (5.1%) | |
| Other | 176 (44.2%) | 104 (53.3%) | |
| Hispanic | 118 (29.6%) | 56 (28.7%) | 0.220 |
| Comorbidity | |||
| Asthma | 25 (6.3%) | 16 (8.2%) | 0.386 |
| Coronary artery disease | 46 (11.6%) | 22 (11.3%) | 0.921 |
| Chronic obstructive pulmonary disease | 25 (6.3%) | 11 (5.6%) | 0.759 |
| Carcinoma | 25 (6.3%) | 9 (4.6%) | 0.412 |
| Chronic kidney disease | 28 (7.0%) | 16 (8.2%) | 0.610 |
| Diabetes | 104 (26.1%) | 58 (29.7%) | 0.354 |
| Hypertension | 170 (42.7%) | 96 (49.2%) | 0.134 |
| Heart failure | 10 (2.5%) | 10 (5.1%) | 0.097 |
| Immunosuppression | 22 (5.5%) | 12 (6.2%) | 0.758 |
| Smoking | 56 (14.1%) | 49 (25.1%) | 0.0009 |
| Laboratory test and vital sign | |||
| Alanine aminotransferase, U/L | 44.01 (48.97) | 48.19 (46.74) | 0.321 |
| C-reactive protein, mg/L | 8.21 (7.54) | 15.44 (10.29) | <0.0001 |
| D-dimer, nmol/L | 864.91 (3863.9) | 939.58 (2103.2) | 0.801 |
| Ferritin, μg/L | 882.11 (1275.8) | 1469.2 (1401.1) | <0.0001 |
| Lactate dehydrogenase, U/L | 340.30 (148.89) | 481.81 (191.99) | <0.0001 |
| Leukocytes×109 /liter | 7.57 (4.06) | 8.73 (4.57) | 0.002 |
| Lymphocytes % | 16.73 (9.57) | 12.25 (8.37) | <0.0001 |
| Procalcitonin, ng/mL | 0.59 (2.61) | 2.66 (15.76) | 0.011 |
| Heart Rate, bpm | 99.23 (48.71) | 107.09 (87.96) | 0.163 |
| Pulse oxygen saturation, % | 94.43 (3.68) | 88.92 (10.52) | <0.0001 |
| Respiratory rate, rate/min | 20.81 (5.78) | 25.03 (8.45) | <0.0001 |
| SBP, mmHg | 128.58 (23.44) | 126.46 (29.25) | 0.342 |
| Temperature, °C | 37.80 (3.34) | 37.75 (0.95) | 0.839 |
P values are based on Chi-square test or two-sample t-test.
Predictive performance of machine learning algorithms
| Algorithms | Mortality | ICU | ||||
|---|---|---|---|---|---|---|
| AUC (SD) | Sensitivity | Specificity | AUC (SD) | Sensitivity | Specificity | |
| Random Forests | 89.0% (1.3%) | 76.4% | 89.5% | 78.1% (3.1%) | 73.4% | 79.6% |
| Xgboost | 88.4% (1.9%) | 30.3% | 96.8% | 78.9% (2.9%) | 54.2% | 86.0% |
| Kernel SVM | 87.8% (4.3%) | 20.8% | 99.5% | 76.1% (2.2%) | 43.3% | 92.3% |
| Neural network | 84.4% (2.6%) | 36.8% | 90.3% | 71.8% (4.4%) | 56.7% | 77.8% |
| Logistic | 81.7% | 26.2% | 90.1% | 66.9% | 55.3% | 78.5% |
Discussion
This study investigated different machine learning algorithms to predict the likelihood of ICU admission and mortality in COVID-19 patients using clinical characteristics and laboratory results at admissions. The top 6 mortality predictors were age, procalcitonin, C-creative protein, lactate dehydrogenase, D-dimer and lymphocytes. The top 6 ICU admission predictors are procalcitonin, lactate dehydrogenase, C-creative protein, pulse oxygen saturation, ferritin and temperature. The best machine learning algorithms predicted mortality with 0.88 AUC and ICU admission with 0.79 AUC.
Significant features of logistic regression (p<0.05) and top 6 features of machine learning algorithms for mortality and ICU models
| Logistic | Random Forests | Xgboost | SVM | Neural Network |
|---|---|---|---|---|
| Mortality | ||||
| Age | Age | Age | Age | CKD |
| Heart Failure | D-Dimer | D-Dimer | D-Dimer | Ferritin |
| LDH | CRP | COPD | Procalcitonin | Age |
| CRP | LDH | CRP | CRP | Respiratory Rate |
| Hypertension | Procalcitonin | Procalcitonin | Lymphocytes | ALT |
| Immunosuppression | Lymphocytes | LDH | LDH | LDH |
| ICU | ||||
| CRP | Procalcitonin | LDH | Procalcitonin | SpO2 |
| LDH | LDH | Procalcitonin | LDH | Heart Rate |
| SpO2 | CRP | CRP | CRP | Age |
| Heart Failure | SpO2 | SpO2 | Ferritin | Ferritin |
| Smoking | Temperature | Temperature | SpO2 | LDH |
| SBP | Ferritin | Ferritin | lymphocytes | CRP |
ALT: Alanine aminotransferase, LDH: lactate dehydrogenase, CRP: C-reactive protein, SBP: systolic blood pressure. COPD: chronic obstructive pulmonary.
Most of the top predictors of mortality and ICU admission overlapped and they were procalcitonin, LDH, CRP, and SpO2. Procalcitonin is usually found to be elevated during bacterial infections, and less so during viral infection. Its elevation in critically ill COVID-19 patients could suggest the occurrence of potential bacterial co-infections or a decreased host immune response, both leading to worse outcome in these patients [23]. LDH reflects tissue damage and has been known to be elevated in COVID-19 infection and other lung infections [2, 3]. Elevated CRP, a blood marker of inflammation, suggests inflammatory response and tissue damage in the body [24]. The surge of inflammation and the associated cytokine storm as a consequence have been associated with worse outcomes in COVID-19. Low SpO2 indicates failure of the lungs to oxygenate blood effectively, leading to hypoxia and respiratory failures that lead to mortality.
Some top predictors also differed for mortality and ICU admission. Age and D-dimer were uniquely associated with mortality whereas temperature and ferritin were uniquely associated with ICU admission. It is not surprising that old age is associated with a higher mortality. D-dimer, a small fragment protein by-product from breaking down blood clot, is indicative of a hyper-coagulability as a result of severe inflammatory reaction [25]. These findings could explain why elevated D-dimer is associated with high mortality rate. On the other hand, high temperature might result in higher likelihood of ICU admission but not mortality. Similarly, elevated ferritin is a marker of aberrant iron metabolism that could render the lungs susceptible to oxidative damage [26]. These findings suggest that abnormal temperature and ferritin are likely to cause severe disease requiring ICU care, but might be “treatable” or reversible as they are not significant predictors of mortality. Further research is needed to elucidate effective treatments.
It is also interesting to note that symptoms, comorbidities, and race/ethnicity were not amongst the 6 top predictors of mortality and ICU admission, although some of these variables have been associated with mortality or critical illness previously [27-30]. Symptoms are subjective and highly variable, thus it is not surprising they were not highly ranked [31]. A few studies have previously reported that patients with multiple comorbidities [27, 29] and certain race/ethnicity [28, 30] showed higher mortality rate or more likely to need escalated care. Comorbidity did not rank high on our cohort relative to other variables, likely because of the small sample sizes or that the clinical variables were indeed more predictive. Note that previous studies did not directly compare comorbidities and other clinical variables, and thus it is not known or not well established whether comorbidities are more predictive of mortality or of the need for escalated care relative to other clinical variables. Our sample sizes on the race and ethnicity cohorts were likely insufficient to reach meaningful conclusion. Further studies are warranted.
Another novelty of our study is that we performed analysis using 5 different predictive models. Random forests and Xgboost showed higher prediction accuracy than SVM and neural network. The random forests algorithms performed better than the neural networks likely due to small sample size. Overall, we found that all models predicted mortality better than ICU admission. Top common predictors were more consistent across different machine-learning models for predicting mortality than ICU admission. This is not unexpected as ICU admission decision were likely more variable because of how each physician practices and of how the pandemic has progressed temporally.
Although all these top predictors have been previously associated with COVID-19 infection [1-3], only a few studies have attempted to develop methods to predict mortality and disease severity. Lu et al. created a three-tiered risk score based on only two variables, age and CRP thresholds, to determine mortality [10], Xie et al. reported age, lymphocyte count, LDH and SpO2 to be independent predictors of mortality [11]. Ji et al. predicted stable versus progressive COVID-19 patients based on whether their conditions worsened during hospitalization [12]. They reported comorbidity, older age, lower lymphocyte and higher LDH at presentation to be independent high-risk factors for COVID-19 progression. A nomogram of these 4 factors yielded a concordance index of 0.86. Jiang et al. found mildly elevated alanine aminotransferase, myalgias, and hemoglobin at presentation to be predictive of severe acute respiratory distress syndrome (ARDS) of COVID-19 with 70% to 80% accuracy [14]. However, this study had small and non-uniform clinical variables from different hospitals. Although some of the predictors of mortality were shared amongst these and our studies, there is currently no consensus as to which clinical variables are most predictive of mortality or the needs for escalated care. These differences in findings could be due to different outcome measures (mortality, ARDS, and disease severity), patient cohorts, different disease severity at admission, hospital environment, and analysis methods employed, among other factors. Our study differed from previously studies in several ways. We employed ML, in contrast to the majority of previous studies which used logistic regression. Our models identified top 6 predictors that accurately predicted both the needs for escalated care and mortality. We also compared different ML methods. Our study included comparatively large sample size and is amongst the few that described a patient cohort in the United States to date.
This study has several limitations. Although our study had a reasonably large sample size from a major academic hospital in New York, a temporal epicenter of the COVID-19 pandemic, it is a retrospective study carried out in a single hospital. These findings need to be replicated in large and multi-institutional settings for generalizability. We only analyzed clinical variables at admission. Longitudinal changes of these clinical variables need to be studied. It is important to note that the COVID-19 pandemic circumstance is unusual and evolving. ICU admission of COVID-19 patients may depend on individual hospital's patient load, practice, and available resources, which also differ amongst countries. Our institution to date has not been constrained by number of ICU beds nor mechanical ventilators in this study time frame. As in all observational studies, other residual confounders may exist that were not accounted for in our analysis. Future prospective studies validating our predictive models are warranted.
Conclusion
We implemented and compared different machine learning algorithms to predict the likelihood of ICU admission and mortality in COVID-19 patients. This approach has the potential to provide frontline physicians with an objective tool to manage COVID-19 patients more effectively in time-sensitive, stressful and potentially resource-constrained environments.
Key points
- Question: What are the top clinical parameters that predict ICU admission and mortality associated with COVID-19 infection?
- Findings: The top 6 mortality predictors were age, procalcitonin, C-creative protein, lactate dehydrogenase, D-dimer and lymphocytes. The top 6 ICU admission predictors are procalcitonin, lactate dehydrogenase, C-creative protein, pulse oxygen saturation, temperature and ferritin. The best machine learning algorithms predicted mortality with 0.88 AUC and ICU admission with 0.79 AUC.
- Meaning: This predictive model accurately predicts ICU admission and mortality in COVID-19 infection. It may prove useful for frontline physicians in clinical decision making.
Acknowledgements
We thank all healthcare professionals for their hard work being at the front line of the pandemic. All authors declare no conflict of interest, including financial interests, activities, relationships, and affiliations. This research did not receive any specific grant from funding agencies in the public, commercial, or not-for-profit sectors.
Competing Interests
The authors have declared that no competing interest exists.
References
1. Li Q, Guan X, Wu P, Wang X, Zhou L, Tong Y. et al. Early Transmission Dynamics in Wuhan, China, of Novel Coronavirus-Infected Pneumonia. N Engl J Med. 2020;382(13):1199-207
2. Zhu N, Zhang D, Wang W, Li X, Yang B, Song J. et al. A Novel Coronavirus from Patients with Pneumonia in China, 2019. N Engl J Med. 2020;382(8):727-33
3. Huang C, Wang Y, Li X, Ren L, Zhao J, Hu Y. et al. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet. 2020;395(10223):497-506
4. https://www.who.int/emergencies/diseases/novel-coronavirus-2019/situation-reports/
5. https://coronavirus.jhu.edu/map.html
6. Leung K, Wu JT, Liu D, Leung GM. First-wave COVID-19 transmissibility and severity in China outside Hubei after control measures, and second-wave scenario planning: a modelling impact assessment. The Lancet. 2020;395(10233):1382-93
7. Rodriguez-Morales AJ, Cardona-Ospina JA, Gutiérrez-Ocampo E, Villamizar-Peña R, Holguin-Rivera Y, Escalera-Antezana JP. et al. Clinical, laboratory and imaging features of COVID-19: A systematic review and meta-analysis. Travel medicine and infectious disease. 2020 p:101623
8. Brown RAC, Barnard J, Harris-Skillman E, Harbinson B, Dunne B, Drake J. et al. Lymphocytopaenia is associated with severe SARS-CoV-2 disease: A Systematic Review and Meta-Analysis of Clinical Data. medRxiv. 2020. 2020 04.14.20064659
9. Cao Y, Liu X, Xiong L, Cai K. Imaging and Clinical Features of Patients With 2019 Novel Coronavirus SARS-CoV-2: A systematic review and meta-analysis. Journal of Medical Virology. 2020;3:03
10. Lu J, Hu S, Fan R, Liu Z, Yin X, Wang Q. et al. ACP risk grade: a simple mortality index for patients with confirmed or suspected severe acute respiratory syndrome coronavirus 2 disease (COVID-19) during the early stage of outbreak in Wuhan, China. medRxiv. 2020
11. Xie J, Hungerford D, Chen H, Abrams ST, Li S, Li X. et al. Development and external validation of a prognostic multivariable model on admission for hospitalized patients with COVID-19. medRxiv. 2020 in press
12. Ji D, Zhang D, Xu J, Chen Z, Yang T, Zhao P. et al. Prediction for Progression Risk in Patients with COVID-19 Pneumonia: the CALL Score. Clin Infect Dis. 2020
13. Hu H, Yao N, Qiu Y. Comparing Rapid Scoring Systems in Mortality Prediction of Critically Ill Patients With Novel Coronavirus Disease. Acad Emerg Med. 2020
14. Jiang X, Coffee M, Bari A, Wang J, Jiang X, Huang J. et al. Towards an Artificial Intelligence Framework for Data-Driven Prediction of Coronavirus Clinical Severity. Computers, Materials & Continua. 2020;63:537-51
15. Deo RC. Machine Learning in Medicine. Circulation. 2015;132(20):1920-30
16. Santos MK, Ferreira Junior JR, Wada DT, Tenorio APM, Barbosa MHN, Marques PMA. Artificial intelligence, machine learning, computer-aided diagnosis, and radiomics: advances in imaging towards to precision medicine. Radiol Bras. 2019;52(6):387-96
17. Hwang TJ, Kesselheim AS, Vokinger KN. Lifecycle Regulation of Artificial Intelligence- and Machine Learning-Based Software Devices in Medicine. JAMA. 2019
18. Killock D. AI outperforms radiologists in mammographic screening. Nat Rev Clin Oncol. 2020;17(3):134
19. Singer AJ, Morley E, Meyers K, Fernandes R, Rowe AL, Viccellio P. et al. Cohort of 4404 Persons Under Investigation for COVID-19 in a NY Hospital and Predictors of ICU Care and Ventilation. Annals of Emergency Medicine. 2020 in press
20. van Buuren S, Groothuis-Oudshoorn K. mice: Multivariate Imputation by Chained Equations in R. Journal of Statistical Software. 2011;45(3):1-67
21. Altmann A, Tolosi L, Sander O, Lengauer T. Permutation importance: a corrected feature importance measure. Bioinformatics. 2010;26(10):1340-7
22. Gedeon TD. Data mining of inputs: analysing magnitude and functional measures. Int J Neural Syst. 1997;8(2):209-18
23. Assicot M, Gendrel D, Carsin H, Raymond J, Guilbaud J, Bohuon C. High serum procalcitonin concentrations in patients with sepsis and infection. Lancet. 1993;341(8844):515-8
24. Gabay C, Kushner I. Acute-phase proteins and other systemic responses to inflammation. N Engl J Med. 1999;340(6):448-54
25. Griffin DO, Jensen A, Khan M, Chin J, Chin K, Saad J. et al. Pulmonary Embolism and Increased Levels of d-Dimer in Patients with Coronavirus Disease. Emerg Infect Dis. 2020 26(8)
26. Mumby S, Upton RL, Chen Y, Stanford SJ, Quinlan GJ, Nicholson AG. et al. Lung heme oxygenase-1 is elevated in acute respiratory distress syndrome. Crit Care Med. 2004;32(5):1130-5
27. Nandy K, Salunke A, Pathak SK, Pandey A, Doctor C, Puj K. et al. Coronavirus disease (COVID-19): A systematic review and meta-analysis to evaluate the impact of various comorbidities on serious events. Diabetes Metab Syndr. 2020;14(5):1017-25
28. Inal J. COVID-19 comorbidities, associated pro-coagulant extracellular vesicles and venous thromboembolisms: a possible link with ethnicity? Br J Haematol. 2020
29. Parveen R, Sehar N, Bajpai R, Bharal Agarwal N. Association of diabetes and hypertension with disease severity in covid-19 patients: a systematic literature review and exploratory meta-analysis. Diabetes Res Clin Pract. 2020 108295
30. Wilder JM. The Disproportionate Impact of COVID-19 on Racial and Ethnic Minorities in the United States. Clin Infect Dis. 2020
31. Leung C. Risk factors for predicting mortality in elderly patients with COVID-19: A review of clinical data in China. Mech Ageing Dev. 2020;188:111255
Author contact
![]() Corresponding authors: E-mail: wei.houedu; or, tim.duongorg.
Corresponding authors: E-mail: wei.houedu; or, tim.duongorg.

 Global reach, higher impact
Global reach, higher impact