Impact Factor
ISSN: 1449-1907
Int J Med Sci 2021; 18(3):785-791. doi:10.7150/ijms.51488 This issue Cite
Research Paper
Over-expression of RALYL suppresses the progression of ovarian clear cell carcinoma through inhibiting MAPK and CDH1 signaling pathways
1. Reproductive Medicine Center, Shuguang Hospital Affiliated to Shanghai University of Traditional Chinese Medicine, Shanghai 201203, China.
2. Department of Hepatobiliary Surgery, Shenzhen Second People's Hospital, Clinical Institute of Guangzhou Medical University, Shenzhen 518035, Guangdong Province, China.
#These authors contributed equally to this work.
Received 2020-8-4; Accepted 2020-11-30; Published 2021-1-1
Abstract
Background: The molecular mechanism in the progression of ovarian clear cell carcinoma (OCCC) remains unclear.
Objective: This study aimed to investigate the potential function of RAYLY in OCCC.
Methods: To validate RAYLY expression, immunohistochemistry, quantitative real-time PCR and western blotting were performed in OCCC tissues and the cell lines of OCCC and epithelial ovarian carcinoma (EOC). Subsequently, the biological effects of RALYL were evaluated through colony formation, and cell proliferation, migration and invasion assays. Finally, RNA-sequencing and gene set enrichment analysis (GSEA) were conducted to explore potential mechanism of RALYL in OCCC.
Results: In our study, RALYL was significantly down-regulated in a majority of OCCC tissues compared to adjacent non-tumorous tissues, and OCCC cells had a lower expression level of RALYL than that of EOC cells. OCCC patients with high RALYL expression had a better pathological stage and prognosis. In vitro, over-expression of RALYL inhibited cell proliferation, migration and invasion in OCCC. GSEA analysis and western blot indicated an enrichment of MAPK and CDH1 signaling pathways in OCCC cells without RALYL over-expression.
Conclusions: RALYL played an important role in the progression of OCCC, and might serve as a potential prognostic biomarker and novel therapeutic target for OCCC.
Keywords: RAYLY, ovarian clear cell carcinoma, MAPK, CDH1
Introduction
Epithelial ovarian carcinoma (EOC) is the second most common gynecologic cancer and the leading cause of death for patients with gynecologic cancer in developed countries [1]. EOC has four major pathological types including serous, clear cell, endometrioid and mucinous. Of the four subtypes, ovarian clear cell carcinoma (OCCC) is the second most common, and characterized by resistance to conventional platinum-based chemotherapy [2, 3]. For recurrent and chemo-resistant OCCC, treatment options are limited to coupling cytotoxic drugs like docetaxel, paclitaxel, and irinotecan with platinum-based therapy [4, 5]. Several clinical trials indicated no improvement of these cytotoxic drug cocktails in the OCCC prognosis [6, 7]. This underlines the urgent need for better and more effective therapeutic alternatives for OCCC.
As the same cellular subtype in pathology, OCCC has a similar gene expression pattern with renal clear cell carcinoma (RCCC) [8]. The effective drug of sunitinib in treating RCCC also demonstrates a definite efficacy in the therapy of OCCC [9-11]. In our previous study, we found a significant decrease of RALYL in RCCC for the first time [12]. RALYL expression negatively correlated with a poor RCCC prognosis, indicating a potential therapeutic target of RALYL in clear cell carcinoma. However, the anti-tumor mechanism remains unclear, as well as its role in OCCC. In this study, we investigated the aberrant expression of RALYL in OCCC and its correlation with clinical characteristics. Additionally, we also performed a series of in vitro experiments to elucidate potential biological function of RALYL in OCCC.
Materials and methods
Tissue sample collection
Tumor and matched adjacent non-tumor tissue samples of 40 OCCC patients were collected from Department of Gynaecology, Shuguang Hospital (Shanghai, China) between April 2010 and September 2019, and immediately stored in liquid nitrogen. All samples were obtained with informed consent, and our study was approved by the ethic committee of Shuguang Hospital, Shanghai University of Traditional Chinese Medicine.
Immunohistochemistry (IHC)
Paraformaldehyde-fixed, paraffin-embedded and sectioned samples were blocked in 5% BSA for 40 min, included with the primary antibody against human RALYL (1:100, Abcam) overnight at 4 °C, and sequentially incubated with CY3-conjugated anti-rabbit secondary antibody (Servicebio, China) for 1 h at room temperature. IHC images were obtained under the microscope (Olympus, Japan), and Image-Pro Plus 6.0 software (Media Cybernetics, USA) was used to quantify the corresponding staining.
Cell culture and transfection
The OCCC cell line ES-2 and the EOC cell line A2780 were purchased from the Cell Resource Center of Shanghai Institutes for Biological Sciences (Shanghai, China). Cells were cultured in DMEM (HyClone, USA) containing 10% FBS (Gibco, USA) and 1% penicillin-streptomycin (HyClone, USA) at 37 °C in a 5% CO2 atmosphere. Full-length RALYL cDNA (915bp) was inserted into pLVX-IRES-NEO vector (pLVX-IRES-NEO-RALYL) (Clontech, USA). Lentiviral vectors packaged with RALYL and negative control vectors were respectively transfected to cells using Lipofectamine 2000 Reagent (Invitrogen, USA).
Quantitative real-time PCR
Total RNA was extracted from tumor tissues and cells using TRIzol reagent (Invitrogen, USA), and then reversely transcribed to cDNA by ReverTra Kit (TOYOBO, Japan). Quantitative real-time PCR was conducted with SYBR Premix Ex Taq Kit (TaKaRa, Japan) in Applied Biosystems 7300 System (Thermo, USA). The expression levels were calculated using the comparative CT method for relative quantification against GAPDH.
Western blotting
Total protein in tumor tissues and cells was extracted using RIPA lysis buffer (Beyotime, China). Protein lysates were separated by SDS-PAGE, transferred to PVDF membranes (Millipore, USA), and blocked with 5% BSA (Biofroxx, Germany). The membranes were incubated with the primary antibodies overnight at 4 °C, and subsequently with the anti-rabbit HRP-conjugated secondary antibody (Servicebio, China). Chemiluminescent signals were detected by Enhanced Chemiluminescence (Thermo, USA).
Colony formation assay
Cells in the logarithmic growth phase were seeded into a 6-well plate at 1000 cells/well, and cultured at 37 °C with 5% CO2 for 2 weeks. Then, the cells were fixed with 4% paraformaldehyde and stained with crystal violet. Colonies with at least 50 cells were counted under the microscope.
Cell proliferation assay
Cell proliferation was detected using CCK8 assay (Sigma, USA). Cells were seeded in a 96-well plate at 5000 cells/well, and incubated at 37 °C for 1, 2, 3, 4 and 5 days respectively. At each time point, the cells were incubated with 0.5% MTT for 4 h, and then lysed with DMSO (Sigma, USA). The absorbance value (OD) at 490 nm was measured using a microplate reader (BioTek, USA).
Cell migration and invasion assay
Cell migration and invasion were determined using Transwell assay (Corning, USA). Cells were diluted with serum-free medium to 1×105/ml. 500 μl of the suspension were transferred to the upper chamber, and 750 μl medium containing 10% FBS was added into the lower chamber. Transwell plate was incubated at 37 °C for 24 h, fixed with 4% paraformaldehyde for 30 min and stained with 5% crystal violet for 30 min. Cells were counted under the microscope for quantification.
RNA-sequencing and GSEA analysis
Illumina HiSeq 4000 (Illumina, USA) was used to perform paired-end transcriptome sequencing in the samples of ES-2 cells with and without RALYL transfection. In quality control, the reads with adapter contamination, undetermined bases and low-quality bases were removed by the Cutadapt software (Martin, 2011). The remaining reads were aligned to the human genome sequences (GRCh38.p12) using the HISAT software (Johns Hopkins University, USA). The expression levels of all transcripts were determined by calculating the FPKM.
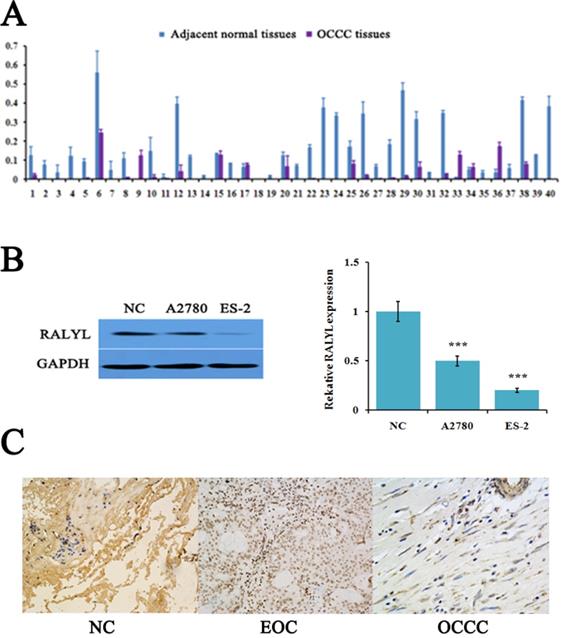
The expression of RALYL in ovarian clear cell carcinoma (OCCC). (A) Relative expression of RALYL in 40 pairs of OCCC tissues. (B) Relative expression of RALYL in the OCCC cell line ES-2 and the epithelial ovarian cancer (EOC) cell line A2780. (C) Immunohistochemistry analysis of RALYL in OCCC tissues. *P<0.05, **P<0.01, ***P<0.001.
To identify potential function of RALYL, gene set enrichment analysis (GSEA) was conducted to detect biological processes enriched in the samples with RALYL highly expressed. False discovery rate (FDR) <0.05 was chosen as the cut-off criteria.
Statistical analysis
All data were presented as mean ± SD unless otherwise indicated. The statistical difference between groups was assessed by unpaired two-tailed Student's t test using GraphPad 6.0 software (GraphPad, USA). Two-tailed P values < 0.05 were considered statistically significant. Differences among three or more groups were assessed by one-way ANOVA test. Pearson's χ2 test was used to evaluate the correlation of clinical characteristics between two groups with differential expression. Survival analysis was carried out using the KM method, and the log-rank method was used to identify the difference in survival curve. P values < 0.05 were considered statistically significant.
Results
RALYL is down-regulated in OCCC
The expression level of RALYL detected by quantitative real-time PCR showed that RALYL was significantly down-regulated in a majority of OCCC tissues compared to adjacent non-tumorous tissues (Figure 1A). Subsequently, we evaluated RALYL expression in the OCCC cell line ES-2 and the EOC cell line A2780. As shown in Figure 1B, RALYL expression was obvious down-regulated in both ovarian cancer cell lines when compared to the normal ovarian cell line. Furthermore, ES-2 cells had a lower expression level of RALYL than that of A2780. In OCCC tissues, IHC analysis indicated RALYL protein was located in the cell membrane and cytoplasm (Figure 1C).
RALYL correlates with the clinical characteristics of OCCC
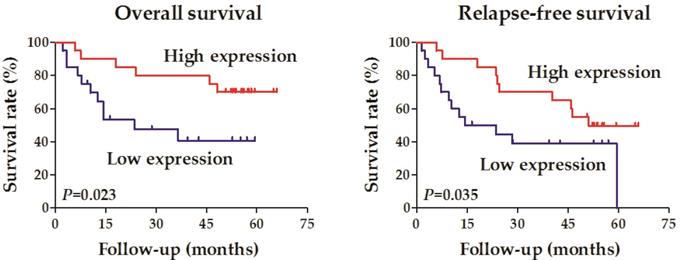
Forty OCCC samples were divided into two groups according to the expression level of RALYL (high vs. low expression). Correlation analysis was conducted to evaluate the relationship between RALYL expression and the clinical characteristics of OCCC, including age, pathological differentiation, tumor depth, lymph node metastasis, distant metastasis, and tumor stage (Table 1). The results showed that RALYL expression had a strong negative correlation with the tumor depth and stage (P<0.05). Survival analysis also indicated a better prognosis in the RALYL high expression group, including both overall survival and relapse-free survival (Figure 2).
RALYL inhibits OCCC cell proliferation
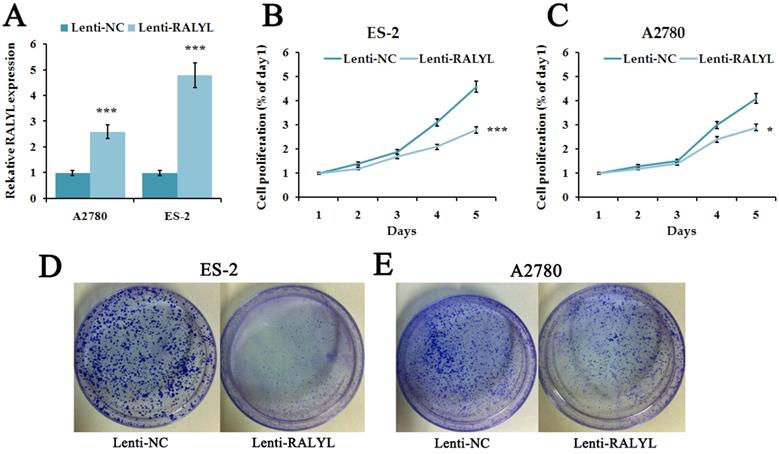
Since RALYL was down-regulated in OCCC, we further investigated its biological function. We over-expressed RALYL in OCCC cells by transfecting lentiviral vectors packaged with RALYL. Quantitative real-time PCR and western blot were conducted to verify the over-expression efficiency of RALYL (Figure 3A). To detect whether RALYL regulated the proliferation of OCCC cells, we performed CCK8 and colony formation assay. The results showed that the growth capacity and colony number of ES-2 cells and A2780 cells dramatically decreased following over-expression of RALYL (Figure 3B-3E). Compared to A2780 cells, the decrease was more significant in ES-2 cells after over-expression of RALYL.
Survival analysis of the association between RALYL expression and the prognosis in ovarian clear cell carcinoma.
Correlation between RALYL expression and clinicopathological characteristics in patients with ovarian clear cell carcinoma
| Characteristics | Number (n=40) | Percent | RALYL | P value | |
|---|---|---|---|---|---|
| High | Low | ||||
| Ages (years) | |||||
| <60 | 11 | 27.5% | 6 | 5 | 0.724 |
| >60 | 29 | 62.5% | 14 | 15 | |
| Pathological differentiation | |||||
| Well and moderate | 17 | 42.5% | 9 | 8 | 0.749 |
| Poor | 23 | 57.5% | 11 | 12 | |
| Tumor depth | |||||
| T1 and T2 | 19 | 47.5% | 13 | 6 | 0.027 |
| T3 and T4 | 21 | 52.5% | 7 | 14 | |
| Lymph node metastasis | |||||
| N0 | 25 | 62.5% | 14 | 11 | 0.327 |
| N1 | 15 | 37.5% | 6 | 9 | |
| Distant metastasis | |||||
| M0 | 39 | 97.5% | 20 | 19 | 0.311 |
| M1 | 1 | 2.5% | 0 | 1 | |
| Tumor stage | |||||
| I and II | 22 | 55.0% | 13 | 9 | 0.027 |
| III and IV | 18 | 45.0% | 7 | 11 | |
RALYL inhibits the migration and invasion of OCCC cells
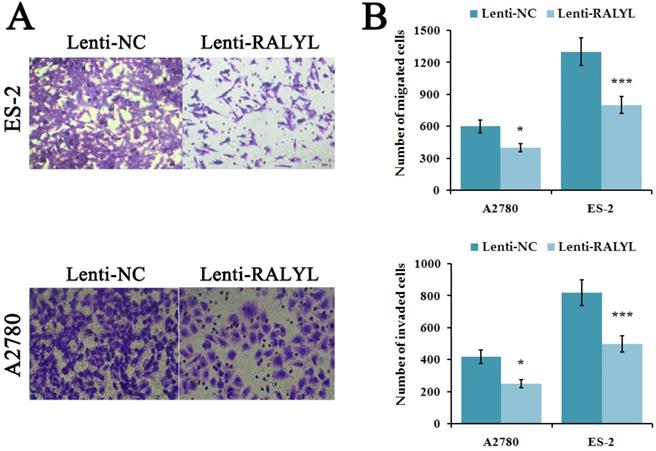
To evaluate the effects of RALYL on metastasis ability of OCCC cells, cell migration and invasion assay were performed. The results indicated a significant decrease of migrated and invaded cells in ES-2 cells and A2780 cells after over-expression of RALYL (Figure 4A-4B). Compared to A2780 cells, the suppression was more significant in ES-2 cells after over-expression of RALYL.
RALYL inhibits MAPK and CDH1 signaling pathways in OCCC
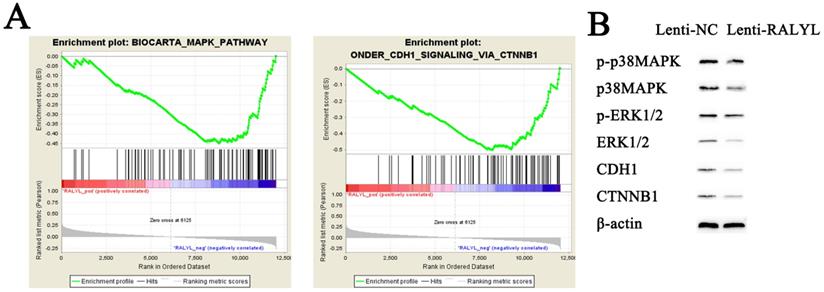
To identify potential function of RALYL in OCCC, we conducted RNA-sequencing of ES-2 cells treated with the RALYL over-expression vector and the control vector. The standardized expression data were used to perform GSEA analysis, which could help detect biological processes enriched in the samples with RALYL over-expression. The results demonstrated an enrichment of MAPK and CDH1 signaling pathways in ES-2 cells after over-expression of RALYL (Figure 5A). Collectively, these findings implied an involvement of RALYL in the OCCC pathogenesis by targeting MAPK and CDH1 signaling pathways, which have been reported to play a key role in the tumorigenesis (13, 14). To verify our speculation, western blot was conducted to investigate the expression levels of proteins in the signaling pathways. The results indicated a significant decrease of p-p38MAPK, p38MAPK, p-ERK1/2, ERK1/2, CDH1 and CTNNB1 in ES-2 cells after over-expression of RALYL (Figure 5B).
Discussion
In this study, we found an obvious down-regulation of RALYL in OCCC tissues, and the OCCC cell line also showed a more significant decrease in RALYL expression than that of the EOC cell line. In clinical analysis, RALYL expression had a strong negative correlation with the tumor depth and stage, and the patients with high RALYL expression demonstrated a better prognosis. These findings indicated an anti-tumor role of RALYL in OCCC. To validate its biological function, CCK8, colony formation assay, cell migration and invasion assay were conducted, and we found a significant inhibition in the OCCC cells after over-expression of RALYL. Furthermore, compared to the EOC cells, the suppression was more significant in OCCC cells after over-expression of RALYL. To investigate the mechanism, we performed a RNA-sequencing and GSEA analysis, and found an enrichment of MAPK and CDH1 signaling pathways in the OCCC cells overexpressing RALYL. Generally, RALYL played an anti-tumor role in OCCC, and might sever as a therapeutic and prognostic biomarker for OCCC.
The effects of RALYL over-expression on OCCC cell proliferation. (A) The transfection efficiency of lentiviral vectors packaged with RALYL in ES-2 and A2780. (B-C) CCK-8 assay analysis of lenti-RALYL transfected ES-2 cells and A2780 cells. (D-E) Colony formation assay analysis of lenti-RALYL transfected ES-2 cells and A2780 cells. *P<0.05, **P<0.01, ***P<0.001.
Effects of RALYL over-expression on the migration and invasion of OCCC cells. (A) Migration and invasion assay analysis of lenti-RALYL transfected ES-2 cells and A2780 cells. (B) Migrated and invaded cells transfected with lenti-RALYL were counted. *P<0.05, **P<0.01, ***P<0.001.
Inhibition of MAPK and CDH1 signaling pathways by RALYL over-expression in OCCC. (A) Gene set enrichment of biological processes associated with RALYL. (B) The effects of RALYL over-expression on MAPK and CDH1 signaling pathways.
RALY RNA binding protein-like (RALYL) belonged to the RALY subfamily, and it contained ten exons and one RRM (RNA recognition motif) domain so as to combine the RNA. The microarray expression date showed that RALYL were highly expressed in the normal adrenal, kidney and brain, and low RALYL expression was correlated with mental disorder, Parkinson's disease, and multiple cancers [15-17].
After searching protein-protein interaction database of STRING (https://string-db.org/), we found that RALYL could interact with multiple proteins, like TIGD3, RALY, RBM22, HNRNPCL1, HNRNPCL2, HNRNPC, CRNKL1, OR52N5, PNN and PPIE. RALY was up-regulated in multiple cancers, and RALY over-expression led to an aggressive biological behavior and a dismal prognosis as well as activating CDH1 signaling pathway [18-20]. By specific binding with RALY, RALYL could inhibit the oncogenic effects of RALY. Furthermore, HNRNPC, CRNKL1, PNN and PPIE were also reported to have a high expression and biological function in several cancers including ovarian cancer [21-24]. RALYL might also play the anti-tumor effects by suppressing these oncogenic genes.
In conclusion, our study revealed thar RALYL was down-regulated in OCCC, and patients with RALYL low expression had a poor pathological stage and prognosis. In addition, over-expression of RALYL showed an obvious anti-tumor effect by inhibiting MAPK and CDH1 signaling pathways.
Acknowledgements
Funding
This research is supported by Outstanding Talent Project of Shanghai Municipal Commission of Health (XBR2011067) and Special project of integrated traditional Chinese and Western medicine (SGZXY-201906).
Competing Interests
The authors have declared that no competing interest exists.
References
1. McGuire V, Jesser CA, Whittemore AS. Survival among U.S. women with invasive epithelial ovarian cancer. Gynecol Oncol. 2002;84:399-403
2. Heintz AP, Odicino F, Maisonneuve P. et al. FIGO 26th Annual Report on the Results of Treatment in Gynecological Cancer. Int J Gynaecol Obstet. 2006;95(Suppl 1):S161-92
3. Anglesio MS, Carey MS, Köbel M. et al. Clear cell carcinoma of the ovary: a report from the first Ovarian Clear Cell Symposium, June 24th, 2010. Gynecol Oncol. 2011;121:407-15
4. Crotzer DR, Sun CC, Coleman RL. et al. Lack of effective systemic therapy for recurrent clear cell carcinoma of the ovary. Gynecol Oncol. 2007;105:404-8
5. Lee YY, Kim TJ, Kim MJ. et al. Prognosis of ovarian clear cell carcinoma compared to other histological subtypes: a meta-analysis. Gynecol Oncol. 2011;122:541-7
6. Vasey PA, Jayson GC, Gordon A. et al. Phase III randomized trial of docetaxel-carboplatin versus paclitaxel-carboplatin as first-line chemotherapy for ovarian carcinoma. J Natl Cancer Inst. 2004;96:1682-91
7. Sugiyama T, Okamoto A, Enomoto T. et al. Randomized Phase III Trial of Irinotecan Plus Cisplatin Compared With Paclitaxel Plus Carboplatin As First-Line Chemotherapy for Ovarian Clear Cell Carcinoma: JGOG3017/GCIG Trial. J Clin Oncol. 2016;34:2881-7
8. Tothill RW, Tinker AV, George J. et al. Novel molecular subtypes of serous and endometrioid ovarian cancer linked to clinical outcome. Clin Cancer Res. 2008;14:5198-208
9. Chen W, Hill H, Christie A. et al. Targeting renal cell carcinoma with a HIF-2 antagonist. Nature. 2016;539:112-7
10. DeVorkin L, Hattersley M, Kim P. et al. Autophagy Inhibition Enhances Sunitinib Efficacy in Clear Cell Ovarian Carcinoma. Mol Cancer Res. 2017;15:250-8
11. Chan JK, Brady W, Monk BJ. et al. A phase II evaluation of sunitinib in the treatment of persistent or recurrent clear cell ovarian carcinoma: An NRG Oncology/Gynecologic Oncology Group Study (GOG-254). Gynecol Oncol. 2018;150:247-52
12. Cui ZW, Xia Y, Ye YW. et al. RALY RNA binding protein-like reduced expression is associated with poor prognosis in clear cell renal cell carcinoma. Asian Pac J Cancer Prev. 2012;13:3403-8
13. Wagner EF, Nebreda AR. Signal integration by JNK and p38 MAPK pathways in cancer development. Nat Rev Cancer. 2009;9:537-49
14. Corso G, Figueiredo J, De Angelis SP. et al. E-cadherin deregulation in breast cancer. J Cell Mol Med. 2020;24:5930-6
15. Shyamsundar R, Kim YH, Higgins JP. et al. A DNA microarray survey of gene expression in normal human tissues.A DNA microarray survey of gene expression in normal human tissues. Genome Biol. 2005;6:R22
16. Lee C, Iafrate AJ, Brothman AR. Copy number variations and clinical cytogenetic diagnosis of constitutional disorders. Nat Genet. 2007;39:S48-54
17. Giordano TJ, Kuick R, Else T. et al. Molecular classification and prognostication of adrenocortical tumors by transcriptome profiling. Clin Cancer Res. 2009;15:668-76
18. Bondy-Chorney E, Baldwin RM, Didillon A. et al. RNA binding protein RALY promotes Protein Arginine Methyltransferase 1 alternatively spliced isoform v2 relative expression and metastatic potential in breast cancer cells. Int J Biochem Cell Biol. 2017;91:124-35
19. Zhu Z, Zhang Y, Huang C. et al. Overexpression of RALY promotes migration and predicts poor prognosis in hepatocellular carcinoma. Cancer Manag Res. 2018;10:5559-72
20. Song G, Guo G, Du T. et al. RALY may cause an aggressive biological behavior and a dismal prognosis in non-small-cell lung cancer. Exp Cell Res. 2020;389:111884
21. Kim JO, Nau MM, Allikian KA. et al. Co-amplification of a novel cyclophilin-like gene (PPIE) with L-myc in small cell lung cancer cell lines. Oncogene. 1998;17:1019-26
22. Li Z, Yao Q, Zhao S. et al. Protein coding gene CRNKL1 as a potential prognostic biomarker in esophageal adenocarcinoma. Artif Intell Med. 2017;76:1-6
23. Kleemann M, Schneider H, Unger K. et al. MiR-744-5p inducing cell death by directly targeting HNRNPC and NFIX in ovarian cancer cells. Sci Rep. 2018;8:9020
24. Mini E, Lapucci A, Perrone G. et al. RNA sequencing reveals PNN and KCNQ1OT1 as predictive biomarkers of clinical outcome in stage III colorectal cancer patients treated with adjuvant chemotherapy. Int J Cancer. 2019;145:2580-93
Author contact
![]() Corresponding author: Guoqing Tong, Tel: +86-21-2025-6688; Fax: +86-021-2025-6426; E-mail: xiaye87com.
Corresponding author: Guoqing Tong, Tel: +86-21-2025-6688; Fax: +86-021-2025-6426; E-mail: xiaye87com.

 Global reach, higher impact
Global reach, higher impact