3.2
Impact Factor
ISSN: 1449-1907
Int J Med Sci 2021; 18(6):1348-1355. doi:10.7150/ijms.52181 This issue Cite
Research Paper
Association of Polymorphism rs1045411 in the HMGB1 Gene with Cancer Risk: Evidence from a Meta-analysis
1. Department of Clinical Laboratory, The Third Affiliated Hospital of Kunming Medical University, Kunming 650118, China.
2. Department of Internal Medicine, The People's Hospital of Guandu District, Kunming 650200, China.
Received 2020-8-19; Accepted 2021-1-4; Published 2021-1-21
Abstract
The high-mobility group box protein 1 (HMGB1) rs1045411 polymorphism has been demonstrated to be associated with cancer risk in some studies. However, the results regarding this topic are inconsistent. A meta-analysis was applied to elucidate the association between the HMGB1 rs1045411 polymorphism and cancer risk. Ten relevant studies were subjected to our analysis, and pooled odds ratios (ORs) and 95% confidence intervals (CIs) were calculated. In total, of 3,918 cases and 5,296 controls were included in this study. The pooled ORs were calculated using a random-effects or fixed-effects model according to the heterogeneity. The pooled results revealed that TT genotype was significantly related to increased cancer risk in the comparisons of TT vs. CC+TC (OR=1.35; 95% CI: 1.09-1.67; p=0.005). Though no statistical significance was achieved between HMGB1 rs1045411 polymorphism and cancer risk in other four genetic models (T vs. C: OR=1.08, 95% CI 0.90-1.30; TC vs. CC: OR=1.01, 95% CI 0.82-1.24; CC vs. TC+TT: OR=0.95, 95% CI 0.77-1.18; TT vs. CC: OR=1.42; 95% CI 0.98-2.05), a trend of increased risk could be drawn. In the subgroup analysis by type of malignancy and ethnicity, no obvious difference was found in the tumour risk regarding the HMGB1 rs1045411 polymorphism amongst the cancer types except for breast cancer (OR=1.94; 95% CI: 1.05-3.59; p=0.03) and hepatocellular carcinoma (OR=1.82; 95% CI: 1.15-2.88; p=0.01), while rs1045411 polymorphism was positively associated with risks of cancer amongst Hans (OR=1.37; 95% CI: 1.11-1.69; p=0.004) rather than Caucasians (OR=0.89; 95% CI: 0.26-3.02; p=0.01). These results suggest that the HMGB1 rs1045411 polymorphism might be associated with increased cancer risk.
Keywords: HMGB1, polymorphisms, meta-analysis
Introduction
Cancer is the most frequently diagnosed disease in the world, and the exact mechanisms of which remain unclear [1, 2]. Most studies have demonstrated that multiple genetic and epigenetic changes are involved in cancer development [3]. Therefore, studying the genetic and molecular mechanisms of cancer can help reveal the development process and predict the risk of cancer [1, 4]. Previously, reports have indicated that genetic variation plays an important role in cancer susceptibility and development. By genotyping single-nucleotide polymorphisms (SNPs), the distribution frequency of SNPs among cases and controls can be compared [5, 6]. Some reports have shown an association between the SNPs rs1045411 and cancer risk; however, the results are controversial [7, 8].
As a highly conserved nuclear protein, HMGB1 functions as a chromatin structural protein in the nucleus or pro-inflammatory cytokine extracellularly [9, 10]. As a non-histone DNA-binding protein, nuclear HMGB1 promotes the assembly of site-specific DNA targets [11]. By contrast, extracellular HMGB1 acts as a damage-associated molecular pattern that serves as a key ingredient in many diseases such as inflammatory diseases and tumors [12]. Previously, we also reported that elevated HMGB1 levels are associated with lung cancer [9]. Additionally, accumulating evidences suggests that high HMGB1 expression is closely related to the development and progression of cancer through its important functions in promoting proliferation, invasion and migration [13-15]. However, little is known regarding the effects of HMGB1 gene variants on cancer.
As HMGB1 rs1045411 polymorphism is closely correlated with altered binding of miR-505-5P in the 3'-UTR of mRNA transcripts, HMGB1 gene polymorphisms could emerge as a crucial player in cancer development through a post-transcriptional mechanism [7]. Chromosomal instability is considered important in the pathogenesis of cancer, and the HMGB1 loss can reduce telomerase activity, decrease telomere length, and increase chromosomal instability [16-19]. Thus, understanding the molecular bases of HMGB1 might be important for exploring its precise role in cancer [7]. Until now, many case-control studies have been carried out to explore the relevance of the HMGB1 polymorphism rs1045411 to cancer. However, due to the limitations of study design, such as a small sample size and lower statistical power, these studies have reported inconsistent results [1, 12, 20-27]. A meta-analysis to summarise the inconsistent results from the relevant studies may provide evidence for the correlation between the HMGB1 rs1045411 polymorphism and cancer risk.
Methods
Literature search and data extraction
Articles published up to April 2020 from PubMed, Embase, Wanfang Data Knowledge Service Platform and China National Knowledge Infrastructure were searched using the terms HMGB1 polymorphisms, with no language restrictions. The studies included in this meta-analysis were original studies that reported odds ratios (ORs) with 95% confidence intervals (CIs) or provided useful data to calculate ORs and 95% CIs. In this meta-analysis, all studies were independently verified against the inclusion and exclusion criteria by two investigators. Useful information was extracted from each included study. Allele frequencies were calculated from the corresponding genotype distributions when they were not given (n T= n TT×2+nCT, n C= n CC×2+nCT). These processes were also carried out independently by two investigators (Xia and Tao).
Statistical analysis
Pooled ORs and 95% CIs were calculated for allele contrast model (T vs. C), heterozygote model (TC vs. CC), homozygote model (TT vs. CC), dominant model (TT vs. CC+TC) and recessive model (CC vs. TC + TT) by using STATA (v. 16.0; STATACORP LP, College Station, TX, USA) and Review Manager Software (v.5.2; The Nordic Cochrane Centre, The Cochrane Collaboration, Copenhagen, Denmark), respectively. Additionally, χ2-based Q statistics and I2 metrics were used to assess the heterogeneity between studies. When I2<50%, a fixed-effects model was used to calculate the pooled OR; otherwise, a random-effects model was used.
Results
A database that included each paper's first author, country, sample size, genotyping method and other useable information was set up based on the information extracted from 10 relevant studies that met the inclusion criteria (Table 1). Our original search yielded a total of 88 articles related to our keywords. Figure 1 summarizes the selection process of this study. After titles, key words and abstracts were screened, 69 of these articles were excluded. The full texts of 19 articles were reviewed, and an additional 9 articles were excluded (with 8 articles excluded for not providing usable data and 1 article excluded due to the duplication of the same article in different languages); thus, 10 studies remained for further review. One study [24] whose distribution of genotype deviated from Hardy-Weinberg equilibrium (HWE) (p HWE < 0.05) in the control was also included in this study but was excluded from the sensitivity analysis.
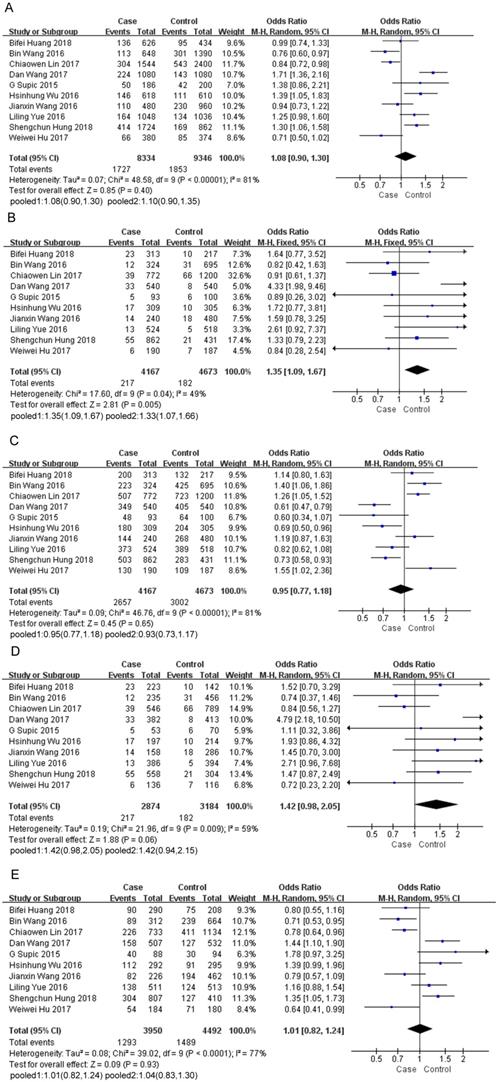
In total, of 3,918 cases and 5,296 controls were included in this study. The pooled ORs were calculated using a random-effects or fixed-effects model in terms of heterogeneity (Table 2). The pooled results demonstrated that HMGB1 rs1045411 polymorphism emerge as a risk factor for cancer, as a significant association between increased cancer risk and TT genotype was indicated in the comparison of the TT vs. CC+TC genotype (OR=1.35; 95% CI: 1.09-1.67; p=0.005). The same result was also detected by excluding one study [24] deviated from HWE in the comparison of the TT vs. CC+TC genotype (OR=1.33; 95% CI: 1.07-1.66; p=0.01) (Figure 2B). For the T vs. C genotype (OR=1.08; 95% CI: 0.90-1.30; p=0.40; Figure 2A) or CC vs. TC+TT genotype (OR=0.95; 95% CI: 0.77-1.18; p=0.65; Figure 2C) or TT vs. CC genotype (OR=1.42; 95% CI: 0.98-2.05; p=0.06; Figure 2D) or TC vs. CC genotype (OR=1.01; 95% CI: 0.82-1.24; p=0.93; Figure 2E), no significant association was detected, although the pooled ORs did not reach statistical significance in these four genetic models, a trend of increased risk could be drawn (Table 2).
Flow diagram of the search and selection process in this study.
Characteristics of case-control studies included in the meta-analysis
| Sample Size | p value for HWE in control | ||||
|---|---|---|---|---|---|
| Study | Country/Area | Case | Control | Genotyping method | rs1045411C >T |
| G Supic 201520 | Caucasian | 93 | 100 | Taqman | 0.33786 |
| Bin Wang 201621 | China | 324 | 695 | Taqman | 0.72217 |
| Liling Yue 201622 | China | 524 | 518 | Ligase-PCR | 0.15262 |
| Hsinhung Wu 201623 | Taiwan | 309 | 305 | Taqman | 0.96957 |
| Jianxin Wang 201624 | China | 240 | 480 | PCR-RFLP | 0.0167 |
| Weiwei Hu 20171 | China | 372 | 379 | Taqman | 0.26819 |
| Dan Wang 201725 | China | 540 | 540 | Ligase-PCR | 0.5826 |
| Chiaowen Lin 201726 | China | 772 | 1200 | Taqman | 0.45078 |
| Bifei Huang 201827 | China | 313 | 217 | Taqman | 0.87461 |
| Shengchun Hung 201812 | Taiwan | 431 | 862 | Taqman | 0.32333 |
HWE, Hardy-Weinberg equilibrium
Meta-analysis of the HMGB1 rs1045411 polymorphism and cancer risk
| Sample size | Studies(n) | Random or fixed-effects model | Test of heterogeneity | |||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Polymorphism | Study | Case | Control | OR (95%CI) | Z | p value | χ2 | p value | I2 | |
| T vs. C | Overalla | 1726 | 1843 | 10 | 1.08(0.90,1.30) | 0.85 | 0.40 | 48.58 | 0.00001 | 81% |
| T vs. C | In HWEb | 1616 | 1623 | 9 | 1.10(0.90,1.35) | 0.92 | 0.36 | 47.66 | 0.00001 | 83% |
| TT vs. CC+TC | Overalla | 217 | 182 | 10 | 1.35(1.09,1.67) | 2.81 | 0.005 | 17.6 | 0.04 | 49% |
| TT vs. CC+TC | In HWEb | 203 | 164 | 9 | 1.33(1.07,1.66) | 2.56 | 0.01 | 17.31 | 0.03 | 54% |
| CC vs. TC+TT | Overalla | 2657 | 3002 | 10 | 0.95(0.77,1.18) | 0.45 | 0.65 | 46.76 | 0.00001 | 81% |
| CC vs. TC+TT | In HWEb | 2513 | 2734 | 9 | 0.93(0.73,1.17) | 0.63 | 0.53 | 45 | 0.00001 | 82% |
| TT vs. CC | Overalla | 217 | 182 | 10 | 1.42(0.98,2.05) | 1.88 | 0.06 | 21.96 | 0.009 | 59% |
| TT vs.CC | In HWEb | 203 | 164 | 9 | 1.42(0.94,2.15) | 1.68 | 0.09 | 21.87 | 0.005 | 63% |
| TC vs. CC | Overalla | 1293 | 1489 | 10 | 1.01(0.82,1.24) | 0.09 | 0.93 | 39.02 | 0.00001 | 77% |
| TC vs. CC | In HWEb | 1211 | 1295 | 9 | 1.04(0.83,1.30) | 0.34 | 0.74 | 36.87 | 0.00001 | 78% |
a All of the studies. b Excluding the study deviated from Hardy-Weinberg equilibrium (HWE).
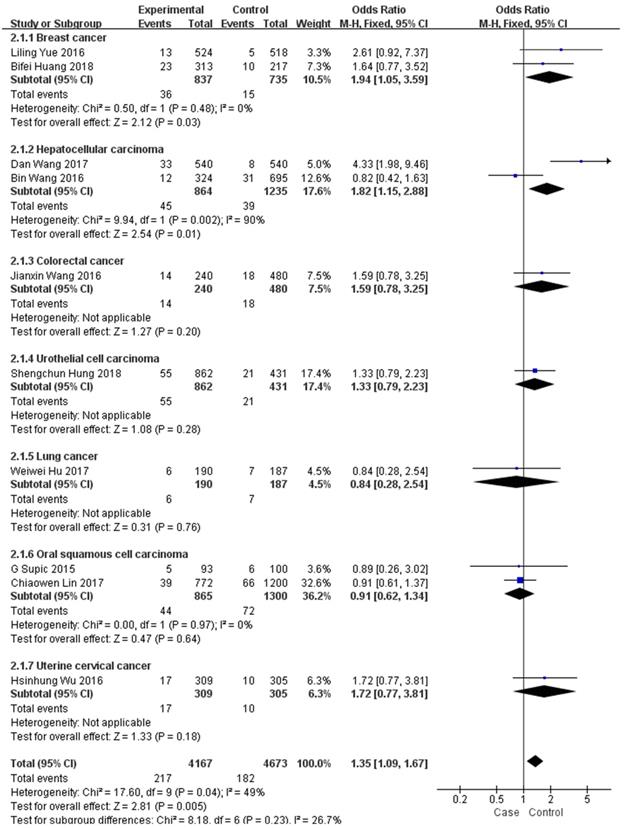
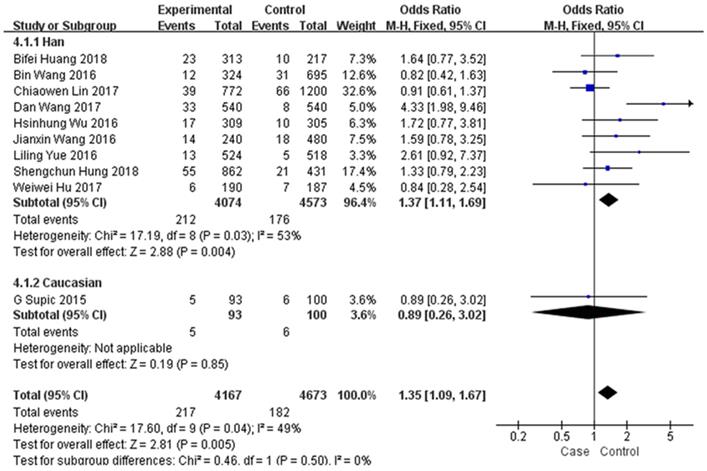
In the subgroup analysis by type of malignancy, no obvious difference was found in the tumour risk regarding the HMGB1 rs1045411 polymorphism amongst the cancer types (Colorectal cancer: OR=1.59; 95% CI: 0.78-3.25; p=0.20; Urothelial cell carcinoma : OR=1.33; 95% CI: 0.79-2.23; p=0.28; Lung cancer: OR=0.84; 95% CI: 0.28-2.54; p=0.76; Oral squamous cell carcinoma: OR=0.91; 95% CI: 0.62-1.34; p=0.64; Uterine cervical cancer: OR=1.72; 95% CI: 0.77-3.81; p=0.18) except for breast cancer (OR=1.94; 95% CI: 1.05-3.59; p=0.03) and hepatocellular carcinoma (OR=1.82; 95% CI: 1.15-2.88; p=0.01) in the dominant model (Figure 3). Next, subgroup analysis of rs1045411 stratified by ethnic groups was also conducted and fixed-effects model was used in the dominant genetic model. Our results demonstrated that rs1045411 polymorphism was positively associated with risks of cancer amongst Hans (OR=1.37; 95% CI: 1.11-1.69; p=0.004) rather than Caucasians (OR=0.89; 95% CI: 0.26-3.02; p=0.85; Figure 4).
Forest plot of the meta-analysis for the association of the HMGB1 rs1045411 allele distribution with cancer risk by comparing T vs. C under the random-effects model (A). TT vs. CC+TC under the fixed-effects model (B). CC vs. TC+TT under the random-effects model(C). TT vs. CC under the random-effects model (D). TC vs. CC under the random-effects model (E). (1) Including all of the 10 studies. (2) Excluding the study deviated from Hardy-Weinberg equilibrium. Abbreviations: CI, confidence interval; IV, inverse variance.
Forest plot of rs1045411 in HMGB1 gene and risk of cancer: subgroup analysis by cancer type using the dominant model.
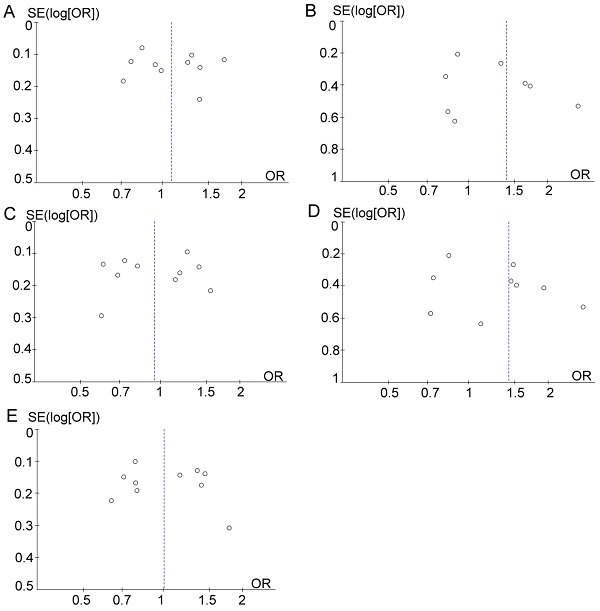
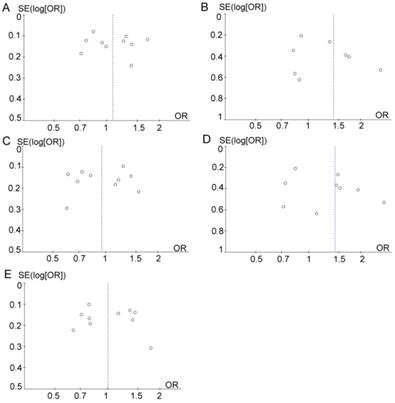
Sensitivity analysis was performed by removing one study at a time to assess the stability of these results. After the removal of Wang et al. study [25], the resulting heterogeneity across studies decreased from moderate heterogeneity (χ2 = 17.31; df= 8; p=0.03; I2= 54%) to low (χ2 = 7.14; df = 7; p=0.41; I2= 2%) in the dominant model (TT vs. CC+TC). However, after eliminating the Wang et al. study [25], the pooled ORs were not distinctly changed, with stable results. Funnel plots were drawn to determine the risk of bias, and they were symmetric (Figure 5), indicating the absence of publication bias. Finally, STATA software was used to perform Egger's test to calculate publication bias. No publication bias was assessed via Egger's test, which was conducted to provide statistical evidence for funnel plot symmetry (p=0.578 for T vs. C; p=0.268 for TT vs. CC+TC; p=0.982 for CC vs. TC+TT; p=0.253 for TT vs. CC; p=0.583 for TC vs. CC).
Discussion
During the past few years, some studies have reported the association between HMGB1 polymorphisms (rs2249825, rs1045411, rs1412125 and rs1360485) and different cancer types [25]. After reviewing lots of literatures on HMGB1 polymorphisms, a great deal of literature and information indicate that the HMGB1 rs1045411 polymorphism might be most likely associated with increased cancer risk, though the results are controversial. Hence, a meta-analysis to summarise the inconsistent results from the relevant studies may provide evidence for the correlation between HMGB1 rs1045411 polymorphism and cancer risk. In total, of 3,918 cases and 5,296 controls were included in this study to reveal the correlation between HMGB1 rs1045411 polymorphism and cancer risk. To our knowledge, this meta-analysis represents the largest study of its kind to date. And our results reveal a positive relationship between HMGB1 rs1045411 polymorphism and cancer risk.
Forest plot of rs1045411 in HMGB1 gene and risk of cancer: subgroup analysis by ethnicity using the dominant model.
Funnel plot analysis to detect publication bias. (A) T vs. C. (B) TT vs. CC+TC. (C) CC vs. TC+TT. (D) TT vs. CC. (E) TC vs. CC.
HMGB1 is a tumour-related gene [28], and its overexpression of HMGB1 is associated with the hallmarks of cancer [29], such as unlimited replicative potential, ability to develop blood vessels, evasion of programmed cell death, self-sufficiency in growth signals, insensitivity to inhibitors of growth, inflammation, tissue invasion and metastasis [30]. Because the HMGB1 rs1045411 polymorphism is closely correlated with altered binding of miR-505-5P in the 3'-UTR of mRNA transcripts, HMGB1 gene polymorphisms could emerge as a crucial player in cancer development through a post-transcriptional mechanism [7, 25]. Furthermore, since the rs1045411 polymorphism resides in the 3'-flanking regions, suggesting a role in mRNA stability as miRNAs can bind the 3'-UTR regions of mRNA transcripts and inhibit HMGB1 expression at the post-transcriptional level [25]. Although most studies have demonstrated that HMGB1 is upregulated in nearly all examined tumours, its role might depend on complex conditions, such as binding partners, diverse locations and different stages [14]. Despite its complexity, the role of HMGB1 in cancer is unquestionable. Thus, further understanding of the mechanisms underlying carcinogenesis is needed to characterize the genetic alterations linked to cancer development. And once the results hold up, HMGB1 SNP rs1045411 might be used as an index of predicting cancer occurrence in the future.
In this study, the significant connection of increased cancer risk and the TT genotype was indicated in the comparisons of TT vs. CC+TC. Though no evidence of association was found between rs1045411 polymorphism and cancer risk in some other genetic models (T vs. C; TC vs. CC; CC vs. TC+TT; TT vs. CC), HMGB1 rs1045411 polymorphism still emerged as a risk factor for cancer. And there were trends towards an association with higher cancer susceptibility, which might become more distinct with a larger sample size. Since the statistically significant differences in cancer risk amongst carriers of this SNP variant compared with non-carriers could not be detected. Whether it was covered up by the counterbalance of its pleiotropic roles in cancer progression or reflected in diversified statistical strategies requires further investigation.
Compared with the former meta-analysis conducted by Kumari T et al. [8], this study have obtained clear conclusions that rs1045411 polymorphism increased cancer risk in some genetic models, especially in the comparison of TT vs. CC+TC while statistical significance was not achieved in any genetic model for all polymorphisms studied by Li XY et al. [7], probably because more studies with larger sample size were included in this meta-analysis and the number of subjects studied was high. Subgroup analysis was also performed by the type of malignancy and ethnicity stratification in the current study, however, no obvious differences were found in the tumour risks in the HMGB1 rs1045411 polymorphism amongst the cancer types except for breast cancer and hepatocellular carcinoma. Hence, more studies are needed for each cancer type. Additionally, though most of the included studies comprised on individuals of Chinese descent except the G. Supic et al. study, in which the subjects were non-Asian, subgroup analysis based on ethnicity was also conducted. But surprisingly, our results demonstrated that rs1045411 polymorphism was positively associated with risks of cancer amongst Hans rather than Caucasians. Therefore, more studies of HMGB1 polymorphism in different ethnic backgrounds, such as, Caucasian, African and others, should be conducted in the future. During the sensitivity analysis, Wang et al. study [25] was found to contribute to the majority of the heterogeneity in this meta-analysis. After carefully reviewing this study, it was found that the percentages of smokers and alcohol drinkers were much higher in patients than controls, which might be confounding factors, however, after the removal of Wang et al. study, the pooled ORs were not distinctly changed, with stable results, which is consistent with previous study [7].
In spite of the considerable efforts to explore the possible relationship between the HMGB1 rs1045411 polymorphism and cancer risk, some limitations of the current meta-analysis should be noted. First, although we tried to gather as much evidence as possible from the present literature, due to the lack of usable data, we could not perform a methodological assessment of certain studies. More studies must be pursued in the future. Second, potential publication bias might arise because several unpublished articles and abstracts were not considered because they were not available. Additionally, due to our language criteria, only studies published in English or Chinese were included; this language restriction might also lead to bias risk and affect the results. Finally, this meta-analysis may have been too underpowered to obtain original data from the included studies.
Despite all the above limitations, by means of investigating associated cases of large samples and analyzing all five genetic models, our study provides new evidence that the HMGB1 rs1045411 polymorphism may be associated with increased cancer risk. However, due to the limitation of heterogeneity and sample size, the results of this study should be interpreted with caution and more work need to be done in the future to validate our findings.
Abbreviations
HMGB1: high-mobility group box protein 1; ORs: odds ratios; Cis: confidence intervals; SNPs: single-nucleotide polymorphisms; HWE: Hardy-Weinberg equilibrium.
Acknowledgements
This work was supported by the Research Foundation of Education Bureau of Yunnan Province, China (Grant No. 2018JS228). And we thank Wangheng Xia and Huoying Chen for assistance in expert technology.
Author Contributions
Juan Xu and Quansong Xia participated in research design; Quansong Xia and Pengzuo Tao participated in the literature research and data analysis; Quansong Xia and Juan Xu participated in the writing of paper.
Competing Interests
The authors have declared that no competing interest exists.
References
1. Hu WW, Liu PY, Yang YC, Chen PC, Su CM, Chao CC. et al. Association of HMGB1 Gene Polymorphisms with Lung Cancer Susceptibility and Clinical Aspects. Int J Med Sci. 2017;14:1197-1202
2. Torre LA, Bray F, Siegel R, Ferlay J, Lortet-Tieulent J, Jemal A. Global cancer statistics, 2012. CA Cancer J Clin. 2015;65:87-108
3. Zhu J, Zeng Y, Xu C, Qin H, Lei Z, Shen D. et al. Expression profile analysis of microRNAs and downregulated miR-486-5p and miR-30a-5p in non-small cell lung cancer. Oncol Rep. 2015;34:1779-86
4. Fang X, Yin Z, Li X, Xia L, Quan X, Zhao Y. et al. Multiple functional SNPs in differentially expressed genes modify risk and survival of non-small cell lung cancer in Chinese female non-smokers. Oncotarget. 2017;8:18924-18934
5. Shastry BS. SNP alleles in human disease and evolution. J Hum Genet. 2002;47:561-6
6. Naykoo NA. et al. Single nucleotide polymorphisms, haplotype association and tumour expression of the vascular endothelial growth factor (VEGF) gene with lung carcinoma. Gene. 2017;608:95-102
7. Li XY, Liang CH, Yang YJ, Liu L, Du YJ, Liang HS. et al. No association between HMGB1 polymorphisms and cancer risk: evidence from a meta-analysis. Biosci Rep. 2018;38(5):BSR20180658
8. Kumari T, Kumar B. High-mobility group box 1 protein (HMGB1) gene polymorphisms and cancer susceptibility: A comprehensive meta-analysis. Clin Chim Acta. 2018;483:170-182
9. Xia Q, Xu J, Chen H, Gao Y, Gong F, Hu L. et al. Association between an elevated level of HMGB1 and non-small-cell lung cancer: a meta-analysis and literature review. Onco Targets Ther. 2016;9:3917-23
10. Wu XJ, Chen YY, Gong CC, Pei DS. The role of high-mobility group protein box 1 in lung cancer. J Cell Biochem. 2018;119:6354-6365
11. Mandke P, Vasquez KM. Interactions of high mobility group box protein 1 (HMGB1) with nucleic acids: Implications in DNA repair and immune responses. DNA Repair (Amst). 2019: 83: 102701.
12. Hung SC, Wang SS, Li JR, Chen CS, Yang CK, Chiu KY. et al. Effect of HMGB1 Polymorphisms on Urothelial Cell Carcinoma Susceptibility and Clinicopathological Characteristics. Int J Med Sci. 2018;15:1731-1736
13. Chen M, Liu Y, Varley P, Chang Y, He XX, Huang H. et al. High-Mobility Group Box 1 Promotes Hepatocellular Carcinoma Progression through miR-21-Mediated Matrix Metalloproteinase Activity. Cancer Res. 2015;75:1645-56
14. Chung HW, Jang S, Kim H, Lim JB. Combined targeting of high-mobility group box-1 and interleukin-8 to control micrometastasis potential in gastric cancer. Int J Cancer. 2015;137:1598-609
15. Huber R, Meier B, Otsuka A, Fenini G, Satoh T, Gehrke S. et al. Tumour hypoxia promotes melanoma growth and metastasis via High Mobility Group Box-1 and M2-like macrophages. Sci Rep. 2016;6:29914
16. Bustin M. Regulation of DNA-dependent activities by the functional motifs of the high-mobility-group chromosomal proteins. Mol Cell Biol. 1999;19:5237-46
17. Kang R, Chen R, Zhang Q, Hou W, Wu S, Cao L. et al. HMGB1 in health and disease. Mol Aspects Med. 2014;40:1-116
18. Polanská E, Dobšáková Z, Dvořáčková M, Fajkus J, Štros M. HMGB1 gene knockout in mouse embryonic fibroblasts results in reduced telomerase activity and telomere dysfunction. Chromosoma. 2012;121:419-31
19. Ke S, Zhou F, Yang H, Gong J, Mei Z, Wu L. et al. Downregulation of high mobility group box 1 modulates telomere homeostasis and increases the radiosensitivity of human breast cancer cells. Int J Oncol. 2015;46:1051-8
20. Supic G, Kozomara R, Zeljic K, Stanimirovic D, Magic M, Surbatovic M. et al. HMGB1 genetic polymorphisms in oral squamous cell carcinoma and oral lichen planus patients. Oral Dis. 2015;21:536-43
21. Wang B, Yeh CB, Lein MY, Su CM, Yang SF, Liu YF. et al. Effects of HMGB1 Polymorphisms on the Susceptibility and Progression of Hepatocellular Carcinoma. Int J Med Sci. 2016;13:304-9
22. Yue L, Zhang Q, He L, Zhang M, Dong J, Zhao D. et al. Genetic predisposition of six well-defined polymorphisms in HMGB1/RAGE pathway to breast cancer in a large Han Chinese population. J Cell Mol Med. 2016;20:1966-73
23. Wu HH, Liu YF, Yang SF, Lin WL, Chen SC, Han CP. et al. Association of single-nucleotide polymorphisms of high-mobility group box 1 with susceptibility and clinicopathological characteristics of uterine cervical neoplasia in Taiwanese women. Tumour Biol. 2016;37:15813-15827
24. Wang JX, Yu HL, Bei SS, Cui ZH, Li ZW, Liu ZJ. et al. Association of HMGB1 Gene Polymorphisms with Risk of Colorectal Cancer in a Chinese Population. Med Sci Monit. 2016;22:3419-3425
25. Wang D, Qi X, Liu F, Yang C, Jiang W, Wei X. et al. A multicenter matched case-control analysis on seven polymorphisms from HMGB1 and RAGE genes in predicting hepatocellular carcinoma risk. Oncotarget. 2017;8:50109-50116
26. Lin CW, Chou YE, Yeh CM, Yang SF, Chuang CY, Liu YF. A functional variant at the miRNA binding site in HMGB1 gene is associated with risk of oral squamous cell carcinoma. Oncotarget. 2017;8:34630-34642
27. Huang BF, Tzeng HE, Chen PC, Wang CQ, Su CM, Wang Y. et al. HMGB1 genetic polymorphisms are biomarkers for the development and progression of breast cancer. Int J Med Sci. 2018;15:580-586
28. Martinotti S, Patrone M, Ranzato E. Emerging roles for HMGB1 protein in immunity, inflammation, and cancer. Immunotargets Ther. 2015;4:101-9
29. Hanahan D, Weinberg RA, Weinberg. Hallmarks of cancer: the next generation. Cell. 2011;144:646-74
30. Venereau E, De Leo F, Mezzapelle R, Careccia G, Musco G, Bianchi ME. HMGB1 as biomarker and drug target. Pharmacol Res. 2016;111:534-544
Author contact
![]() Corresponding author: Juan Xu, Department of Internal Medicine, The People's Hospital of Guandu District, 63# Yinfeng Road, Kunming 650200, China. E-mail: kmxujuancom
Corresponding author: Juan Xu, Department of Internal Medicine, The People's Hospital of Guandu District, 63# Yinfeng Road, Kunming 650200, China. E-mail: kmxujuancom

 Global reach, higher impact
Global reach, higher impact