Impact Factor
ISSN: 1449-1907
Int J Med Sci 2021; 18(1):120-127. doi:10.7150/ijms.47193 This issue Cite
Research Paper
Risk factors related to the severity of COVID-19 in Wuhan
1. Department of Pediatric, Union Hospital, Tongji Medical College, Huazhong University of Science and Technology, 1277 Jiefang Road, Wuhan, 430022, P.R. China.
2. Huazhong University of Science and Technology Hostipal. Luoyu Road 1037, Wuhan, 430074, P.R China.
Received 2020-4-21; Accepted 2020-10-13; Published 2021-1-1
Abstract
Objective: To evaluate the characteristics at admission of patients with moderate COVID-19 in Wuhan and to explore risk factors associated with the severe prognosis of the disease for prognostic prediction.
Methods: In this retrospective study, moderate and severe disease was defined according to the report of the WHO-China Joint Mission on COVID-19. Clinical characteristics and laboratory findings of 172 patients with laboratory-confirmed moderate COVID-19 were collected when they were admitted to the Cancer Center of Wuhan Union Hospital between February 13, 2020 and February 25, 2020. This cohort was followed to March 14, 2020. The outcomes, being discharged as mild cases or developing into severe cases, were categorized into two groups. The data were compared and analyzed with univariate logistic regression to identify the features that differed significantly between the two groups. Based on machine learning algorithms, a further feature selection procedure was performed to identify the features that can contribute the most to the prediction of disease severity.
Results: Of the 172 patients, 112 were discharged as mild cases, and 60 developed into severe cases. Four clinical characteristics and 18 laboratory findings showed significant differences between the two groups in the statistical test (P<0.01) and univariate logistic regression analysis (P<0.01). In the further feature selection procedure, six features were chosen to obtain the best performance in discriminating the two groups with a linear kernel support vector machine. The mean accuracy was 91.38%, with a sensitivity of 0.90 and a specificity of 0.94. The six features included interleukin-6, high-sensitivity cardiac troponin I, procalcitonin, high-sensitivity C-reactive protein, chest distress and calcium level.
Conclusions: With the data collected at admission, the combination of one clinical characteristic and five laboratory findings contributed the most to the discrimination between the two groups with a linear kernel support vector machine classifier. These factors may be risk factors that can be used to perform a prognostic prediction regarding the severity of the disease for patients with moderate COVID-19 in the early stage of the disease.
Keywords: COVID-19, moderate cases, severe prognosis, risk factors, severity, prognostic prediction, machine learning
Introduction
COVID-19 was initially reported in Wuhan, China, in December 2019 and rapidly spread to all other provinces in China and throughout the world [1, 2]. Without specific treatment or prevention options for COVID-19, such as targeted antiviral drugs and vaccines, China has focused on isolation, quarantine, social distancing, and community containment to contain the outbreak [3]. By May 18, 2020, there were 84,494 confirmed cases of COVID-19 and 4,645 deaths in China and 4,534,327 confirmed cases and 307,202 deaths outside of China [4]. The pandemic of COVID-19 has raised wide public concern and imposed a heavy burden on global health care systems because approximately 15-20% of patients develop severe interstitial pneumonia [5]. In addition, COVID-19 patients admitted to ICUs experienced higher mortality (38%) than non-ICU patients (4%) [6]. A mortality rate of 50-60% was reported in patients developing ARDS and requiring invasive mechanical ventilation therapy in the ICU [7].
Patients with COVID-19 were divided into mild, moderate, severe, and critical cases [8]. Because of the high mortality rate in severe or critical patients [6, 7], early identification of patients' risk of developing into severe or critical cases is important so that patients with a poor prognosis can receive timely intervention and minimize the progression of the disease [9]. Therefore, prognostic tools and biomarkers are urgently needed [10, 11]. However, most studies have focused mainly on identifying the factors related to death and recovery [12-14]. Although some prognostic information has been revealed by using univariate- or multivariate analyses based on prior clinical knowledge or evidence [9, 15-21], these studies have not paid enough attention to feature selection in multivariate prognostic prediction modeling. As a result, the published prognostic prediction tools may not make the most of patient data to perform prognostic prediction modeling.
This study built a multivariate prognostic prediction model to predict the risk of developing severe cases among patients with moderate COVID-19. With the patients' characteristics at admission and outcomes, a feature selection procedure based on machine learning algorithms was conducted to identify the features contributing the most to distinguishing between the two groups. These features were then chosen as the risk factors on which to build the prognostic prediction model. We believe that this multivariate prognostic prediction tool will be of considerable value for patients with moderate COVID-19 in isolation or self-quarantine so that they can receive timely intervention and active intensive care to minimize progression of the disease and so that health care agencies can prioritize their services, especially in resource-constrained areas.
Materials and methods
The ethics committees of Union Hospital, Tongji Medical College, Huazhong University of Science and Technology approved this retrospective study. The requirement for informed consent was waived. This study was conducted in accordance with the Declaration of Helsinki.
Patients
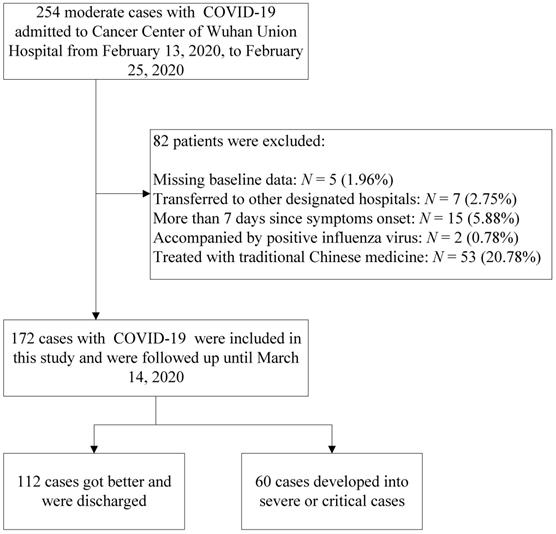
In this retrospective, single-center study, a search for patient data in the electronic record system was performed for patients admitted to the Cancer Center of Wuhan Union Hospital between February 13, 2020 and February 25, 2020. According to the report of the WHO-China Joint Mission on COVID-19, patients with COVID-19 were divided into mild (laboratory confirmed, without pneumonia), moderate (laboratory confirmed and with pneumonia), severe (dyspnea, respiratory frequency ≥30 beats per minute (bpm), oxygen saturation (SpO2) ≤93%, PaO2/FiO2 ratio <300, and/or lung infiltrates >50% of the lung field within 24-48 hours), and critical (respiratory failure requiring mechanical ventilation, shock, or other organ failure that requires intensive care) cases [8]. Our institution was a designated hospital capable of receiving patients with moderate, severe and critical cases of COVID-19. The inclusion criteria in this study were as follows: (1) patients with laboratory-confirmed COVID-19 according to viral nucleic acid detection using RT-PCR with samples from pharynx swabs; (2) patients who underwent complete laboratory tests (routine blood tests, biochemistry analysis, cytokine tests, immunology tests, and L subset tests) and clinical recording at admission; and (3) patients diagnosed with moderate COVID-19 at admission. The flow diagram of the exclusion criteria is shown in Figure 1.
Treatments
The number of patients with COVID-19 is rapidly growing worldwide, and specialized treatment has not been available in the early stage of the global outbreak. Patients in this study were moderate cases, and their treatments followed the therapeutic principles based on the 2019-nCoV guidelines (Trial Version 5) proposed by the China National Health Commission [22]. The basic treatment included antiviral treatment (abidor 200 mg three times daily, orally), antibacterial treatment (moxifloxacin 400 mg once daily, orally), recombinant human interferon α2b (aerosol inhalation) and symptomatic treatment. Some of the moderate cases were treated with traditional Chinese medicine. However, they were not included in this study, as shown in Figure 1.
Data collection
This cohort was followed to March 14, 2020. Patient data were obtained at admission, including demographics, comorbidities, signs and symptoms, and laboratory findings. The assessed comorbidities included hypertension, cardiovascular disease, diabetes, malignancy, cerebrovascular disease, COPD, chronic kidney disease, chronic liver disease, HIV infection, rheumatic disease and hyperuricemia. The laboratory findings were obtained through the complete laboratory tests mentioned earlier. Finally, 172 patients with moderate COVID-19 at admission were included in this study. In addition, 112 cases were discharged as mild cases, whereas 60 cases developed into severe or critical cases.
Statistical analysis and prognostic prediction modeling
Descriptive statistics were used to describe the demographics, comorbidities, signs and symptoms, and laboratory findings of the 172 moderate cases. Between the two groups, categorical data were compared by using the chi-squared test (Fisher's exact test if the expected count was fewer than 5 for at least one cell). Continuous variables were compared using the independent variable t-test (Mann-Whitney U test if the data were not normally distributed). Univariate logistic regression models were also built to identify the potential risk factors related to the severe prognosis of COVID-19. As a result, the features that differed significantly between the two groups in the above statistics test (P<0.01) and were significant in univariate logistic regression (P<0.01) were chosen as candidates for further processing as follows.
First, multivariate logistic regression analysis with L1 regularization was performed with feature standardization. The regression aimed to identify a subset of features from the aforementioned candidate features that could contribute the most to the discrimination between the two groups [23]. In addition, a parameter sweep on the C parameter was performed in the mentioned L1 regularization. Second, an SVM classifier with a linear kernel was adopted to measure the prediction performance of the top k features from the aforementioned subset. The top k features were selected according to their coefficients in the previous multivariate logistic regression (k ranged from 1 to the size of the subset). Finally, the top k features with the highest 5-fold SVM classification accuracy were chosen as the most predictive features to perform the clinical prognostic prediction modeling with a linear kernel SVM model. The predicted probability (pi) of a moderate case developing into a severe one was calculated with the following equation: ln [pi/(1-pi)] = a0 + a1 * xi1 +…+ ak * xik. For the ith individual, xik was the kth indicator variable in the prognostic prediction model, and aik was the weight for the kth feature. a0 is the intercept.
The statistical analysis was performed using SPSS 22 (SPSS, Inc., Chicago, IL, USA). SVM modeling and multivariate logistic regression modeling with L1 regularization used Python (Version 3.7) and the scikit-learn package (Version 0.22.1) [24].
Results
Clinical characteristics
The basic information about the two groups is summarized in Table 1. The median age of the patients was 65 years (IQR 57-71 years). Patients in the severe group were significantly older than those in the mild group (P<0.001), with an average age of 70.6 (SD 11.6) and a median age of 64 (IRQ 50-67), respectively. Among the 172 cases, 52.3% of the patients were female. There was a higher female proportion in the mild group than in the severe group (P=0.01). Comorbidities were present in 55.2% of the patients, but the difference between the two groups was not significant. Hypertension and diabetes were the most common comorbidities. For each kind of comorbidity, there was no significant difference between the two groups.
As shown in Table 2, 15 signs or symptoms were recorded in these moderate cases at admission. Fever, dry cough, and fatigue were the most common initial symptoms (63.4%, 55.2%, and 58.1%, respectively). But fever and dry cough showed no significant difference between the two groups, while fatigue was significantly different. Chest distress and anorexia were significantly more common in the severe group than in the mild group (P<0.001). In the univariate logistic regression analysis, age, chest distress, fatigue, and anorexia showed significant differences regarding to the outcomes (P<0.01), as shown in Table 4.
Comparison of epidemiology and baseline characteristics between mild and severe outcomes of patients with COVID-19
| Characteristics | Total cases (n=172) | Mild (n=112) | Severe (n=60) | P value |
|---|---|---|---|---|
| Demographics | ||||
| Age (years) | 65 (57-71) | 64 (50-67) | 70.6 (11.6) | <0.001a |
| Gender | ||||
| Female | 90 (52.3%) | 67 (59.8%) | 23 (38.3%) | 0.01b |
| Male | 82 (47.7%) | 45 (40.2%) | 37 (61.7%) | |
| Comorbidities | ||||
| Background Diseases | 95 (55.2%) | 57 (50.9%) | 38 (63.3%) | 0.107b |
| Hypertension | 63 (36.6%) | 38 (33.9%) | 25 (41.7%) | 0.318b |
| Cardiovascular disease | 17 (9.9%) | 7 (6.3%) | 10 (16.7%) | 0.033b |
| Diabetes | 27 (15.7%) | 12 (10.7%) | 15 (25.0%) | 0.016b |
| Malignancy | 7 (4.1%) | 4 (3.6%) | 3 (5.0%) | 0.694 |
| Cerebrovascular disease | 5 (2.9%) | 1 (0.9%) | 4 (6.7%) | 0.049 |
| COPD | 4 (2.3%) | 2 (1.8%) | 2 (3.3%) | 0.609 |
| Chronic kidney disease | 3 (1.7%) | 1 (0.9%) | 2 (3.3%) | 1.000 |
| Chronic liver disease | 3 (1.7%) | 2 (1.8%) | 1 (1.7%) | 1.000 |
| HIV infection | 1 (0.6%) | 1 (0.9%) | 0 (0) | 1.000 |
| rheumatic disease | 3 (1.7%) | 3 (2.7%) | 0 (0) | 0.552 |
| hyperuricemia | 4 (2.3%) | 1 (0.9%) | 3 (5.0%) | 0.119 |
Note: Categorical data are n (%). Continuous data are the mean (SD) if following a normal distribution according to the Shapiro-Wilk test (P<0.01); otherwise, the median (lower quartile-upper quartile) is shown.
a: Mann-Whitney U test;
b: chi-squared test; otherwise, Fisher's exact test, owing to an expected count less than five for at least one cell.
Study population.
Comparison of signs and symptoms between mild and severe outcomes of patients with COVID-19
| Signs and symptoms | Total cases (n=172) | Mild (n=112) | Severe (n=60) | P value |
|---|---|---|---|---|
| Fever | 109 (63.4%) | 71 (63.4%) | 38 (63.3%) | 1a |
| Dry cough | 95 (55.2%) | 64 (57.1%) | 31 (51.7%) | 0.523a |
| Expectoration | 30 (17.4%) | 19 (17.0%) | 11 (18.3%) | 0.836a |
| Pharyngalgia | 9 (5.2%) | 6 (5.3%) | 3 (5.0%) | 1 |
| Runny nose | 7 (4.1%) | 4 (3.6%) | 3 (5.0%) | 0.696 |
| Headache | 7 (4.1%) | 5 (4.5%) | 2 (3.3%) | 1 |
| Chest distress | 59 (34.3%) | 24 (21.4%) | 35 (58.3%) | <0.001 |
| Pectoralgia | 4 (2.3%) | 3 (2.7%) | 1 (1.7%) | 1 |
| Myalgia | 38 (22.1%) | 24 (21.4%) | 14 (23.3%) | 0.848a |
| Fatigue | 100 (58.1%) | 55 (49.1%) | 45 (75%) | 0.001a |
| Anorexia | 75 (43.6%) | 37 (33.0%) | 38 (63.3%) | <0.001a |
| Diarrhea | 15 (8.7%) | 9 (8.0%) | 6 (10%) | 0.778a |
| Emesis | 7 (4.1%) | 5 (4.5%) | 2 (3.3%) | 1 |
| Ageusia | 21 (12.2%) | 13 (11.6%) | 8 (13.3%) | 0.808a |
| Anosmia | 32 (18.6%) | 19 (17.0%) | 13 (21.7%) | 0.538a |
Note: Categorical data are n (%);
a: Chi-squared test; otherwise, Fisher's exact test, owing to expected count less than five for at least one cell.
Laboratory findings
Laboratory findings on hospital admission are summarized in Table 3. Among the 172 patients, 19 laboratory findings showed significant differences between the two groups. Patients in the severe group demonstrated significantly increased WBC, N count, N percentage, AST, LDH, TNI, CK, CK-MB, CysC, ESR, CRP, PCT, IL-6, and IL-10 but significantly decreased L count, L percentage, A/G, ALB, and Ca (P<0.01). In the univariate logistic regression analysis, these laboratory findings also showed significant differences regarding to the outcomes (P<0.01), except CK-MB, as shown in Table 4.
Prediction model for severe prognosis
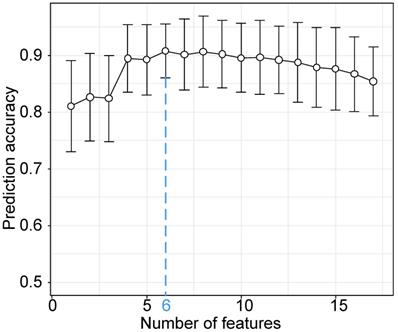
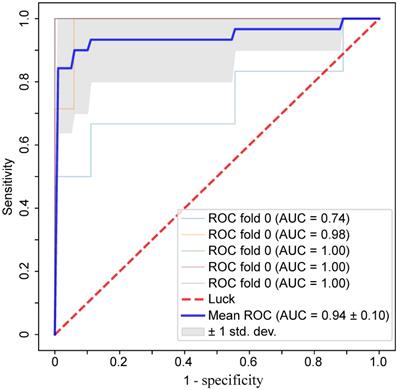
Based on the results of statistical analysis and univariate logistic regression, four clinical characteristics and 18 laboratory findings differed significantly between the two groups (P<0.01) and were significant in univariate logistic regression (P<0.01), as shown in Table 4. In the further feature selection procedure, these features were used to perform multivariate logistic regression with L1 regularization and feature standardization. With a parameter C=3.999 in the regularization, 17 features were finally selected as shown in Table 4. Eventually, the top six features ranked by regression coefficients were chosen as the most predictive features with which to build a prognostic prediction model for severity for patients with moderate COVID-19. The highest prediction accuracy of 91.38% was reached with the selected six features and a linear kernel SVM with 5-fold cross validation, as shown in Figure 2. The SVM model for prognostic prediction had a sensitivity of 0.90 and a specificity of 0.94, with a mean area under the ROC curve of 0.94, as shown in Figure 3. In the formula for the multivariate prognostic prediction model, the intercept a0 was -0.14, and the feature weights for the six most predictive features are shown in Table 4.
Comparison of laboratory findings between mild and severe outcomes of patients with COVID-19
| Laboratory findings | Total cases (n=172) | Mild (n=112) | Severe (n=60) | P value |
|---|---|---|---|---|
| Hematologic parameters | ||||
| WBC (G/L) | 5.8 (4.5-7.3) | 5.4 (4.4-6.9) | 6.5 (5.1-7.9) | 0.006 |
| N percentage (%) | 64.9 (56.2-73) | 60.6 (54.6-66.6) | 74.8 (65.5-86.6) | <0.001 |
| N (G/L) | 3.56 (2.75-5.02) | 3.32 (2.52-4.31) | 4.69 (3.38-6.51) | <0.001 |
| L percentage (%) | 22.8 (10.8) | 26.8 (8.9) | 15.4 (10.1) | <0.001a |
| L (G/L) | 1.29 (0.86-1.67) | 1.51 (1.11-1.77) | 0.81 (0.53-1.29) | <0.001 |
| Hb (g/L) | 120 (15) | 121 (12) | 119 (19) | 0.535a |
| PLT (G/L) | 216 (165-280) | 224 (180-291) | 208 (92) | 0.036 |
| Biochemical parameters | ||||
| TB (µmol/L) | 11.5 (9.1-14.1) | 11.2 (3.5) | 12.2 (9.8-18) | 0.01 |
| ALT (U/L) | 28 (18-49) | 27 (18-46) | 34 (18-52) | 0.429 |
| AST (U/L) | 26 (20-37) | 23 (20-30) | 32 (24-47) | 0.001 |
| A/G | 1.20 (1.10-1.55) | 1.40 (1.20-1.70) | 1.10 (0.90-1.28) | <0.001 |
| ALB (g/L) | 36.0 (32.1-40.3) | 38.2 (4.6) | 31.1 (28.0-35.0) | <0.001 |
| GLB (g/L) | 27.8 (24.6-32.2) | 28.0 (5.1) | 30.3 (6.2) | 0.01a |
| ALP (U/L) | 77 (63-99) | 76 (62-98) | 84 (69-106) | 0.096 |
| LDH (U/L) | 200 (167-273) | 181 (164-230) | 273 (211-407) | <0.001 |
| TNI (ng/L) | 2.80 (1.40-8.18) | 1.80 (1.00-3.75) | 10.1 (3.1-170) | <0.001 |
| CK (U/L) | 56.5 (39.8-87.3) | 51 (39-73) | 75 (41-142) | 0.003 |
| CK-MB (ng/mL) | 0.50 (0.30-1.00) | 0.40 (0.30-0.68) | 0.95 (0.50-2.80) | <0.001 |
| CysC (mg/L) | 1.13 (0.95-1.39) | 1.08 (0.91-1.23) | 1.25 (1.02-1.78) | 0.001 |
| BUN (mmol/L) | 4.30 (3.50-5.38) | 4.20 (3.49-5.00) | 4.50 (3.66-9.61) | 0.012 |
| CREA (µmol/L) | 66 (57-81) | 65 (56-75) | 75 (58-92) | 0.027 |
| TG (mmol/L) | 1.38 (1.01-1.86) | 1.41 (1.02-1.91) | 1.32 (1.00-1.77) | 0.389 |
| Na (mmol/L) | 139 (137-141) | 140 (138-141) | 139 (6) | 0.161 |
| K (mmol/L) | 4.14 (3.72-4.50) | 4.15 (0.45) | 4.12 (0.86) | 0.753a |
| Ca (mmol/L) | 2.38 (0.17) | 2.30 (0.15) | 2.13 (0.16) | <0.001a |
| ESR (mm/h) | 38 (20-59) | 37.4 (22.3) | 51.3 (27.1) | 0.004a |
| CRP (mg/L) | 4.2 (2.9-36.8) | 3.1 (1.0-8.1) | 76.4 (14.0-132.8) | <0.001 |
| PCT (µg/L) | 0.13 (0.09-0.13) | 0.13 (0.06-0.13) | 0.13 (0.13-0.21) | <0.001 |
| Cytokine levels | ||||
| IL-6 (pg/ml) | 11.1 (5.9-33.1) | 8.1 (5.4-11.5) | 54.7 (27.5-88.4) | <0.001 |
| TNF-α (pg/ml) | 3.55 (2.73-5.47) | 3.72 (2.79-5.80) | 3.28 (2.51-4.90) | 0.181 |
| IL-4 (pg/ml) | 2.83 (2.08-3.76) | 3.14 (2.11-3.85) | 2.53 (2.06-3.34) | 0.063 |
| IL-2 (pg/ml) | 2.97 (2.56-3.97) | 3.12 (2.60-4.06) | 2.87 (2.56-3.66) | 0.242 |
| IL-10 (pg/ml) | 4.27 (3.43-5.22) | 4.10 (3.09) | 4.71 (3.66-6.10) | 0.006 |
| IFN-γ (pg/ml) | 2.87 (2.13-3.50) | 2.97 (2.09-3.51) | 2.63 (2.15-3.50) | 0.635 |
| Lymphocyte subpopulation | ||||
| CD4+/CD8+ | 1.93 (1.49-2.84) | 1.86 (1.47-2.68) | 2.16 (1.53-3.30) | 0.101 |
| CD3+ (%) | 75.5 (68.2-80.8) | 73.9 (9.4) | 75.2 (66.2-81.6) | 0.826 |
| CD4+ (%) | 45.4 (10.7) | 45.2 (9.3) | 45.6 (13.1) | 0.824a |
| CD8+ (%) | 23.7 (17.8-29.3) | 24.7 (8.3) | 19.8 (15.0-30.3) | 0.078 |
| B lymphocyte (%) | 11.1 (8.2-15.5) | 12.1 (5.2) | 10.1 (8.3-16.2) | 0.650 |
| NK (%) | 7.6 (4.7-12.0) | 7.2 (4.9-12.0) | 9.5 (3.7-12.0) | 0.491 |
| Immuneglobulin | ||||
| IgG (g/L) | 10.9 (9.3-12.8) | 11.5 (9.8-13.2) | 10.2 (8.1-11.6) | 0.052 |
| IgA (g/L) | 2.43 (1.70-3.04) | 2.56 (1.15) | 2.56 (1.35) | 0.991a |
| IgM (g/L) | 1.01 (0.75-1.33) | 1.05 (0.80-1.36) | 0.86 (0.56-1.10) | 0.092 |
| IgE (IU/ml) | 33.8 (13.4-102.5) | 30.3 (11.3-64.0) | 47.9 (16.8-182.7) | 0.039 |
| Complement | ||||
| C3 (g/L) | 0.87 (0.80-0.98) | 0.86 (0.13) | 0.92 (0.21) | 0.203a |
| C4 (g/L) | 0.21 (0.07) | 0.20 (0.06) | 0.24 (0.10) | 0.027 |
Note: Continuous data are the mean (SD) if following a normal distribution according to the Shapiro-Wilk test (P<0.01); otherwise, median (lower quartile-upper quartile);
a: Independent variable t-test; otherwise, Mann-Whitney U test.
Classification accuracy of the prognostic prediction model with a linear kernel SVM model and the top k features. The classification accuracy was measured with 5-fold cross validation. The blue dotted vertical line was drawn at the optimal number 6 with the parameter C=3.999 when the classification accuracy reached the highest point 91.38%. The six most predictive features used in the prognostic prediction model were IL-6, TNI, PCT, CRP, chest distress and Ca.
Risk factors associated with the severity of patients with COVID-19
| Demographics and clinical characteristics | Univariable OR (95% CI) | P value | Regression coefficient | Feature weight |
|---|---|---|---|---|
| Age, years* | 1.086 (1.050-1.124) | <0.001 | 0.08 | |
| Chest distress | 5.133 (2.592-10.168) | <0.001 | 1.05a | 0.41 |
| Fatigue | 3.109 (1.557-6.210) | 0.001 | 0.55 | |
| Anorexia | 3.501 (1.816-6.749) | <0.001 | 0.20 | |
| Laboratory findings | ||||
| WBC (G/L)* | 1.27 (1.08-1.49) | 0.003 | ||
| N percentage (%)* | 1.10 (1.07-1.14) | <0.001 | 0.19 | |
| N (G/L)* | 1.27 (1.09-1.48) | 0.002 | 0.29 | |
| L percentage (%)* | 0.88 (0.84-0.92) | <0.001 | 0.72 | |
| L (G/L)* | 0.10 (0.05-0.24) | <0.001 | 0.46 | |
| TB (µmol/L)* | 1.125 (1.047-1.209) | 0.001 | ||
| TNI (ng/L)* | 1.113 (1.048-1.181) | <0.001 | 3.87a | 0.74 |
| CysC (mg/L)* | 3.079 (1.441-6.579) | 0.004 | 0.33 | |
| A/G* | 0.031 (0.008-0.114) | <0.001 | 0.43 | |
| ALB (g/L)* | 0.800 (0.738-0.867) | <0.001 | 0.32 | |
| Ca (mmol/L)* | 0.001 (0.000-0.012) | <0.001 | 0.82a | -0.38 |
| CK (U/L)* | 1.007 (1.003-1.012) | 0.002 | 0.45 | |
| LDH (U/L)* | 1.012 (1.007-1.017) | <0.001 | ||
| ESR (mm/h)* | 1.024 (1.007-1.041) | 0.005 | ||
| PCT (µg/L)* | 1.2e4 (22-7.4e6) | 0.004 | 1.19a | 0.96 |
| CRP (mg/L)* | 1.061 (1.038-1.085) | <0.001 | 1.17a | 1.13 |
| IL-6 (pg/ml)* | 1.115 (1.074-1.157) | <0.001 | 11.97a | 2.13 |
| IL-10 (pg/ml)* | 1.474 (1.166-1862) | 0.001 |
Note. *Per 1 for unit increase. The regression coefficient indicated the weights of the selected features in the multivariate logistic regression with L1 regularization and feature standardization. The parameter C had a value of 3.999 in this regression with L1 regularization.
a: The features that were chosen for a prognostic prediction of severity for patients with moderate COVID-19. The feature weight indicated the weight for each feature used in the probability calculation formula for the prediction of severity in the prognostic model of a linear kernel SVM classifier.
Discussion
The number of patients infected with COVID-19 is still increasing rapidly worldwide. However, specialized and effective treatment is not yet available. Therefore, the early identification of a moderate case's risk developing into a severe or critical one is of great importance due to the high mortality rate among severe and critical cases [6, 7]. Timely intervention and active intensive care can help to minimize the progression of the disease. Therefore, paying attention to prognostic prediction for moderate cases can be important. We enrolled 172 patients hospitalized with laboratory-confirmed COVID-19 and diagnosed with moderate COVID-19 at admission. The patient data were systematically analyzed. As a result, six risk factors were identified as the most predictive ones with which to perform prognostic prediction of severity for patients with moderate COVID-19.
The ROC curves for the prognostic prediction model with a linear kernel SVM and 5-fold cross validation. With the six most predictive features, the mean area under the ROC curve was 0.94, with a sensitivity of 0.90 and a specificity of 0.94.
Consistent with previous research results, patients in the severe group were significantly older than those in the mild group. Older patients presented more comorbidities and were more likely to develop severe or critical COVID-19 [5, 6, 25, 26]. The proportion of severe cases in males was 61.7%, which was significantly higher than that in females (38.3%), as reported in a recent study [9]. Several studies concerning comorbid disease with COVID-19 suggested that adequate attention should be paid to comorbidity [18, 26, 27]. In our study, the most common comorbidity was hypertension (36.6%, 63/172), followed by diabetes (15.7%, 27/172) and cardiovascular disease (9.9%, 17/172). Due to the limited sample size, there were no significant differences in comorbidities between the two groups. For all the moderate cases in this study, chest distress, fatigue, and anorexia were significantly different between the two groups, which was consistent with previous reports [28, 29].
In addition, in laboratory findings of the moderate cases at admission, significantly higher levels of WBC, N count, N percentage, AST, LDH, TNI, CK, CK-MB, CysC, ESR, CRP, PCT, IL-6, and IL-10 were found in the severe group compared with the mild group. L count, L percentage, A/G, ALB, and Ca, however, were found to be at lower levels in the severe group. The severity of COVID-19 infection may activate neutrophils to produce an immune response to the virus and cause a cytokine storm. Furthermore, considering that aging is related to decreased immune competence [30], elderly patients who died consequently may be due to a weak immune response [16]. There were no significant differences in the L subsets between the two groups, possibly because of the limited sample size. However, some impressive different results were reported in previous research, indicating that CD3+ and CD4+ T cells might protect patients from developing ARDS [16].
In this study, a feature selection procedure based on machine learning algorithms was performed to identify the most predictive features for a multivariate prognostic prediction of severity. Feature selection was an essential step for multivariate modeling. In previous studies, this was mainly implemented by performing univariate statistical analysis or with prior clinical knowledge or evidence [9, 15, 17, 19]. In our study, a two-step feature selection procedure was performed based completely on patient data. Finally, one clinical characteristic (chest distress) and five laboratory findings (IL-6, TNI, PCT, CRP, Ca) were identified as the most predictive risk factors for prognostic prediction of severity for patients with moderate COVID-19. Without adequate knowledge about COVID-19, information completely extracted from the data may be of great value to perform valid prognostic prediction.
The level of IL-6 was mildly elevated or within the normal range in the mild group but markedly elevated in the severe group. Elevated cytokines were likely produced by highly inflammatory macrophages that were implicated in a cytokine storm [31]. Myocardial damage with biomarker elevations was a prominent feature in COVID-19 and was related to a worse prognosis [32, 33]. TNI, as one of the most predictive factors in our prognostic prediction model, was used to evaluate patients with suspected acute coronary syndrome [34]. TNI level in plasma and CRP level were positively and significantly related [35]. This implies that the pathological process of myocardial damage and inflammation might have a close relationship in the course of COVID-19 disease. Chest distress was an important factor that could contribute to the prognostic prediction of severity in this study and was reported as one of the most common symptoms in COVID-19 [28]. Increasing PCT level was reported in the discrimination between mild and moderate cases [36]. In our study, PCT was identified as a risk factor for prognostic prediction of severity. Lower level of Ca was recognized as a predictor of severity in our prognostic prediction model. This was an interesting result that has not yet been reported.
There are also some limitations in our research. First, a larger sample size may result in a more convincing prognostic prediction model with the feature selection procedure based on machine learning algorithms. Due to the limited size, the comorbidities and some signs and symptoms showed no significant difference between the two groups. Moreover, age was not one of the most predictive factors related to severity in this cohort of moderate cases, but age was an important risk factor in prognosis in previous research [9]. Second, the temporal changes in patient characteristics from admission to outcome were not included in this study because most of the tests were performed only once at admission. The prognosis may benefit from information on temporal changes [19]. Third, the patients' stage of disease progression at admission, such as time since symptom onset and exposure history, was not considered as a candidate predictor. This information relied on patients' memory and might have been affected by recall bias. Fourth, the data in this study were from an outbreak, so these might differ in a nonoutbreak situation. However, since we included as many COVID-19 cases as possible in our hospital, we believe our study population is representative of cases diagnosed and treated in Wuhan.
Conclusions
With the data collected at admission, one clinical characteristic (chest distress) and five laboratory findings (IL-6, TNI, PCT, CRP, Ca) showed the best performance in discriminating between the two groups with a linear kernel SVM model. They may be risk factors that can be used to perform a prognostic prediction of severity for patients with moderate COVID-19 in the early stage of the disease and thus help minimize the progression of the disease.
Abbreviations
COVID-19: Coronavirus Disease 2019; ICU: intensive care unit; ARDS: acute respiratory distress syndrome; WHO: World Health Organization; RT-PCR: quantitative real-time reverse transcription polymerase chain reaction; COPD: chronic obstructive pulmonary disease; HIV: human immunodeficiency virus; SVM: support vector machine; IQR: interquartile range; SD: standard deviation; WBC: white blood cell; N: neutrophil; AST: aspartate aminotransferase; LDH: lactate dehydrogenase; TNI: high-sensitivity cardiac troponin I; CK: creatine kinase; CK-MB: creatine kinase-MB; CysC: cystatin C; ESR: erythrocyte sedimentation rate; CRP: high-sensitivity C-reactive protein; PCT: procalcitonin; IL: interleukin; L: lymphocyte; A/G: albumin to globulin ratio; ALB: albumin concentration; Ca: calcium; ROC: receiver operating characteristic curve; Hb: hemoglobin; PLT: platelet count; TB: total bilirubin; ALT: alanine aminotransferase; GLB: globulin; ALP: alkaline phosphatase; BUN: blood urea nitrogen; CREA: creatinine; TG: triglyceride; Na: sodium; K: Potassium; TNF: Tumor necrosis factor-α; NK: natural killer cell; Ig: Immune globulin; ROC: Receiver operating characteristic; AUC: area under curve.
Acknowledgements
We are grateful to the many frontline medical staff for their dedication in the face of this outbreak.
Author Contributions
Chen Zhao, Yan Bai, Cencen Wang, Yanyan Zhong, Na Lu, Li Tian, Fucheng Cai, and Runming Jin collected the clinical data. Chen Zhao and Yan Bai performed the data analysis and drafted the manuscript. Chen Zhao, Yan Bai and Runming Jin revised the manuscript and approved the final manuscript.
Competing Interests
The authors have declared that no competing interest exists.
References
1. Li Q, Guan XH, Wu P, Wang XY, Zhou L, Tong YQ. et al. Early Transmission Dynamics in Wuhan, China, of Novel Coronavirus-Infected Pneumonia. New England Journal of Medicine. 2020;382:1199-207
2. Chinazzi M, Davis JT, Ajelli M, Gioannini C, Litvinova M, Merler S. et al. The effect of travel restrictions on the spread of the 2019 novel coronavirus (COVID-19) outbreak. Science. 2020;368:395-400
3. Wu ZY, McGoogan JM. Characteristics of and Important Lessons From the Coronavirus Disease 2019 (COVID-19) Outbreak in China Summary of a Report of 72 314 Cases From the Chinese Center for Disease Control and Prevention. Jama-Journal of the American Medical Association. 2020;323:1239-42
4. WHO. Coronavirus disease (COVID-19) Situation Report - 119. 2020. WHO; 2020. https://www.who.int/docs/default-source/coronaviruse/situation-reports/20200518-covid-19-sitrep-119.pdf?sfvrsn=4bd9de25_4
5. Guan W, Ni Z, Hu Y, Liang W, Ou C, He J. et al. Clinical Characteristics of Coronavirus Disease 2019 in China. New England Journal of Medicine. 2020;382:1708-20
6. Huang CL, Wang YM, Li XW, Ren LL, Zhao JP, Hu Y. et al. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet. 2020;395:497-506
7. Yang XB, Yu Y, Xu JQ, Shu HQ, Xia JA, Liu H. et al. Clinical course and outcomes of critically ill patients with SARS-CoV-2 pneumonia in Wuhan, China: a single-centered, retrospective, observational study. Lancet Respiratory Medicine. 2020;8:475-81
8. WHO. Report of the WHO-China Joint Mission on Coronavirus Disease 2019 (COVID-19). WHO; 2020. https://www.who.int/docs/default-source/coronaviruse/who-china-joint-mission-on-covid-19-final-report.pdf
9. Xu PP, Tian RH, Luo S, Zu ZY, Fan B, Wang XM. et al. Risk factors for adverse clinical outcomes with COVID-19 in China: a multicenter, retrospective, observational study. Theranostics. 2020;10:6372-83
10. Wynants L, Van Calster B, Bonten MMJ, Collins GS, Debray TPA, De Vos M. et al. Prediction models for diagnosis and prognosis of covid-19 infection: systematic review and critical appraisal. Bmj-British Medical Journal. 2020 369
11. Phua J, Weng L, Ling L, Egi M, Lim CM, Divatia JV. et al. Intensive care management of coronavirus disease 2019 (COVID-19): challenges and recommendations. Lancet Respiratory Medicine. 2020;8:506-17
12. Wang Y, Lu XF, Li YS, Chen H, Chen TG, Su N. et al. Clinical Course and Outcomes of 344 Intensive Care Patients with COVID-19. American Journal of Respiratory and Critical Care Medicine. 2020;201:1430-4
13. Deng Y, Liu W, Liu K, Fang YY, Shang J, Zhou L. et al. Clinical characteristics of fatal and recovered cases of coronavirus disease 2019 in Wuhan, China: a retrospective study. Chinese Medical Journal. 2020;133:1261-7
14. Chen T, Wu D, Chen HL, Yan WM, Yang DL, Chen G. et al. Clinical characteristics of 113 deceased patients with coronavirus disease 2019: retrospective study. Bmj-British Medical Journal. 2020 368
15. Zhou F, Yu T, Du RH, Fan GH, Liu Y, Liu ZB. et al. Clinical course and risk factors for mortality of adult inpatients with COVID-19 in Wuhan, China: a retrospective cohort study. Lancet. 2020;395:1054-62
16. Wu CM, Chen XY, Cai YP, Xia JA, Zhou X, Xu S. et al. Risk Factors Associated With Acute Respiratory Distress Syndrome and Death in Patients With Coronavirus Disease 2019 Pneumonia in Wuhan, China. Jama Internal Medicine. 2020;180:934-43
17. Pan F, Yang L, Li YC, Liang B, Li L, Ye TH. et al. Factors associated with death outcome in patients with severe coronavirus disease-19 (COVID-19): a case-control study. International Journal of Medical Sciences. 2020;17:1281-92
18. Li X, Xu S, Yu M, Wang K, Tao Y, Zhou Y. et al. Risk factors for severity and mortality in adult COVID-19 inpatients in Wuhan. Journal of Allergy and Clinical Immunology. 2020
19. Li LL, Yang L, Gui S, Pan F, Ye TH, Liang B. et al. Association of clinical and radiographic findings with the outcomes of 93 patients with COVID-19 in Wuhan, China. Theranostics. 2020;10:6113-21
20. Du RH, Liang LR, Yang CQ, Wang W, Cao TZ, Li M. et al. Predictors of mortality for patients with COVID-19 pneumonia caused by SARS-CoV-2: a prospective cohort study. European Respiratory Journal. 2020 55
21. Chen R, Liang W, Jiang M, Guan W, Zhan C, Wang T. et al. Risk factors of fatal outcome in hospitalized subjects with coronavirus disease 2019 from a nationwide analysis in China. Chest. 2020
22. National Health Commission of the People's Republic of China. Notice on the issuance of a new coronavirus pneumonia diagnosis and treatment plan (try the fifth revised version). National Health Commission of the People's Republic of China; 2020. http://www.nhc.gov.cn/yzygj/s7653p/202002/d4b895337e19445f8d728fcaf1e3e13a.shtml
23. Li LL, Fan WL, Li J, Li QL, Wang J, Fan Y. et al. Abnormal brain structure as a potential biomarker for venous erectile dysfunction: evidence from multimodal MRI and machine learning. European Radiology. 2018;28:3789-800
24. Pedregosa F, Varoquaux G, Gramfort A, Michel V, Thirion B, Grisel O. et al. Scikit-learn: Machine Learning in Python. Journal of Machine Learning Research. 2011;12:2825-30
25. Wang DW, Hu B, Hu C, Zhu FF, Liu X, Zhang J. et al. Clinical Characteristics of 138 Hospitalized Patients With 2019 Novel Coronavirus-Infected Pneumonia in Wuhan, China. Jama-Journal of the American Medical Association. 2020;323:1061-9
26. Chen NS, Zhou M, Dong X, Qu JM, Gong FY, Han Y. et al. Epidemiological and clinical characteristics of 99 cases of 2019 novel coronavirus pneumonia in Wuhan, China: a descriptive study. Lancet. 2020;395:507-13
27. Wang T, Du Z, Zhu F, Cao Z, An Y, Gao Y. et al. Comorbidities and multi-organ injuries in the treatment of COVID-19. Lancet. 2020;395:E52
28. Mo P, Xing Y, Xiao Y, Deng L, Zhao Q, Wang H. et al. Clinical characteristics of refractory COVID-19 pneumonia in Wuhan, China. Clinical infectious diseases: an official publication of the Infectious Diseases Society of America. 2020
29. Han R, Huang L, Jiang H, Dong J, Peng H, Zhang D. Early Clinical and CT Manifestations of Coronavirus Disease 2019 (COVID-19) Pneumonia. American Journal of Roentgenology. 2020;215:338-43
30. Goronzy JJ, Fang FQ, Cavanagh MM, Qi Q, Weyand CM. Naive T Cell Maintenance and Function in Human Aging. Journal of Immunology. 2015;194:4073-80
31. Pedersen SF, Ho Y-C. SARS-CoV-2: a storm is raging. Journal of Clinical Investigation. 2020;130:2202-5
32. Shi SB, Qin M, Shen B, Cai YL, Liu T, Yang F. et al. Association of Cardiac Injury With Mortality in Hospitalized Patients With COVID-19 in Wuhan, China. JAMA Cardiology. 2020;5:802-10
33. Ruan QR, Yang K, Wang WX, Jiang LY, Song JX. Clinical predictors of mortality due to COVID-19 based on an analysis of data of 150 patients from Wuhan, China. Intensive Care Medicine. 2020;46:846-8
34. Chapman AR, Lee KK, McAllister DA, Cullen L, Greenslade JH, Parsonage W. et al. Association of High-Sensitivity Cardiac Troponin I Concentration With Cardiac Outcomes in Patients With Suspected Acute Coronary Syndrome. Jama-Journal of the American Medical Association. 2017;318:1913-24
35. Guo T, Fan YZ, Chen M, Wu XY, Zhang L, He T. et al. Cardiovascular Implications of Fatal Outcomes of Patients With Coronavirus Disease 2019 (COVID-19). JAMA Cardiology. 2020;5:811-8
36. Qiu HY, Wu JH, Hong L, Luo YL, Song QF, Chen D. Clinical and epidemiological features of 36 children with coronavirus disease 2019 (COVID-19) in Zhejiang, China: an observational cohort study. Lancet Infectious Diseases. 2020;20:689-96
Author contact
![]() Corresponding author: Yan Bai, Department of Pediatric, Union Hospital, Tongji Medical College, Huazhong University of Science and Technology, 1277 Jiefang Road, Wuhan, 430022, P.R. China. Tel.: +86 18872283653; E-mail: yanbaixhedu.cn.
Corresponding author: Yan Bai, Department of Pediatric, Union Hospital, Tongji Medical College, Huazhong University of Science and Technology, 1277 Jiefang Road, Wuhan, 430022, P.R. China. Tel.: +86 18872283653; E-mail: yanbaixhedu.cn.

 Global reach, higher impact
Global reach, higher impact