Impact Factor
ISSN: 1449-1907
Int J Med Sci 2021; 18(3):660-671. doi:10.7150/ijms.52706 This issue Cite
Research Paper
Meta-analysis of the Diagnostic Performance of Circulating MicroRNAs for Pancreatic Cancer
1. Department of Hepatopancreatobiliary Surgery, Third Xiangya Hospital, Central South University, Changsha 410013, Hunan, China.
2. Center for Medical Experiments, Third Xiangya Hospital, Central South University, Changsha 410013, Hunan, China.
3. Department of Endocrinology, Third Xiangya Hospital, Central South University, Changsha 410013, Hunan, China.
Received 2020-9-1; Accepted 2020-11-23; Published 2021-1-1
Abstract
Background: Numerous studies have suggested that differentially expressed miRNAs may be promising diagnostic markers for pancreatic cancer (PC), but the results are inconsistent. We aimed to summarize the diagnostic accuracy of circulating miRNAs, carbohydrate antigen 19-9 (CA19-9), and the combination of miRNAs and CA19-9.
Material and Methods: A literature search of online databases including PubMed, EMBASE, Cochrane Library, China National Knowledge Infrastructure (CNKI) and WanFang was conducted. Relative data were extracted from eligible included studies, and a meta-analysis was performed.
Results: A total of 46 studies involving 4,326 PC patients and 4,277 non-PC controls were included. The pooled sensitivity (SEN), specificity (SPE) and AUC of the circulating miRNAs for differentiating PC patients from non-PC controls were 0.79 (0.77-0.81), 0.77 (0.75-0.79), and 0.85 (0.81-0.87), respectively. The combination of miRNAs and CA19-9 greatly improved the SEN, SPE and AUC to 0.84 (0.80-0.87), 0.91 (0.89-0.93) and 0.94 (0.92-0.96), respectively. Moreover, circulating miRNAs also yielded an acceptable diagnostic accuracy for early-stage PC with a SEN of 0.79 (0.76-0.82), a SPE of 0.74 (0.68-0.79) and an AUC of 0.81 (0.77-0.84).
Conclusion: Circulating miRNAs exhibited satisfactory diagnostic performance for PC and even early-stage PC. The combination of circulating miRNAs and CA19-9 can further improve the diagnostic accuracy, providing a novel strategy for PC diagnosis.
Keywords: pancreatic cancer, microRNAs, diagnosis, meta-analysis, circulating
Background
Pancreatic cancer (PC) is a highly malignant digestive tract cancer characterized by strong invasiveness, a high recurrence rate and a poor prognosis. In 2018, there were approximately 458,918 new cases of PC worldwide, accounting for 2.5% of all new cases of cancer. Moreover, 432,242 patients died of PC, making it the seventh most common cause of cancer-related death[1].
According to the recommendation of the US Preventive Services Task Force (USPSTF), screening for PC in asymptomatic individuals is currently not recommended[2]. However, early detection is valuable for individuals with risk factors (such as a familial history) for PC, as it can increase the resection rates and result in longer median survival[3]. Overall, the 5-year survival rate for PC is 9.3%, but it is largely determined by the stage at which PC is diagnosed. For PC patients with metastatic disease at the time of diagnosis, the 5-year survival rate is 2.9%. If regional disease is present, the 5-year survival rate is 12.4%. For patients with localized PC, the 5-year survival rate can increase to 37.4%[4].
Conventional imaging methods, such as computed tomography (CT), magnetic resonance imaging (MRI), and ultrasound (US), and serum marker carbohydrate antigen 19-9 (CA19-9) have been widely used for the diagnosis of PC, but the diagnostic accuracy of these modalities may be suboptimal[5], especially for early-stage PC [6]. Endoscopic ultrasonography (EUS) has gradually come to be considered to be the most accurate diagnostic tool since it not only has higher sensitivity (SEN) and specificity (SPE) but can also facilitate the retrieval of specimens of suspected tissue by EUS-guided fine-needle aspiration (EUS-FNA) for pathological confirmation. However, its invasiveness and the risks attendant on sedation make it only suitable for selected individuals[7, 8]. In this clinical setting, liquid biopsy is of great interest from both scientific and clinical perspectives because of its noninvasiveness, higher sensitivity and cost-efficiency[9]. Numerous biomarkers derived from PC, including circulating tumor cells (CTCs), cell-free circulating tumor DNAs (cfDNAs), circulating microRNAs (miRNAs), long noncoding RNAs (lncRNAs), proteins and metabolites, circulating tumor extracellular vesicles (e.g., exosomes) and tumor-educated platelets (TEPs), can be detected by liquid biopsy[6, 9].
MiRNAs are a class of noncoding RNAs 19-25 nucleotides in length that regulate protein synthesis at the posttranscriptional level and play an indispensable role in cancer initiation, proliferation, progression, metastasis and chemo-resistance[10]. Since 2010, many studies on the application of circulating miRNAs for the diagnosis of PC have been published. The purpose of the present study is to summarize these original studies and evaluate the diagnostic performance of circulating miRNAs for PC.
Material and Methods
Literature search and study selection
The process of the literature search and study selection was performed in accordance with the PRISMA guidelines[11]. A combination of MeSH terms and entry terms was used to search the mainstream databases, including PubMed, EMBASE and Cochrane Library. We also searched Chinese databases, including the China National Knowledge Infrastructure (CNKI) and WanFang databases. In addition, we conducted a manual search for potentially eligible studies based on the identified review articles' reference lists. The search terms we used included (1) circulating, circulatory, serum, plasma, blood; (2) microRNAs, miRNAs, miR, panel; (3) Pancreatic Neoplasms, Pancreatic Intraductal Neoplasms, pancreatic cancer, cancer of pancreas, pancreatic cancer, carcinoma, pancreas, pancreatic ductal adenocarcinoma or PDAC; and (4) screen, diagnosis, diagnostic, prediction, predict, monitor, detection, detect, predictor, marker, sensitivity, specificity, AUC. For example, our electronic search strategy for PubMed is detailed in the Search strategy Section at the end of this paper.
We obtained a substantial number of retrieved records through the database search and manual search. First, duplicated publications were removed by Endnote X9 software, and then we checked again to ensure that there were no duplicate records. The remaining articles were evaluated based on their titles and abstracts and were included for full-text assessment if they met all eligibility criteria based on the PICOS principle: (1) Participants: patients with PC; (2) Interventions: the detection of circulating miRNAs; (3) Comparisons: non-PC controls; (4) Outcomes: diagnostic SEN and specificity SPE, or the number of true positive (TP), false positive (FP), true negative (TN) and false negative (FN) results of the diagnostic test; and (5) Study design: diagnostic research. Any article was excluded during the full-text assessment if the data were found to be insufficient.
Quality assessment
The quality of the included studies was assessed using the QUADAS-2 (Quality Assessment of Diagnostic Accuracy Studies 2) tool, which has been widely used since its publication in 2011[12]. The QUADAS-2 tool contains four domains, namely, "patient selection", "index test", "reference standard" and "flow and timing", which are used to objectively evaluate the risk of bias and concerns about the applicability of the included studies. The process of quality assessment and mapping was performed with RevMan 5.3 software.
Data extraction and statistical analysis
The process of data extraction was independently completed by two researchers, with one extracting the data and another rechecking the data. The original data were extracted with a standardized form that included the following items: (1) general information about the article: the name of the first author, publication year, country; (2) research content: specimen type, conference test, the studied miRNAs or other markers and their corresponding expression levels in PC patients, normalization control; and (3) the data for the meta-analysis: the number of PC patients and non-PC controls, the composition of the control population, diagnostic SEN and SPE or the number of true positive (TP), false positive (FP), true negative (TN) and false negative (FN) results for the standard diagnostic test, if available.
The extracted original data were regrouped according to the research purpose. Then, we performed statistical analyses in STATA 14.0 software to obtain the pooled SEN, SPE, positive likelihood ratio (PLR), negative likelihood ratio (NLR), diagnostic odds ratio (DOR) and their corresponding 95% confidence intervals (CIs). We also plotted the summary receiver operating characteristics (sROC) curve to obtain the value of the area under the curve (AUC) and the corresponding 95% CI.
An I2 value greater than 50% was suggestive of substantial heterogeneity, and then subgroup analysis was performed to identify the source of heterogeneity based on professional knowledge. The existence of a threshold effect was detected by Meta-DiSc software. Publication bias was assessed using Deeks' funnel plots. A sensitivity analysis was used to confirm the stability of the results. A P-value <0.05 was considered statistically significant.
Results
Characteristics and quality of the included studies
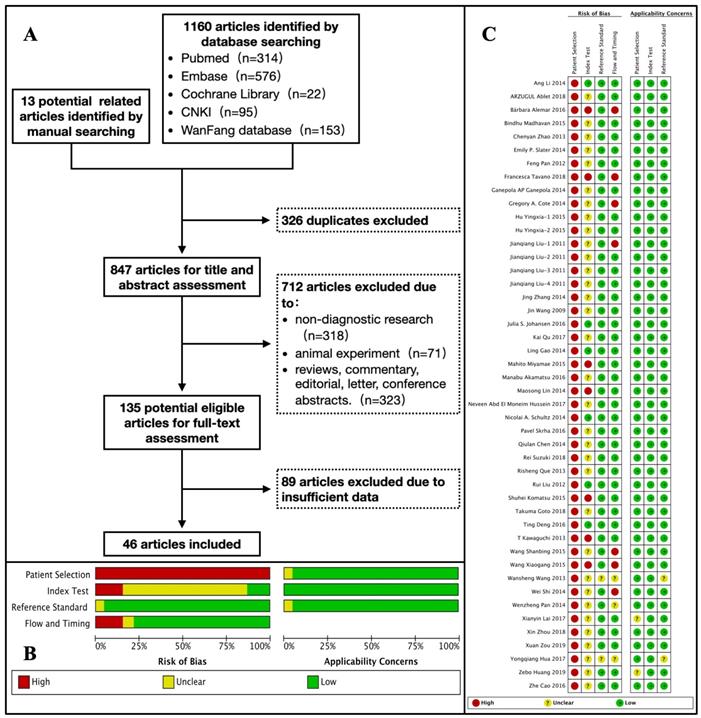
After duplicate removal, title and abstract assessment, and full-text evaluation, we finally included 46 studies involving 4,326 PC patients and 4,277 non-PC controls. The characteristics of the included studies are listed in Table 1. Among these original studies, 34 studies were conducted in Asia[13-46], 6 in Europe[47-52], 4 in North America[53-56], 1 in Africa[57], and 1 in South America[58]. The publication years were 2019 (n=2), 2018 (n=5), 2017 (n=4), 2016 (n=6), 2015 (n=7), 2014 (n=11), 2013 (n=4), 2012 (n=2), 2011 (n=4), and 2009 (n=1). The flow diagram of the literature search and study selection is detailed in Figure 1 (A).
Study selection and quality assessment. (A) Flow diagram of the literature search and study selection process. (B) The review authors' judgment about each domain of risk of bias and applicability concerns presented as percentages across the included studies. (C) The review authors' judgment about each domain of risk of bias and applicability concerns for each included study.
Characteristics of the included studies
| Author | Year | Region | Specimen | Conference test | Markers and expression level in PC patients | Normalization controls | PC patients | Non-PC controls | ||
|---|---|---|---|---|---|---|---|---|---|---|
| No. | Population | No. | Population | |||||||
| Xuan Zou | 2019 | China | serum | Histopathology | let-7b-5p ↑, miR-192-5p ↑, 19a-3p ↑, 19b-3p ↑, 223-3p ↑, 25-3p ↑ | cel-miR-34 | 159 | PC | 137 | HC |
| Zebo Huang | 2019 | China | serum | Histopathology | miR-16 ↑ | cel-miR-39 | 155 | PC | 137 | HC |
| Takuma Goto | 2018 | Japan | serum | Imaging | miR-191 ↑, 21 ↑, 451a ↑, CEA ↑, CA19-9 ↑ | unclear | 32 | PC | 22 | GBP (4), Chronic gastritis (3), Gallbladder stone (2), ADM (2), Liver cyst (1), IBS (1), Accessory spleen (1), Only symptom (7) |
| Francesca Tavano | 2018 | Italy | plasma | Histopathology or Imaging | miR-1290 ↑, CA19-9 ↑ | unclear | 167 | PC | 267 | HC |
| Rei Suzuki | 2018 | Japan | serum | Histopathology | miR-let-7d ↓, CEA ↑, CA19-9 ↑ | unclear | 45 | PC | 42 | CP (18), Biliary stone (20), others (4) |
| Jin Wang | 2018 | USA | plasma | Histopathology | miR-21 ↑, 210 ↑, 155 ↑, 196a ↑ | miR-16 | 49 | PC | 36 | HC |
| Xin Zhou | 2018 | China | plasma | Histopathology | miR-122-5p ↑, 125b-5p ↑, 192-5p ↑, 193b-3p ↑, 221-3p ↑, 27b-3p ↑ | miR-103a | 216 | PC | 220 | HC |
| Arzugul Ablet | 2018 | China | plasma | Histopathology | miR-21 ↑, 155 ↑ | U6 | 42 | PC | 84 | CP (42), HC (42) |
| Xianyin Lai | 2017 | China | plasma | Histopathology | miR-10b ↑, 20a ↑, 21 ↑, 30c ↑, 106b ↑, 181a ↑, let-7a ↓, 122 ↑ | miR-425-5p | 29 | PC | 6 | HC |
| Kai Qu | 2017 | China | serum | Histopathology | miR-21-5p ↑ | cel-miR-39 | 56 | PC | 15 | HC |
| Yongqiang Hua | 2017 | China | serum | Unclear | miR-373 ↓ | U6 | 103 | PC | 50 | HC |
| Neveen Abd EI Moneim Hussein | 2017 | Egypt | plasma | Histopathology | miR-22-3p ↑, 643b-3p ↑, 885-5p ↑, CA19-9 ↑ | miR-3196 | 35 | PC | 15 | HC |
| Ting Deng | 2016 | China | serum | Histopathology | miR-25 ↑ | unclear | 303 | PC | 760 | HC (600), CP (40), gastric cancer (20), breast cancer (20), lung cancer (20), liver cancer (20), esophageal cancer (20), colorectal cancer (20) |
| Bárbara Alemar | 2016 | Brazil | serum | Histopathology | miR-21 ↑, 34a ↑ | cel-miR-39 | 24 | PC | 9 | HC |
| Zhe Cao | 2016 | China | plasma | Histopathology | miR-486-5p ↓, 126-3p ↓, 106-3p ↓, 938 ↓, 26b-3p ↓, 1285 ↓, CA19-9 ↑ | U6 | 185 | PC | 158 | CP (73), OPN (85) |
| Pavel Skrha | 2016 | Czech Republic | serum | Histopathology | miR-196 ↑, 200 ↑, CA19-9 ↑ | miR-191, 454 | 77 | PC | 64 | HC |
| Manabu Akamatsu | 2016 | Japan | serum | Histopathology | miR-7 ↑, 34a ↑, 181d ↑, 193b ↑ | cel-miR-39 | 69 | PC | 15 | AIP |
| Julia S. Johansen | 2016 | Denmark | serum | Histopathology | miR-16 ↑, 18a ↓, 24 ↓, 25 ↓, 27a ↓, 30a-5p ↓, 323-3p ↓, 20a ↑, 29c ↓, 191 ↓, 345 ↓, 483-5p ↑, CA19-9 ↑ | unclear | 417 | PC | 340 | PAC (33), CP (59), HC (248) |
| Bindhu Madhavan | 2015 | Germany | serum | Histopathology | miR-1246 ↑, 4644 ↑, 3976 ↑, 4306 ↑ | U43, U6, 18S and 5S rRNA | 87 | PC | 51 | CP (17), BPT (14), HC (20) |
| Shuhei Komatsu | 2015 | Japan | plasma | Histopathology | miR-223 ↑ | cel-miR-39 | 71 | PC | 67 | HC |
| Mahito Miyamae | 2015 | Japan | plasma | Histopathology | miR-744 ↑ | cel-miR-39 | 94 | PC | 68 | HC |
| Hu Yingxia | 2015 | China | plasma | Histopathology | miR-196a ↑, 210 ↑, CA19-9 ↑ | U6 | 60 | PC | 30 | CP (20), HC (10) |
| Hu Yingxia | 2015 | China | plasma | Histopathology | miR-210 ↑, CA19-9 ↑, CA242 ↑, CEA ↑ | U6 | 60 | PC | 30 | CP (20), HC (10) |
| Wang Xiaogang | 2015 | China | serum | Histopathology or Imaging | miR-155 ↑, CA19-9 ↑ | cel-miR-39 | 110 | PC | 70 | CP |
| Wang Shanbing | 2015 | China | plasma | Histopathology or Imaging | miR-21 ↑, 483-3p ↑, 155 ↑, CA19-9 ↑ | miR-16 | 43 | PC | 21 | HC |
| Ling Gao | 2014 | China | plasma | Histopathology | CA19-9 ↑, miR-16 ↑ | cel-miR-39 | 70 | PC | 120 | HC (50), CP (70) |
| Gregory A. Cote | 2014 | USA | plasma | Histopathology | miR-10b ↑, 30c ↑, 106b ↑, 155 ↑, 212 ↑ | miR-425-5p | 40 | PC | 54 | CP (30), BBD (24) |
| Maosong Lin | 2014 | China | serum | Histopathology | miR-492 ↓, 663a ↓ | cel-miR-39 | 49 | PC | 27 | HC |
| Qiulan Chen | 2014 | China | plasma | Histopathology | miR-182 ↑, CA19-9 ↑ | U6 | 109 | PC | 38 | CP |
| Ang Li | 2014 | USA | serum | Histopathology | miR-1290 ↑, 628-3p ↑, 550 ↑, 1825 ↑, 24 ↑, 134 ↑, 146a ↑, 200c ↑, 378 ↑, 484 ↑, 625 ↑, 22 ↑, 210 ↑, 744 ↑, CA19-9 ↑ | miR-16 | 41 | PC | 72 | HC (19), CP (35), pNET (18) |
| Nicolai A. Schultz | 2014 | Denmark | serum | Histopathology | miR-145 ↑, 150 ↓, 223 ↑, 636 ↓, 26b ↑, 34a ↑, 122 ↑, 126 ↑, 145 ↑, 150 ↑, 223 ↑, 505 ↑, 636 ↑, 885-5p ↑, CA19-9 ↑ | ath-miR-159a | 409 | PC | 347 | HC (322), CP (25) |
| Emily P. Slater | 2014 | Germany | serum | Histopathology | miR-196a ↑, 196b ↑ | miR-24 | 24 | PC | 20 | CP (10), HC (10) |
| Ganepola AP Ganepola | 2014 | USA | plasma | Histopathology | miR-885-5p ↑, 22-3p ↑, 642b-3p ↑, CA19-9 ↑ | miR-3196 | 11 | PC | 22 | HC |
| Jing Zhang | 2014 | China | serum | Histopathology | miR-192 ↑, 194 ↑ | U6 | 70 | PC | 40 | HC |
| Wenzheng Pan | 2014 | China | plasma | Histopathology | miR-210 ↑, 25 ↑, CA19-9 ↑ | cel-miR-39 | 30 | PC | 26 | HC |
| Wei Shi | 2014 | China | plasma | Histopathology or Imaging | miR-155 ↑, 196a ↑, CA19-9 ↑, CA242 ↑, CEA ↑ | U6 | 60 | PC | 30 | CP (20), HC (10) |
| Risheng Que | 2013 | China | serum | Histopathology | miR-17-5p ↑, 21 ↑ | U6 | 22 | PC | 27 | AC (6), BPN (7), CP (6), HC (8) |
| T Kawaguchi | 2013 | Japan | plasma | Histopathology | miR-221 ↑ | U6 | 47 | PC | 9 | BPN |
| Wansheng Wang | 2013 | China | serum | Unclear | miR-27a-3p ↑, CA19-9 ↑ | U6 | 129 | PC | 163 | BPD (103), HC (60) |
| Chenyan Zhao | 2013 | China | serum | Histopathology | miR-192 ↑ | U6 | 80 | PC | 40 | HC |
| Rui Liu | 2012 | China | serum | Histopathology | miR-20a ↑, 21 ↑, 24 ↑, 25 ↑, 99a ↑, 185 ↑, 191 ↑, CA19-9 ↑, CEA ↑ | serum volume | 123 | PC | 61 | HC (52), CP (9) |
| Feng Pan | 2012 | China | plasma | Histopathology | miR-451 ↑, 409-3p ↑ | cel-miR-39 | 24 | PC | 24 | HC |
| Jianqiang Liu | 2011 | China | plasma | Histopathology or Imaging | miR-16 ↑, 196a ↑, CA19-9 ↑ | cel-miR-39 | 138 | PC | 175 | HC (68), CP (107) |
| Jianqiang Liu | 2011 | China | plasma | Histopathology | miR-181a ↑, 181b ↑, 210 ↑, CA19-9 ↑ | cel-miR-39 | 55 | PC | 96 | HC (39), CP (57) |
| Jianqiang Liu | 2011 | China | plasma | Histopathology | miR-21 ↑ | cel-miR-39 | 45 | PC | 75 | HC (30), CP (45) |
| Jianqiang Liu | 2011 | China | plasma | Histopathology | miR-155 ↑ | cel-miR-39 | 62 | PC | 97 | HC (36), CP (61) |
Abbreviations: PC, pancreatic cancer; HC, healthy control; GBP, gallbladder cholesterol polyp; ADM, adenomyomatosis; IBS, irritable bowel syndrome; CP, chronic pancreatitis; OPN, other pancreatic neoplasms; AIP, autoimmune pancreatitis; PAC, periampullary cancers; BBD, benign biliary disorders; pNET, pancreatic neuroendocrine tumor; AC, ampullary carcinoma; BPN, benign pancreatic neoplasms; BPD, benign pancreatic disease
We found that there was a high risk of bias in the domain of "Patient Selection" after the quality assessment using the QUADAS-2 tool. According to the statement of the QUADAS-2 group, an ideal diagnostic study should enroll a proportion of suspected patients ("difficult-to-diagnose patients") to reduce the risk of bias[12]. However, all our included studies included patients with a definitive diagnosis, which resulted in a high risk of bias in this domain. In addition, there was a large proportion of studies with an unclear risk of bias in the domain of the "Index Test" because the researchers of these included studies did not describe how they determined the threshold. The risk of bias was low in the domains of "Reference Test" and "Flow and Timing". All domains exhibited low concerns regarding their applicability. The results of the quality assessment are shown in Figure 1 (B-C).
Diagnostic performance of circulating miRNAs
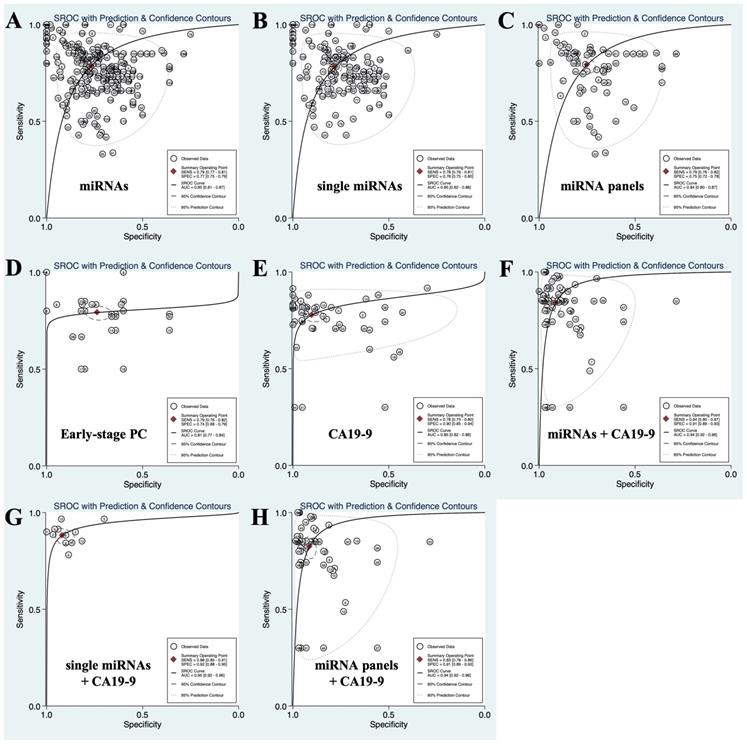
Circulating single miRNAs, which means that only one kind of miRNA was used for diagnosis, distinguished PC patients from non-PC controls with a SEN of 0.78 (0.76-0.81) and a SPE of 0.78 (0.75-0.80), and the PLR, NLR, DOR and AUC were 3.55 (3.13-4.02), 0.28 (0.25-0.31), 12.78 (10.19-16.03) and 0.85 (0.82-0.88), respectively. The circulating miRNA panel, which means multiple miRNAs were applied for diagnosis, discriminated PC patients from non-PC controls with a SEN of 0.79 (0.76-0.82), a SPE of 0.75 (0.72-0.78), a PLR of 3.16 (2.74-3.65), a NLR of 0.28 (0.23-0.33), a DOR of 11.40 (8.55-15.20), and an AUC of 0.84 (0.80-0.87). There was no significant difference in the diagnostic efficacy between single miRNAs and miRNA panels. Overall, the SEN, SPE, PLR, NLR, DOR and AUC of circulating miRNAs (including single miRNAs and miRNA panels) in differentiating patients with PC from non-PC controls were 0.79 (0.77-0.81), 0.77 (0.75-0.79), 3.38 (3.08-3.72), 0.28 (0.25-0.31), 12.22 (10.23-14.60) and 0.85 (0.81-0.87), respectively. The results are shown in Table 2 and Figure 2 (A-C).
In addition, we also summarized the SEN, SPE, PLR, NLR, DOR and AUC of miRNAs in distinguishing PC patients from healthy controls (HC) or patients with chronic pancreatitis (CP). The data are listed in Table 2. In general, the diagnostic accuracy of miRNAs for discriminating PC from HC was higher than that for discriminating PC from CP.
A total of 58 different single miRNAs and 23 miRNA panels were involved in the 46 included studies. For the single miRNAs and miRNA panels being studied in one data set, we extracted the diagnostic SEN, SPE, PLR, NLR and DOR from the original literature. For those being studied in more than 2 data sets, we performed a meta-analysis and obtained pooled diagnostic SEN, SPE, PLR, NLR and DOR values. The results are listed in Table S1 and Table S2. Among the single miRNAs, miR-122, 212, 22-3p, 483-3p, 642b-3p and 885-5p yielded a high SEN of more than 90%, and the SPE values of miR-25, 223, 17-5p, 223-3p, 30c and 409-3p were greater than 90%. The SEN and SPE of miR-451, miR-106b, miR-10b, miR-181a, miR-196b, miR-20a and let-7a were all greater than 90%. For miRNA panels, the SEN of the combination of let-7b-5p, miR-192-5p, 19a-3p, 19b-3p, 223-3p and 25-3p exceeded 90%, while the SPE of the combination of miR-1246, 4464, 3976 and 4306 was over 90%. The combination of miR-196a and 196b and the combination of miR-451 and 409-3p, as well as the combination of 885-5p, 22-3p and 642b-3p, all exhibited high diagnostic accuracy, with SEN and SPE values greater than 90%.
Circulating miRNAs for the diagnosis of early-stage PC
Early-stage PC was defined as stage 0-IIa based on the TNM system[18, 49, 51, 55]. For this group of patients, the SEN, SPE, PLR, NLR, DOR and AUC of circulating miRNAs were 0.79 (0.76-0.82), 0.74 (0.68-0.79), 2.60 (2.19-3.10), 0.35 (0.30-0.41), 8.14 (5.85-11.33) and 0.81 (0.77-0.84), respectively. MiR-196b and the combination of miR-196a and 196b exhibited high diagnostic accuracy with SEN and SPE values greater than 90%. The results are listed in Figure 2 (D) and Table 3.
Diagnostic performance of conventional biomarkers
In addition to circulating miRNAs, some researchers also evaluated the diagnostic efficacy of conventional biomarkers, such as CA19-9, CEA, and CA242. Among these conventional biomarkers, CA19-9 was the most frequently studied[59, 60]. The SEN, SPE, PLR, NLR, DOR and AUC of CA19-9 for discriminating PC patients from non-PC controls were 0.78 (0.75-0.80), 0.90 (0.85-0.94), 7.90 (5.14-12.13), 0.25 (0.22-0.28), 31.89 (18.96-53.62), and 0.85 (0.82-0.88), respectively. The SEN of CEA and CA242 was similar to that of CA19-9, but the SPE was significantly lower than that of CA19-9. CEA distinguished PC patients from non-PC controls with a SEN and a SPE of 0.79 (0.39-0.96) and 0.32 (0.08-0.72), respectively. The PLR, NLR, DOR and AUC of CEA were 1.17 (0.82-1.65), 0.65 (0.26-1.60), 1.80 (0.55-5.88) and 0.59 (0.54-0.63), respectively. The SEN, SPE, PLR, NLR, DOR and AUC of CA242 were 0.79 (0.52-0.93), 0.46 (0.21-0.74), 1.47 (0.95-2.27), 0.45 (0.21-0.97), 3.25 (1.14-9.32) and 0.68 (0.63-0.71), respectively. The results are listed in Figure 2 (E) and Table 2.
SROC curves describing the diagnostic performance of circulating miRNAs, CA19-9 and the combination of miRNAs and CA19-9 in discriminating PC from non-PC controls. (A) Circulating miRNAs; (B) circulating single miRNAs; (C) circulating miRNA panels; (D) circulating miRNAs for the diagnosis of early-stage PC; (E) CA19-9; (F) the combination of circulating miRNAs and CA19-9; (G) the combination of circulating single miRNAs and CA19-9; (H) the combination of circulating miRNA panels and CA19-9.
The results of meta-analysis
| SEN (95% CI) | SPE (95% CI) | PLR (95% CI) | NLR (95% CI) | DOR (95% CI) | AUC (95% CI) | Number of data sets | Number of PC patients | Number of controls | |
|---|---|---|---|---|---|---|---|---|---|
| 1 miRNAs | |||||||||
| PC vs non-PC | 0.79 (0.77-0.81) | 0.77 (0.75-0.79) | 3.38 (3.08-3.72) | 0.28 (0.25-0.31) | 12.22 (10.23-14.60) | 0.85 (0.81-0.87) | 228 | 13554 | 14474 |
| PC vs CP | 0.77 (0.74-0.80) | 0.67 (0.62-0.71) | 2.32 (2.01-2.69) | 0.35 (0.30-0.40) | 6.72 (5.10-8.86) | 0.79 (0.75-0.82) | 48 | 2554 | 1435 |
| PC vs HC | 0.83 (0.80-0.85) | 0.81 (0.78-0.83) | 4.29 (3.67-5.02) | 0.22 (0.18-0.26) | 19.94 (14.73-26.98) | 0.88 (0.85-0.91) | 102 | 5828 | 5983 |
| 1.1 single miRNAs | |||||||||
| PC vs non-PC | 0.78 (0.76-0.81) | 0.78 (0.75-0.80) | 3.55 (3.13-4.02) | 0.28 (0.25-0.31) | 12.78 (10.19-16.03) | 0.85 (0.82-0.88) | 148 | 7107 | 6426 |
| PC vs CP | 0.73 (0.68-0.78) | 0.68 (0.63-0.73) | 2.28 (1.94-2.69) | 0.39 (0.32-0.48) | 5.80 (4.18-8.03) | 0.76 (0.82-0.80) | 26 | 1081 | 824 |
| PC vs HC | 0.81 (0.77-0.85) | 0.81 (0.77-0.84) | 4.21 (3.46-5.12) | 0.23 (0.19-0.29) | 17.98 (12.38-26.10) | 0.88 (0.85-0.90) | 72 | 3756 | 2686 |
| 1.2 miRNA panel | |||||||||
| PC vs non-PC | 0.79 (0.76-0.82) | 0.75 (0.72-0.78) | 3.16 (2.74-3.65) | 0.28 (0.23-0.33) | 11.40 (8.55-15.20) | 0.84 (0.80-0.87) | 80 | 6447 | 8048 |
| PC vs CP | 0.80 (0.77-0.83) | 0.65 (0.56-0.73) | 2.30 (1.79-2.95) | 0.30 (0.25-0.37) | 7.58 (4.91-11.70) | 0.82 (0.78-0.85) | 22 | 1473 | 611 |
| PC vs HC | 0.86 (0.83-0.88) | 0.81 (0.76-0.85) | 4.47 (3.43-5.81) | 0.18 (0.14-0.22) | 25.43 (16.02-40.37) | 0.90 (0.88-0.93) | 30 | 2072 | 3297 |
| 2 miRNAs combined with CA19-9 | |||||||||
| PC vs non-PC | 0.84 (0.80-0.87) | 0.91 (0.89-0.93) | 9.77 (7.65-12.47) | 0.17 (0.14-0.22) | 56.01 (37.70-83.20) | 0.94 (0.92-0.96) | 65 | 6121 | 8124 |
| PC vs CP | 0.82 (0.76-0.87) | 0.82 (0.73-0.89) | 4.61 (2.87-7.40) | 0.22 (0.15-0.32) | 21.12 (9.59-46.51) | 0.89 (0.86-0.91) | 16 | 1280 | 562 |
| PC vs HC | 0.86 (0.81-0.91) | 0.96 (0.94-0.97) | 19.52 (14.92-25.53) | 0.14 (0.10-0.20) | 136.75 (91.16-205.15) | 0.97 (0.96-0.98) | 20 | 1725 | 3106 |
| 2.1 single miRNAs combined with CA19-9 | |||||||||
| PC vs non-PC | 0.88 (0.85-0.91) | 0.92 (0.88-0.95) | 10.80 (7.12-16.38) | 0.13 (0.10-0.17) | 84.16 (47.15-150.25) | 0.95 (0.92-0.96) | 12 | 965 | 830 |
| PC vs CP | 0.83 (0.79-0.87) | 0.88 (0.83-0.92) | 7.18 (4.87-10.58) | 0.19 (0.14-0.24) | 38.29 (22.55-65.00) | 0.92 (0.90-0.94) | 4 | 349 | 198 |
| PC vs HC | 0.92 (0.87-0.96) | 0.94 (0.87-0.97) | 15.30 (6.88-34.01) | 0.08 (0.05-0.14) | 189.00 (89.48-399.17) | 0.97 (0.95-0.98) | 5 | 357 | 379 |
| 2.2 miRNA panel combined with CA19-9 | |||||||||
| PC vs non-PC | 0.83 (0.78-0.86) | 0.91 (0.89-0.93) | 9.49 (7.14-12.61) | 0.19 (0.15-0.25) | 49.60 (31.15-78.98) | 0.94 (0.92-0.96) | 53 | 5156 | 7294 |
| PC vs CP | 0.81 (0.70-0.88) | 0.79 (0.66-0.88) | 3.88 (2.12-7.08) | 0.24 (0.14-0.42) | 16.04 (5.41-47.54) | 0.87 (0.84-0.90) | 12 | 931 | 364 |
| PC vs HC | 0.83 (0.75-0.89) | 0.96 (0.94-0.97) | 20.40 (15.17-27.45) | 0.17 (0.12-0.26) | 116.62 (71.44-190.38) | 0.97 (0.95-0.98) | 15 | 1368 | 2727 |
| 3 Conventional biomarker (PC vs non-PC) | |||||||||
| CA19-9 | 0.78 (0.75-0.80) | 0.90 (0.85-0.94) | 7.90 (5.14-12.13) | 0.25 (0.22-0.28) | 31.89 (18.96-53.62) | 0.85 (0.82-0.88) | 51 | 3787 | 4508 |
| CEA | 0.79 (0.39-0.96) | 0.32 (0.08-0.72) | 1.17 (0.82-1.65) | 0.65 (0.26-1.60) | 1.80 (0.55-5.88) | 0.59 (0.54-0.63) | 10 | 500 | 237 |
| CA242 | 0.79 (0.52-0.93) | 0.46 (0.21-0.74) | 1.47 (0.95-2.27) | 0.45 (0.21-0.97) | 3.25 (1.14-9.32) | 0.68 (0.63-0.71) | 5 | 300 | 90 |
| CA19-9, CEA, CA242 | 0.77 (0.61-0.88) | 0.66 (0.42-0.85) | 2.29 (1.15-4.58) | 0.35 (0.18-0.67) | 6.61 (1.92-22.77) | 0.79 (0.75-0.82) | 5 | 300 | 90 |
Abbreviations: PC, pancreatic cancer; HC, healthy control; CP, chronic pancreatitis; SEN, sensitivity; SPE, specificity; PLR, positive likelihood ratio; NLR, negative likelihood ratio; DOR, diagnostic odds ratio; AUC, area under the curve.
Diagnostic performance of circulating miRNAs combined with CA19-9
The combination of circulating miRNAs and CA19-9 for the diagnosis of PC exhibited a significantly higher diagnostic accuracy than that of circulating miRNAs or CA19-9 alone. The SEN, SPE, PLR, NLR, DOR, and AUC of miRNAs combined with CA19-9 for differentiating PC patients from non-PC controls were 0.84 (0.80-0.87), 0.84 (0.80-0.87), 9.77 (7.65-12.47), 0.17 (0.14-0.22), 56.01 (37.70-83.20) and 0.94 (0.92-0.96), respectively. The results are listed in Figure 2 (F-H) and Table 2.
The combination of miR-196, miR-200 and CA19-9 exhibited a high SEN of more than 90%. There were 5 combinations of circulating miRNAs and CA19-9 with diagnostic specificity (SPE) values exceeding 90%: the combination of miR-1290 and CA19-9; the combination of miR-16 and CA19-9; the combination of miR-16, 196a and CA19-9; the combination of miR-145, 150, 223, 636 and CA19-9; and the combination of miR-26b, 34a, 122, 126, 145, 150, 223, 505, 636, 885-5p and CA19-9. There were 4 combinations with SEN and SPE values exceeding 90%, which were the combination of miR-210 and CA19-9; the combination of miR-25 and CA19-9; the combination of miR-196a, 210 and CA19-9; and the combination of miR-181a, 181b, 210 and CA19-9. The results are listed in Table S3.
Subgroup analysis and threshold effect analysis
Since significant heterogeneity was identified in our meta-analysis (I2>50%), a random-effects model was applied for the pooled analysis. Moreover, subgroup analyses of five potential sources of heterogeneity, namely, region, conference test, miRNA profiling, non-PC control population and specimen, were conducted to identify the source of heterogeneity. However, the results suggested that the I2 value of most subgroups was still greater than 50%, indicating that these factors were not associated with the heterogeneity. The results are listed in Table S4.
The value of the Spearman correlation coefficient was -0.276 (P=0.000) in the threshold effect analysis, suggesting the existence of a threshold effect, which might be the main source of heterogeneity in the present meta-analysis.
Sensitivity analysis and publication bias
A sensitivity analysis was performed to validate the reliability of our results. The removal of any of the original studies did not have a significant impact on the results and corresponding 95% CI, suggesting that the results were stable. Deeks' funnel plots provided no evidence of publication bias (P>0.05).
Discussion
Although the incidence of PC is not high compared with that of other cancers, it is one of the most lethal cancers because of its high invasiveness and rapid progression[61]. It is difficult to diagnose early-stage PC due to the lack of specific clinical manifestations in patients and the absence of auxiliary examination modalities with high sensitivity and specificity. Approximately 50-60% of patients present with distant metastases at the time of diagnosis with PC[62], which leads to a relatively low five-year survival rate of less than 3%. These data suggest that the prognosis of pancreatic cancer is closely related to the clinical stage at diagnosis[63]. CA19-9 is a tumor antigen that was first discovered in 1979 and has been serving as a PC biomarker for decades[61]. However, a meta-analysis of 19 studies showed insufficient diagnostic accuracy of CA19-9, with pooled SEN and SPE values of 0.78 (0.75-0.81) and 0.73 (0.69-0.76), respectively[64]. Moreover, CA19-9 also exhibited FP results for some non-PC cancers (gastric cancer, ovarian cancer, etc.) and even some benign disorders[65]. In the clinical setting, liquid biopsy has been very popular in recent years because it may complement conventional diagnostic methods. The rationale for liquid biopsy is that tumors can release various forms of substances into body fluids, providing us with an opportunity to detect tumors[66]. Circulating miRNA is one of the biomarkers used in liquid biopsies, and many diagnostic studies on circulating miRNAs are published each year.
In the present meta-analysis, we found that the SEN, SPE and AUC of circulating single miRNAs for discriminating PC patients from non-PC controls were 0.78 (0.76-0.81), 0.78 (0.75-0.80) and 0.85 (0.82-0.88), respectively. The diagnostic performance of the miRNA panels was not significantly improved compared with the performance of single miRNAs. The SEN, SPE and AUC were 0.79 (0.76-0.82), 0.75 (0.72-0.78) and 0.84 (0.80-0.87), respectively.
The diagnostic performance of circulating miRNAs for early-stage PC
| MiRNAs | TNM stage | Number of data sets | Number of PC | Number of non-PC | SEN (95% CI) | SPE (95% CI) | PLR (95% CI) | NLR (95% CI) | DOR (95% CI) |
|---|---|---|---|---|---|---|---|---|---|
| miR-196a | 0 | 2 | 10 | 20 | 1.00 (0.69-1.00) | 0.60 (0.36-0.81) | 2.24 (1.32-3.81) | 0.14 (0.02-0.95) | 15.89 (1.73-145.79) |
| miR-196b | 0 | 2 | 10 | 20 | 0.90 (0.56-1.00) | 1.00 (0.83-1.00) | 18.26 (2.64-126.12) | 0.21 (0.06-0.71) | 107.46 (7.99-1444.70) |
| miR-196a, 196b | 0 | 2 | 10 | 20 | 0.90 (0.56-1.00) | 1.00 (0.83-1.00) | 18.26 (2.64-126.12) | 0.21 (0.06-0.71) | 107.46 (7.99-1444.70) |
| miR-1290 | I | 5 | 30 | 198 | 0.83 (0.65-0.94) | 0.78 (0.71-0.83) | 3.45 (2.39-4.99) | 0.22 (0.10-0.49) | 18.21 (6.31-52.56) |
| miR-191 | I-IIa | 1 | 9 | 22 | 0.67 | 0.84 | 4.22 | 0.40 | 10.67 |
| miR-21 | I-IIa | 1 | 9 | 22 | 0.67 | 0.81 | 3.51 | 0.41 | 8.54 |
| miR-451a | I-IIa | 1 | 9 | 22 | 0.67 | 0.86 | 4.66 | 0.39 | 12.00 |
| miR-145, 150, 223, 636 | I-IIa | 9 | 420 | 2082 | 0.77 (0.73-0.81) | 0.64 (0.62-0.66) | 1.83 (1.54-2.18) | 0.40 (0.33-0.48) | 4.65 (3.26-6.64) |
| miR-26b, 34a, 122, 126, 145, 150, 223, 505, 636, 885-5p | I-IIa | 9 | 420 | 2082 | 0.80 (0.76-0.84) | 0.80 (0.79-0.82) | 3.23 (2.55-4.09) | 0.33 (0.23-0.48) | 10.20 (6.03-17.26) |
| Overall | 0-IIa | 32 | 927 | 4488 | 0.79 (0.76-0.82) | 0.74 (0.68-0.79) | 2.60 (2.19-3.10) | 0.35 (0.30-0.41) | 8.14 (5.85-11.33) |
Abbreviations: PC, pancreatic cancer; SEN, sensitivity; SPE, specificity; PLR, positive likelihood ratio; NLR, negative likelihood ratio; DOR, diagnostic odds ratio.
Overall, the pooled SEN, SPE and AUC of circulating miRNAs (including single miRNAs and miRNA panels) were 0.79 (0.77-0.81), 0.77 (0.75-0.79) and 0.85 (0.81-0.87), respectively. In addition, we also summarized the data for CA19-9 in the included studies and found that the SEN, SPE and AUC of CA19-9 for distinguishing between PC and non-PC were 0.78 (0.75-0.80), 0.90 (0.85-0.94) and 0.85 (0.82-0.88), respectively. The AUC is an indicator that comprehensively reflects the diagnostic efficacy of a biomarker. An AUC of 0.8-0.9 is generally considered to indicate that the diagnostic efficacy is acceptable. An AUC above 0.9 represents a high diagnostic efficacy[67]. The AUCs of both circulating miRNAs and CA19-9 were above 0.8, suggesting that their diagnostic efficacy was acceptable. A promising finding was that the combination of miRNAs and CA19-9 greatly improved the diagnostic accuracy. The pooled SEN, SPE and AUC of the combination were 0.84 (0.80-0.87), 0.91 (0.89-0.93) and 0.94 (0.92-0.96), respectively. Therefore, we concluded that the combination of circulating miRNAs and CA19-9 may be a novel and better strategy for the diagnosis of PC. In addition to the pooled analysis, we also summarized the diagnostic accuracy of all the single miRNAs, miRNA panels and the combinations of miRNAs and CA19-9 involved in the included studies. Some miRNAs and combinations exhibited excellent diagnostic performance. For these miRNAs or combinations, their diagnostic efficacy should be further verified, and their association with the development, progression and prognosis of PC may also be valuable future research topics. Although circulating miRNAs hold promise for the accurate diagnosis of PC and many other tumors, it is not a widely accessible technique in the clinic. Two challenges need to be overcome before large-scale clinical application. (1) Technical challenge: Circulating miRNAs are more difficult to isolate and purify than intracellular miRNAs[68]. In addition, the quantitative methods of circulating miRNAs include reverse transcription-polymerase chain reaction (RT-PCR), microarray and next-generation sequencing (NGS)[69]. Therefore, the technical protocols need to be optimized and standardized. (2) Mechanistic challenge: The functions and regulatory networks of circulating miRNAs in PC remain unclear, and more investigations are needed before clinical application[70, 71].
The early diagnosis of PC has been a problem for a long time. As we mentioned above, the prognosis of patients with PC is related to the stage at diagnosis. The earlier the stage, the higher the 5-year survival rate[4]. In addition, patients with PC who were incidentally diagnosed during imaging examination for unrelated diseases have a longer median survival time than those who are symptomatic[72]. Therefore, it is of great clinical significance to explore the methods of early detection of PC since it is the key issue for improving the prognosis of this aggressive disease. Of the 46 included studies, 4 original studies[18, 49, 51, 55] proposed the concept of “Early stage PC” and defined it as stage 0-IIa in the TNM staging system. The diagnostic efficacy of relevant circulating miRNAs in this subgroup of PC patients was evaluated. The results showed that circulating miRNAs also exhibited satisfactory diagnostic efficacy in early-stage PC patients, which were defined as PC patients at stage 0-IIa based on the TNM system. The AUC was 0.81 (0.77-0.84), and the SEN and SPE were 0.79 (0.76-0.82) and 0.74 (0.68-0.79), respectively. MiR-196b and the combination of miR-196a and 196b exhibited high diagnostic accuracy, with SEN and SPE values greater than 90%.
Heterogeneity, which is common in diagnostic meta-analyses, is the result of variations among the different included studies[73]. These variations mainly include differences in the study population, study design, interventions and interpretations of results. In general, heterogeneity is derived from the threshold effect and non-threshold effect. Since heterogeneity existed in the present meta-analysis, we first performed a threshold effect analysis, in which the Spearman correlation coefficient was -0.276 (P=0.000), indicating the existence of a threshold effect. In addition, we further explored heterogeneous sources of non-threshold effects through subgroup analyses. Based on the available data, we explored the region, conference test, miRNA profiling, non-PC control population and specimen. Unfortunately, the results of the subgroup analysis negated the hypothesis that heterogeneity was caused by these five factors. In summary, we believe that the heterogeneity may be derived from the following aspects: (1) Threshold effect: different circulating miRNAs were involved in the included studies; more importantly, the diagnostic cut-off values also varied, leading to some heterogeneity. (2) Variation in normalization controls: currently, there is no consensus on the selection of the normalization controls when performing the PCR quantification of miRNAs. (3) Location: most of the included studies were conducted in Asia, which may also introduce bias.
The advantages of the present meta-analysis are as follows: (1) we conducted the literature search, study selection and quality assessment in strict accordance with the PRISMA guidelines and ultimately included a total of 46 high-quality studies, and the results were representative; (2) we scientifically grouped the original data according to clinical applicability, making the results more instructive for clinical practice; and (3) we generated a detailed summary in addition to the pooled analysis. The diagnostic accuracy of 58 single miRNAs, 23 miRNA panels and 18 combinations of miRNAs and CA19-9 was summarized, providing evidence-based support for further clinical applications and basic research. However, some limitations also existed in the present meta-analysis: (1) heterogeneity was found in our study, which may affect the reliability of the results to some extent, and (2) not all the included studies avoided using a case-control study design, which is a classic but suboptimal diagnostic study model. According to the statement made by the QUADAS-2 group, a high-accuracy diagnostic study should also enroll some “difficult-to-diagnose” patients; otherwise, the diagnostic performance may be overestimated[12]. Researchers should avoid this issue in subsequent diagnostic studies.
Conclusion
The results of the present meta-analysis showed that circulating miRNAs yielded a high diagnostic accuracy for PC. More importantly, they also exhibited a satisfactory diagnostic performance for early-stage PC, meeting the urgent need for an ideal biomarker for early-stage PC in clinical settings. The combination of circulating miRNAs and the traditional marker CA19-9 can further improve the diagnostic efficacy, which may be a novel strategy for PC diagnosis. However, the diagnostic efficacy still needs further validation by more high-quality and large-scale diagnostic research.
Search strategy
(((((((((circulating[Title/Abstract]) OR circulatory[Title/Abstract]) OR serum[Title/Abstract]) OR plasma[Title/Abstract]) OR blood[Title/Abstract]))) AND ((((((((((((Pancreatic Neoplasms[MeSH Terms]) OR Carcinoma, Pancreatic Ductal[MeSH Terms]) OR Pancreatic Intraductal Neoplasms[MeSH Terms]) OR Pancreatic Neoplasms[Title/Abstract]) OR Carcinoma, Pancreatic Ductal[Title/Abstract]) OR Pancreatic Intraductal Neoplasms[Title/Abstract]) OR pancreatic cancer[Title/Abstract]) OR cancer of pancreas[Title/Abstract]) OR pancreatic carcinoma[Title/Abstract]) OR carcinoma of pancreas[Title/Abstract]) OR pancreatic ductal adenocarcinoma[Title/Abstract]) OR PDAC[Title/Abstract])) AND ((((((microRNA[Title/Abstract]) OR microRNAs[Title/Abstract]) OR miRNA[Title/Abstract]) OR miRNAs[Title/Abstract]) OR miR[Title/Abstract]) OR panel[Title/Abstract])) AND (((((((((((((diagnostic[Title/Abstract]) OR diagnosis[Title/Abstract]) OR screen[Title/Abstract]) OR monitor[Title/Abstract]) OR detect[Title/Abstract]) OR predict[Title/Abstract]) OR predictor[Title/Abstract]) OR prediction[Title/Abstract]) OR specificity[Title/Abstract]) OR sensitivity[Title/Abstract]) OR marker[Title/Abstract]) OR AUC[Title/Abstract]) OR detection[Title/Abstract])
Abbreviations
PC: pancreatic cancer; USPSTF: US Preventive Services Task Force; CT: computed tomography; MRI: magnetic resonance imaging; US: ultrasound; CA19-9: carbohydrate antigen 19-9; EUS: endoscopic ultrasonography; EUS-FNA: EUS-guided fine-needle aspiration; CTCs: circulating tumor cells; cfDNAs: cell-free circulating tumor DNAs; miRNAs: microRNAs; lncRNAs: long noncoding RNAs; TEPs: tumor educated platelets; CNKI: China National Knowledge Infrastructure; QUADAS-2: Quality Assessment of Diagnostic Accuracy Studies 2; TP: true positive; TN: true negative; FP: false positive; FN: false negative; SEN: sensitivity; SPE: specificity; PLR: positive likelihood ratio; NLR: negative likelihood ratio; DOR: diagnostic odds ratio; CI: confidence interval; sROC: summary receiver operating characteristics curve; AUC: area under the curve; RT-PCR: reverse transcription-polymerase chain reaction; NGS: next-generation sequencing.
Supplementary Material
Supplementary tables.
Acknowledgements
This project was supported by the National Natural Science Foundation of China (NSFC) (81873589). Additionally, we would like to thank American Journal Experts for polishing this manuscript and the authors of the original studies for sharing their data.
Author Contributions
PC primarily drafted the article, wrote the manuscript, and performed the meta-analysis; LZQ extracted the data and assessed the study quality; HLH conducted revised the manuscript; WJL provided essential technical support and assistance for the statistical analysis; GWZ and LYF designed the search strategy and performed the searching; LX established the inclusion and exclusion criteria; and YX contributed to the concept design, critical revision, and finalization of the manuscript.
Competing Interests
The authors have declared that no competing interest exists.
References
1. Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68:394-424
2. Owens DK, Davidson KW, Krist AH, Barry MJ, Cabana M, Caughey AB. et al. Screening for Pancreatic Cancer: US Preventive Services Task Force Reaffirmation Recommendation Statement. Jama. 2019;322:438-44
3. Lu C, Xu CF, Wan XY, Zhu HT, Yu CH, Li YM. Screening for pancreatic cancer in familial high-risk individuals: A systematic review. World J Gastroenterol. 2015;21:8678-86
4. National Cancer Institute (NCI). Cancer Stat Facts: pancreatic cancer. NCI website. https://seer.cancer.gov/statfacts/html/pancreas.html. Assessed June 12. 2019
5. Moutinho-Ribeiro P, Macedo G, Melo SA. Pancreatic Cancer Diagnosis and Management: Has the Time Come to Prick the Bubble? Front Endocrinol (Lausanne). 2018;9:779
6. Qian L, Li Q, Baryeh K, Qiu W, Li K, Zhang J. et al. Biosensors for early diagnosis of pancreatic cancer: a review. Transl Res. 2019;213:67-89
7. Berry W, Lundy J, Croagh D, Jenkins BJ. Reviewing the Utility of EUS FNA to Advance Precision Medicine in Pancreatic Cancer. Cancers (Basel). 2018 10
8. Kandel P, Wallace MB. Advanced EUS Guided Tissue Acquisition Methods for Pancreatic Cancer. Cancers (Basel). 2018 10
9. Buscail E, Maulat C, Muscari F, Chiche L, Cordelier P, Dabernat S. et al. Liquid Biopsy Approach for Pancreatic Ductal Adenocarcinoma. Cancers (Basel). 2019 11
10. Bracken CP, Scott HS, Goodall GJ. A network-biology perspective of microRNA function and dysfunction in cancer. Nat Rev Genet. 2016;17:719-32
11. Moher D, Liberati A, Tetzlaff J, Altman DG. Preferred reporting items for systematic reviews and meta-analyses: the PRISMA statement. PLoS Med. 2009;6:e1000097
12. Whiting PF, Rutjes AW, Westwood ME, Mallett S, Deeks JJ, Reitsma JB. et al. QUADAS-2: a revised tool for the quality assessment of diagnostic accuracy studies. Ann Intern Med. 2011;155:529-36
13. Akamatsu M, Makino N, Ikeda Y, Matsuda A, Ito M, Kakizaki Y. et al. Specific MAPK-Associated MicroRNAs in Serum Differentiate Pancreatic Cancer from Autoimmune Pancreatitis. PLoS One. 2016;11:e0158669
14. Cao Z, Liu C, Xu J, You L, Wang C, Lou W. et al. Plasma microRNA panels to diagnose pancreatic cancer: Results from a multicenter study. Oncotarget. 2016;7:41575-83
15. Chen Q, Yang L, Xiao Y, Zhu J, Li Z. Circulating microRNA-182 in plasma and its potential diagnostic and prognostic value for pancreatic cancer. Med Oncol. 2014;31:225
16. Deng T, Yuan Y, Zhang C, Zhang C, Yao W, Wang C. et al. Identification of Circulating MiR-25 as a Potential Biomarker for Pancreatic Cancer Diagnosis. Cell Physiol Biochem. 2016;39:1716-22
17. Gao L, He SB, Li DC. Effects of miR-16 plus CA19-9 detections on pancreatic cancer diagnostic performance. Clin Lab. 2014;60:73-7
18. Goto T, Fujiya M, Konishi H, Sasajima J, Fujibayashi S, Hayashi A. et al. An elevated expression of serum exosomal microRNA-191, - 21, -451a of pancreatic neoplasm is considered to be efficient diagnostic marker. BMC Cancer. 2018;18:116
19. Hua Y, Chen H, Wang L, Wang F, Wang P, Ning Z. et al. Low serum miR-373 predicts poor prognosis in patients with pancreatic cancer. Cancer Biomark. 2017;20:95-100
20. Huang Z, Chen W, Du Y, Guo Q, Mao Y, Zhou X. et al. Serum miR-16 as a potential biomarker for human cancer diagnosis: results from a large-scale population. J Cancer Res Clin Oncol. 2019;145:787-96
21. Kawaguchi T, Komatsu S, Ichikawa D, Morimura R, Tsujiura M, Konishi H. et al. Clinical impact of circulating miR-221 in plasma of patients with pancreatic cancer. Br J Cancer. 2013;108:361-9
22. Komatsu S, Ichikawa D, Miyamae M, Kawaguchi T, Morimura R, Hirajima S. et al. Malignant potential in pancreatic neoplasm; new insights provided by circulating miR-223 in plasma. Expert Opin Biol Ther. 2015;15:773-85
23. Lai X, Wang M, McElyea SD, Sherman S, House M, Korc M. A microRNA signature in circulating exosomes is superior to exosomal glypican-1 levels for diagnosing pancreatic cancer. Cancer Lett. 2017;393:86-93
24. Lin MS, Chen WC, Huang JX, Gao HJ, Sheng HH. Aberrant expression of microRNAs in serum may identify individuals with pancreatic cancer. Int J Clin Exp Med. 2014;7:5226-34
25. Liu J, Gao J, Du Y, Li Z, Ren Y, Gu J. et al. Combination of plasma microRNAs with serum CA19-9 for early detection of pancreatic cancer. Int J Cancer. 2012;131:683-91
26. Liu R, Chen X, Du Y, Yao W, Shen L, Wang C. et al. Serum microRNA expression profile as a biomarker in the diagnosis and prognosis of pancreatic cancer. Clin Chem. 2012;58:610-8
27. Miyamae M, Komatsu S, Ichikawa D, Kawaguchi T, Hirajima S, Okajima W. et al. Plasma microRNA profiles: identification of miR-744 as a novel diagnostic and prognostic biomarker in pancreatic cancer. Br J Cancer. 2015;113:1467-76
28. Qu K, Zhang X, Lin T, Liu T, Wang Z, Liu S. et al. Circulating miRNA-21-5p as a diagnostic biomarker for pancreatic cancer: evidence from comprehensive miRNA expression profiling analysis and clinical validation. Sci Rep. 2017;7:1692
29. Que R, Ding G, Chen J, Cao L. Analysis of serum exosomal microRNAs and clinicopathologic features of patients with pancreatic adenocarcinoma. World J Surg Oncol. 2013;11:219
30. Suzuki R, Asama H, Waragai Y, Takagi T, Hikichi T, Sugimoto M. et al. Fibrosis-related miRNAs as serum biomarkers for pancreatic ductal adenocarcinoma. Oncotarget. 2018;9:4451-60
31. Wang WS, Liu LX, Li GP, Chen Y, Li CY, Jin DY. et al. Combined serum CA19-9 and miR-27a-3p in peripheral blood mononuclear cells to diagnose pancreatic cancer. Cancer Prev Res (Phila). 2013;6:331-8
32. Zhang J, Zhao CY, Zhang SH, Yu DH, Chen Y, Liu QH. et al. Upregulation of miR-194 contributes to tumor growth and progression in pancreatic ductal adenocarcinoma. Oncol Rep. 2014;31:1157-64
33. Zhao C, Zhang J, Zhang S, Yu D, Chen Y, Liu Q. et al. Diagnostic and biological significance of microRNA-192 in pancreatic ductal adenocarcinoma. Oncol Rep. 2013;30:276-84
34. Zhou X, Lu Z, Wang T, Huang Z, Zhu W, Miao Y. Plasma miRNAs in diagnosis and prognosis of pancreatic cancer: A miRNA expression analysis. Gene. 2018;673:181-93
35. Zou X, Wei J, Huang Z, Zhou X, Lu Z, Zhu W. et al. Identification of a six-miRNA panel in serum benefiting pancreatic cancer diagnosis. Cancer Med. 2019;8:2810-22
36. ARZUGUL A, Peng Y, NUERMAIMAITI Y, Ding Y. Expression of miR-21 and miR-155 in plasma of patients with pancreatic cancer and its clinical significance. Journal of Clinical and Experimental Medicine. 2018;17:1375-8
37. Hu Y, Zhang H, Shi W, Deng W, Liu W, Tang J. et al. Early diagnostic value of plasma miR-155, miR-196a, miR-21 and miR-210 in patients with pancreatic cancer. Tumor. 2015;35:1135-43
38. Hu Y, Zhang H, Shi W, Tang J. Diagnostic value of plasma miR-210 combined with serum tumor markers in pancreatic cancer. World Chinese Journal of Digestology. 2015;23:147-52
39. Liu J, Gao J, Du Y, Li Z, Ren Y, Wang X. et al. Diagnostic value of combined detection of plasma microRNA in pancreatic cancer. Chinese Journal of Digestion. 2011;31:777-9
40. Liu J, Gao J, Li Z, Ren Y, Wang X, Wang W. et al. Diagnostic value of plasma miR-155 for pancreatic cancer. Chinese Journal of Pancreatology. 2011;11:79-81
41. Liu J, Gao J, Ren Y, Wang X, Wang W, Lu H. Diagnostic value of plasma miR-21 in pancreatic cancer. World Chinese Journal of Digestology. 2011;19:860-3
42. Pan F, Wen Y, Ma S, Cao W, Dong J, Zhao Y. et al. Correlation of serum microRNA profiling with pancreatic cancer risk. ACTA UNIVERSITATIS MEDICINALIS NANJING (Natural Science). 2012;32:1541-4
43. Pan W. A study of plasma miRNA associated with pancreatic cancer diagnosis. Shandong University. 2014
44. Shi W. Research of abnormal expression of miR-155, miR-196a in the early diagnosis of pancreatic cancer [硕士]: Kunming Medical University; 2014
45. Wang S, Liu J, Lei K, Jia Y. Aberrant overexpressions of microRNAs in plasma and their correlations with the clinical features of patients with pancreatic cancer. Tumor. 2015;35:905-10
46. Wang X, Tong Z, Jin G. Value of serum miR-155 in the diagnosis and prognosis of pancreatic cancer Chinese Journal of Hepatobiliary Surgery. 2015; 21: 189-93.
47. Johansen JS, Calatayud D, Albieri V, Schultz NA, Dehlendorff C, Werner J. et al. The potential diagnostic value of serum microRNA signature in patients with pancreatic cancer. Int J Cancer. 2016;139:2312-24
48. Madhavan B, Yue S, Galli U, Rana S, Gross W, Muller M. et al. Combined evaluation of a panel of protein and miRNA serum-exosome biomarkers for pancreatic cancer diagnosis increases sensitivity and specificity. Int J Cancer. 2015;136:2616-27
49. Schultz NA, Dehlendorff C, Jensen BV, Bjerregaard JK, Nielsen KR, Bojesen SE. et al. MicroRNA biomarkers in whole blood for detection of pancreatic cancer. Jama. 2014;311:392-404
50. Skrha P, Horinek A, Pazourkova E, Hajer J, Fric P, Skrha J. et al. Serum microRNA-196 and microRNA-200 in pancreatic ductal adenocarcinoma of patients with diabetes mellitus. Pancreatology. 2016;16:839-43
51. Slater EP, Strauch K, Rospleszcz S, Ramaswamy A, Esposito I, Kloppel G. et al. MicroRNA-196a and -196b as Potential Biomarkers for the Early Detection of Familial Pancreatic Cancer. Transl Oncol. 2014;7:464-71
52. Tavano F, Gioffreda D, Valvano MR, Palmieri O, Tardio M, Latiano TP. et al. Droplet digital PCR quantification of miR-1290 as a circulating biomarker for pancreatic cancer. Sci Rep. 2018;8:16389
53. Cote GA, Gore AJ, McElyea SD, Heathers LE, Xu H, Sherman S. et al. A pilot study to develop a diagnostic test for pancreatic ductal adenocarcinoma based on differential expression of select miRNA in plasma and bile. Am J Gastroenterol. 2014;109:1942-52
54. Ganepola GA, Rutledge JR, Suman P, Yiengpruksawan A, Chang DH. Novel blood-based microRNA biomarker panel for early diagnosis of pancreatic cancer. World J Gastrointest Oncol. 2014;6:22-33
55. Li A, Yu J, Kim H, Wolfgang CL, Canto MI, Hruban RH. et al. MicroRNA array analysis finds elevated serum miR-1290 accurately distinguishes patients with low-stage pancreatic cancer from healthy and disease controls. Clin Cancer Res. 2013;19:3600-10
56. Wang J, Chen J, Chang P, LeBlanc A, Li D, Abbruzzesse JL. et al. MicroRNAs in plasma of pancreatic ductal adenocarcinoma patients as novel blood-based biomarkers of disease. Cancer Prev Res (Phila). 2009;2:807-13
57. Hussein NA, Kholy ZA, Anwar MM, Ahmad MA, Ahmad SM. Plasma miR-22-3p, miR-642b-3p and miR-885-5p as diagnostic biomarkers for pancreatic cancer. J Cancer Res Clin Oncol. 2017;143:83-93
58. Alemar B, Izetti P, Gregorio C, Macedo GS, Castro MA, Osvaldt AB. et al. miRNA-21 and miRNA-34a Are Potential Minimally Invasive Biomarkers for the Diagnosis of Pancreatic Ductal Adenocarcinoma. Pancreas. 2016;45:84-92
59. Zhou B, Xu JW, Cheng YG, Gao JY, Hu SY, Wang L. et al. Early detection of pancreatic cancer: Where are we now and where are we going? Int J Cancer. 2017;141:231-41
60. Zhu H, Li T, Du Y, Li M. Pancreatic cancer: challenges and opportunities. BMC Med. 2018;16:214
61. Jelski W, Mroczko B. Biochemical diagnostics of pancreatic cancer - Present and future. Clin Chim Acta. 2019;498:47-51
62. Kleeff J, Korc M, Apte M, La Vecchia C, Johnson CD, Biankin AV. et al. Pancreatic cancer. Nat Rev Dis Primers. 2016;2:16022
63. Siegel RL, Miller KD, Jemal A. Cancer statistics, 2018. CA Cancer J Clin. 2018;68:7-30
64. Meng Q, Shi S, Liang C, Liang D, Xu W, Ji S. et al. Diagnostic and prognostic value of carcinoembryonic antigen in pancreatic cancer: a systematic review and meta-analysis. Onco Targets Ther. 2017;10:4591-8
65. Parra JL, Kaplan S, Barkin JS. Elevated CA 19-9 caused by Hashimoto's thyroiditis: review of the benign causes of increased CA 19-9 level. Dig Dis Sci. 2005;50:694-5
66. Samandari M, Julia MG, Rice A, Chronopoulos A, Del Rio Hernandez AE. Liquid biopsies for management of pancreatic cancer. Transl Res. 2018;201:98-127
67. Walter SD. Properties of the summary receiver operating characteristic (SROC) curve for diagnostic test data. Stat Med. 2002;21:1237-56
68. Lee I, Baxter D, Lee MY, Scherler K, Wang K. The Importance of Standardization on Analyzing Circulating RNA. Mol Diagn Ther. 2017;21:259-68
69. Qi J, Wang J, Katayama H, Sen S, Liu SM. Circulating microRNAs (cmiRNAs) as novel potential biomarkers for hepatocellular carcinoma. Neoplasma. 2013;60:135-42
70. Cui M, Wang H, Yao X, Zhang D, Xie Y, Cui R. et al. Circulating MicroRNAs in Cancer: Potential and Challenge. Front Genet. 2019;10:626
71. Wang WT, Chen YQ. Circulating miRNAs in cancer: from detection to therapy. J Hematol Oncol. 2014;7:86
72. Poruk KE, Firpo MA, Adler DG, Mulvihill SJ. Screening for pancreatic cancer: why, how, and who? Ann Surg. 2013;257:17-26
73. Lijmer JG, Bossuyt PM, Heisterkamp SH. Exploring sources of heterogeneity in systematic reviews of diagnostic tests. Stat Med. 2002;21:1525-37
Author contact
![]() Corresponding authors: Xiao Yu and Zhiqiang Li, Department of Hepatopancreatobiliary Surgery, Third Xiangya Hospital, Central South University, Tongzipo Road No. 138, Changsha 410013, Hunan, China. E-mail addresses: yuxiaoyx4com (Xiao Yu), 1214905254com (Zhiqiang Li).
Corresponding authors: Xiao Yu and Zhiqiang Li, Department of Hepatopancreatobiliary Surgery, Third Xiangya Hospital, Central South University, Tongzipo Road No. 138, Changsha 410013, Hunan, China. E-mail addresses: yuxiaoyx4com (Xiao Yu), 1214905254com (Zhiqiang Li).

 Global reach, higher impact
Global reach, higher impact