Impact Factor
ISSN: 1449-1907
Int J Med Sci 2020; 17(18):3190-3199. doi:10.7150/ijms.47003 This issue Cite
Research Paper
Combined use of murine double minute-2 promoter methylation and serum AFP improves diagnostic efficiency in hepatitis B virus-related hepatocellular carcinoma
1. Department of Hepatology, Qilu Hospital of Shandong University, Jinan 250012, China.
2. Institute of Hepatology, Shandong University, Jinan 250012, China.
Received 2020-4-14; Accepted 2020-10-7; Published 2020-10-23
Abstract
Objective: Hepatocellular carcinoma (HCC) accounts for approximately 85% of all cases of liver cancer. In China, chronic hepatitis B virus-related HCC (HBV-related HCC) is the most common type of HCC. However, the majority of HBV-related HCC patients are asymptomatic, and the best opportunities for treating these patients are missed. The precise diagnosis of HBV-related HCC is crucial. The main purpose of this study was to evaluate the diagnostic value of murine double minute-2 (MDM2) promoter methylation in HBV-related HCC patients.
Methods: The methylation status of the MDM2 promoter was detected by methylation-specific PCR. The MDM2 expression levels were validated by quantitative real-time PCR. Enzyme-linked immunosorbent assay was used to determine the levels of interleukin-6 (IL-6) and tumor-necrosis factor-α (TNF-α) in plasma.
Results: The methylation frequency of the MDM2 promoter was decreased in HBV-related HCC patients. The MDM2 mRNA levels of patients with HBV-related HCC were higher than those of patients with liver cirrhosis and chronic hepatitis B. The plasma levels of IL-6 and TNF-α were significantly higher in HBV-related HCC patients than that in liver cirrhosis and chronic hepatitis B patients. The TNF-α levels were higher in the unmethylated MDM2 promoter group than in the methylated MDM2 promoter group in HBV-related HCC patients. Moreover, the combination of MDM2 promoter methylation and alpha-fetoprotein (AFP) improved the diagnosis of HBV-related HCC.
Conclusions: Our study indicates, for the first time, that MDM2 promoter hypomethylation is present in HBV-related HCC patients. The combination of MDM2 promoter methylation and AFP can greatly improve diagnostic efficiency in HBV-related HCC, which might provide a new method for HBV-related HCC diagnosis.
Keywords: HBV-related HCC, MDM2, DNA methylation
Introduction
Hepatocellular carcinoma (HCC) is the major type of primary liver cancer, accounting for 85% of liver cancer cases worldwide [1]. HCC is the third leading cause of death from malignant tumors in the world, and the development of HCC is closely related to alteration in the structure or expression of certain oncogenes [2-4]. In China, chronic hepatitis B virus (HBV) infection is one of major causes of HCC [5]. To date, alpha-fetoprotein (AFP) has been used as HCC marker for more than 50 years, but its sensitivity and specificity are limited. It is particularly important to improve the diagnostic efficiency of HBV-related HCC. Therefore, there is an urgent need to search for a potential biomarker to diagnose HBV-related HCC.
Murine double minute-2 (MDM2) is an E3 ubiquitin ligase, and the gene encodes a negative regulator of the p53 tumor suppressor [6]. MDM2 binds to the transcriptional activation domain of p53 to regulate the stabilization and activation of p53 [7,8]. Under normal physiological conditions, MDM2 and p53 form a self-regulating negative feedback loop to ensure a balance between them [9]. However, the negative feedback loop is often observed to be broken in various tumors. MDM2 overexpression, in particular, has been implicated in several tumors, such as sarcoma, colon cancer and gastric cancer [10-12]. Previous studies have shown that MDM2 is an optimal target for therapy in tumors carrying wild-type tumor protein 53 [13-15]. Furthermore, MDM2 can regulate the target protein interleukin-6 (IL-6) and tumor-necrosis factor-α (TNF-α), which play an important role in regulating the mitogen-activated protein kinase (MAPK) and nuclear factor-kappa B (NF-κB) pathways [16].
Epigenetic modification, including aberrant DNA methylation, is one of the important events in carcinogenesis [17]. DNA methylation is a heritable change in gene expression without DNA sequence change [18]. DNA methylation is of great importance for gene expression and can have dire consequences for cells [19]. The overexpressed oncoprotein MDM2 not only binds to p53 and negatively regulates p53 but also contributes to HCC development, regardless of p53 status. A previous study has suggested that MDM2 hypomethylation and overexpression take place in pterygium [20]. DNA methylation has been widely investigated in a variety of tumors, such as gastric cancer and lung cancer [21,22]. Multiple studies have demonstrated that DNA methylation of some genes can be used as biomarkers for diagnosis, prognosis or therapeutic strategies [23,24].
However, studies associating MDM2 methylation in diseases are scarce. To the best of our knowledge, the methylation status of MDM2 in HBV-related HCC has not yet been explored. In this study, we detected the methylation status of MDM2, and we quantitatively analyzed the mRNA levels of MDM2 in peripheral blood mononuclear cells (PBMCs). Moreover, we determined the IL-6 and TNF-α levels in plasma. We aimed to determine the value of MDM2 methylation in the diagnosis of HBV-related HCC.
Materials and methods
Subjects
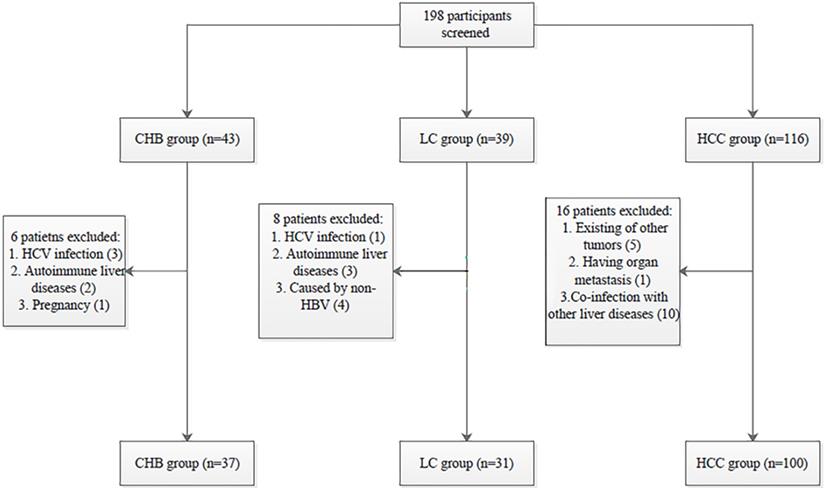
One hundred patients with HBV-related HCC, 31 patients with liver cirrhosis (LC), and 37 patients with chronic hepatitis B (CHB) were recruited from June 2016 to February 2018 at the Department of Hepatology, Qilu Hospital of Shandong University. CHB patients were selected according to the practice guidelines for managing CHB established in the 2018 update of the American Association for the Study of Liver Diseases [25]. The inclusion criteria for LC patients followed the evidence-based clinical practice guidelines for liver cirrhosis 2015 [26]. All HBV-related HCC patients were selected according to the 2010 update of the American Association for the Study of Liver Diseases Practice Guidelines for Management of hepatocellular carcinoma [27].
All procedures performed in the studies involving human participants were in accordance with the Declaration of Helsinki and the ethical standards of the Medical Ethics Committee of Shandong University Qilu Hospital (NO. 2019058). Informed consent was obtained from all individual participants included in the study. The selection process of participants is shown in Fig. 1.
Genomic DNA extraction and sodium bisulfite modification
Genomic DNA was extracted from PBMCs using the QIAamp DNA Mini Kit (Qiagen, Hilden, Germany). Genomic DNA modification was performed using EZ DNA Methylation-Gold kit (Zymo Research, Irvine, CA, USA), and the procedure was conducted in accordance with the instructions. Bisulfite-converted DNA (20 μL) was used for methylation-specific polymerase chain reaction (MSP) or stored in a refrigerator at -80 ºC for standby application.
Methylation-specific PCR (MSP)
The modified DNA was amplified by MSP. The MDM2 promoter structure and MSP primer are shown in Fig. 2. The MDM2 primers for MSP were shown as follows: methylated forward primer 5'- TAACGGTTAAAGGAGTGTTATAGCG -3' and reverse primer 5'- GAAATAAAAATATTAACCGCGAAC -3'; unmethylated forward primer 5'-AATGGTTAAAGGAGTGTTATAGT-3' and reverse primer 5'-CAAAATAAAAATATTAACCACAAAC-3'. Each MSP contained 1 μL modified DNA, 0.5 μL forward and reverse primers (10 μM), 12.5 μL Premix Taq (Zymo Research, CA, USA), and 10.5 μL nuclease-free water. The PCR program included initial denaturation at 95 ºC for 5 min, followed by 35 cycles of 95 ºC for the 30 sec, 58.5 ºC for 30 sec, and 72 ºC for 30 sec; and a final extension at 72 ºC for 10 min. Water without DNA was used as the negative control. The separation of the 10 μL PCR products was conducted by electrophoresis on a 2% agarose gel, which was stained with Gel Red (Biotium, California, USA), and visualized under UV illumination. The MDM2 methylation status was determined positive if the MDM2 methylated primer or MDM2 methylated primer and MDM2 unmethylated primer both produced a band of expected size; otherwise it was negative.
RNA extraction and quantitative real-time polymerase chain reaction (RT-qPCR)
Total RNA was extracted from 5 ml peripheral venous blood by TRIzol (Invitrogen, Carlsbad, CA, USA). The RNA was transcribed into cDNA using first-strand cDNA synthesis kit (Fermentas, Vilnius, Lithuania). The levels of MDM2 were measured by RT-qPCR, using the SYBR Green PCR mix (Takara, Japan), the Agilent Mx3005P (Agilent Technologies, USA). β-actin was used as the internal reference gene. The primer sequences were described as follows: MDM2: forward primer 5'-GGGAGTGATCAAAAGGAC-3' and reverse primer 5'-CCAAATGTGAAGATGAAGGTTTC-3' [28]; β-actin: forward primer 5'-ATGGGTCAGAAGGATTCCTATGTG-3' and reverse primer 5'- CTTCATGAGGTAGTCAGTCAGGTC-3'. The total volume of 20 μL PCR solution contained 10 μL SYBR Green premix, 8.2 μL nuclease-free water, 1 μL cDNA and 0.4 μM of each primer. Each PCR program consisted of denaturation at 95 ºC for the 30 sec, followed by 40 cycles of 95 ºC for 5 sec, 60 ºC for 30 sec and 72 ºC for 30 sec. The mRNA level was determined using the comparative (2-ΔΔCt) method.
Enzyme-linked immunosorbent assay (ELISA) for detection of IL-6 and TNF-α in plasma
Plasma was obtained from peripheral venous blood by centrifugation (3000 r/min, 10 min). The plasma IL-6 and TNF-α levels were measured using the Human IL-6 Immunoassay Quantikine ELISA kit (R&D San Diego, CA, USA) and Human TNF-α Immunoassay Quantikine ELISA kit (R&D San Diego, CA, USA) according to the manufacturer's instructions.
Patient selection process. HBV: hepatitis B virus; HCV: hepatitis C virus; CHB: chronic hepatitis B; LC: liver cirrhosis; HBV-related HCC: hepatitis B virus-related hepatocellular carcinoma.
MSP amplification regions and TSS. MSP amplified regions (288-380) are indicated and TSS (1477) is indicated. MSP: methylation-specific PCR; TSS: transcription start site.

Statistical analysis
Statistical analysis was performed using SPSS version 19.0 software (SPSS, Chicago, IL, USA) and GraphPad Prism 6.0 (San Diego, CA, USA). The quantitative variables were expressed as median (centile 25; centile 75), and the differences between groups were compared using Mann-Whitney U-test or Kruskal-Wallis test. The categorical variables were expressed as number (%), and differences between groups were compared with Chi-squared tests. Associations of the MDM2 expression levels with the clinicopathological characteristics of patients were analyzed by the Spearman correlation test. In order to assess the diagnostic value of MDM2 promoter methylation, we combined MDM2 promoter methylation and the AFP to measure the area under the receiver operating characteristic (ROC) curve. The results were considered statistically significant when P<0.05.
Results
Clinical characteristics
In this study, we included a total of 168 subjects. The clinicopathological variables are shown in Table 1. The clinical differences between the patients in the MDM2 methylated group and those in the MDM2 unmethylated group are shown in Table 2. The significant gender and HBe Ag differences between the patients with LC the MDM2 methylated group and MDM2 unmethylated group are shown. Among the CHB and LC patients, the AFP levels in the MDM2 unmethylated group were higher than those in MDM2 methylated group. However, there was no statistically significant difference in the patients with HCC.
Baseline characteristics of the patients
| Variable | CHB (n=37) | LC (n=31) | HCC (n=100) | P value |
|---|---|---|---|---|
| Gender (M/F) | 25/12 | 24/7 | 80/20 | 0.309b |
| Age (years) | 41 (31-53) | 48 (39-55) | 54 (49-62) | <0.001a |
| Log10[HBV DNA] | 6.33 (4.29-7.04) | 4.90 (3.52-7.14) | 2.94 (2.00-4.35) | <0.001 a |
| HBe Ag (+/-) | 26/11 | 20/11 | 30/70 | <0.001 b |
| ALT (U/I) | 88 (58.00-101.50) | 71.00 (48.00-97.00) | 78.50 (59.00-103.25) | 0.597 a |
| AST (U/I) | 84.00 (53.00-116.50) | 63.00 (50.00-90.00) | 82.50 (65.25-120.75) | 0.048 a |
| TBIL (μmol/L) | 18.70 (12.10-45.35) | 37.20 (18.30-83.80) | 23.95 (16.55-52.08) | 0.106 a |
| ALB (g/L) | 42.40 (38.85-45.30) | 38.60 (28.70-42.40) | 39.90 (34.83-43.05) | 0.015 a |
| PT (s) | 12.10 (11.40-13.55) | 13.90 (12.70-16.70) | 13.10 (11.83-14.78) | <0.001 a |
| AFP (ng/ml) | 12.75 (3.83-33.26) | 16.82 (2.83-83.44) | 28.41 (5.99-1214.65) | 0.010 a |
| Child-Pugh class | 0.619 a | |||
| A | NA | 16 | 61 | |
| B | NA | 11 | 27 | |
| C | NA | 4 | 12 | |
| Antiviral agent | 0.604 a | |||
| Adefovir | 10 | 6 | 27 | |
| Entecavir | 17 | 18 | 52 | |
| Tenofovir | 1 | 2 | 1 | |
| Lamivudine | 1 | 2 | 3 | |
| No | 8 | 3 | 17 | |
| Treatment duration | 0.582 a | |||
| < 6 months | 18 | 16 | 40 | |
| 6-12 months | 3 | 1 | 12 | |
| 12-24 months | 2 | 2 | 10 | |
| > 24 months | 6 | 9 | 21 | |
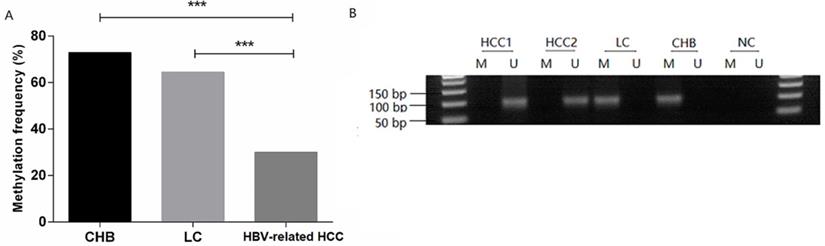
| MDM2 methylation | 27 (72.97%) | 20 (64.52%) | 30 (30.00%) | <0.001 b |
CHB: chronic hepatitis B; LC: liver cirrhosis; HCC: hepatocellular carcinoma; HBe Ag: hepatitis B e antigen; ALT: alanine aminotransferase; AST: aminotransferase aspartate; TBIL: total bilirubin; ALB: albumin; PT: prothrombin time; NA: not available.
a: Kruskal-Wallis H test.
b: Chi-square test.
The clinical differences between patients in the MDM2 methylated group and the MDM2 unmethylated group
| Variable | CHB (n=37) | P value | LC (n=31) | P value | HBV-related HCC (n=100) | P value | |||
|---|---|---|---|---|---|---|---|---|---|
| Methylated | Unmethylated | Methylated | Unmethylated | Methylated | Unmethylated | ||||
| Gender (M/F) | 17/10 | 8/2 | 0.326b | 13/7 | 11/0 | 0.026 b | 24/6 | 56/14 | 1.000b |
| Age (years) | 41 (31-54) | 48.5 (31.75-52.5) | 0.768a | 48 (40.5-55.75) | 44 (31-55) | 0.364 a | 52.5 (46.75-63) | 54 (50-62) | 0.377a |
| Log10[HBV DNA] | 6.03 (4.16-7.17) | 6.39 (4.13-7.06) | >0.05a | 4.61 (3.02-7.05) | 6.68 (4.18-7.42) | 0.182 a | 2.84 (2.00-4.69) | 3.04 (2.00-4.24) | 0.977a |
| HBe Ag (+/-) | 21/6 | 5/5 | 0.101 b | 10/10 | 10/1 | 0.023b | 7/23 | 23/47 | 0.341b |
| ALT (U/I) | 81.00 (57.00-101.00) | 90.00 (49.25-103.50) | 0.756 a | 61.50 (41.75-96.75) | 84.00 (59.00-100.00) | 0.256 a | 75.00 (58.25-98.50) | 79.00 (59.00-105.25) | 0.566a |
| AST (U/I) | 84.00 (54.00-117.00) | 76.00 (48.75-111.25) | 0.794 a | 62.00 (47.50-88.75) | 81.00 (50.00-113.00) | 0.256 a | 91.00 (69.25-138.50) | 82.00 (64.75-114.75) | 0.511a |
| TBIL (μmol/L) | 16.70 (10.50-39.80) | 24.70 (15.85-123.60) | 0.253 a | 25.95 (16.50-75.35) | 47.70 (20.60-109.5) | 0.208 a | 20.70 (16.88-49.25) | 24.95 (16.48-59.98) | 0.743a |
| ALB (g/L) | 42.80 (40.40-45.10) | 39.60 (37.10-45.85) | 0.473a | 40.35 (29.98-42.30) | 35.90 (28.70-43.10) | 0.847 a | 38.80 (34.78-41.90) | 40.40 (34.50-43.95) | 0.227a |
| PT (s) | 12.10 (11.20-13.60) | 12.00 (11.55-13.70) | 0.743 a | 13.70 (12.58-16.93) | 14.40 (12.70-16.60) | 0.677 a | 13.45 (12.38-14.60) | 12.70 (11.80-14.43) | 0.124a |
| AFP (ng/ml) | 7.44 (2.77-15.48) | 43.54 (30.86-69.95) | <0.001a | 4.56 (1.95-16.61) | 91.05 (46.27-297.30) | <0.001a | 93.63 (6.34-1719.69) | 16.30 (5.84-1213.87) | 0.528a |
CHB: chronic hepatitis B; LC: liver cirrhosis; HBV-related HCC: hepatitis B virus-related hepatocellular carcinoma; HBe Ag: hepatitis B e antigen; ALT: alanine aminotransferase; AST: aminotransferase aspartate; TBIL: total bilirubin; ALB: albumin; PT: prothrombin time; AFP: alpha-fetoprotein.
a: Kruskal-Wallis H test.
b: Chi-square test.
MDM2 promoter methylation status in patients. (A) MDM2 promoter methylation frequency of HBV-related HCC, LC, and CHB patients. (B) Methylation-specific PCR results of the MDM2 promoter. CHB: chronic hepatitis B; LC: liver cirrhosis; HBV-related HCC: hepatitis B virus-related hepatocellular carcinoma; M: methylated sequence; U: unmethylated sequence; NC: negative control. ***P<0.001.
Association between clinicopathological features and the methylation of MDM2 promoter in patients with HBV-related HCC
| Variable | Total number | Methylated | Unmethylated | P value |
|---|---|---|---|---|
| Gender | 1.000 | |||
| Male | 80 | 24 (30.00%) | 56(70.00%) | |
| Female | 20 | 6 (30.00%) | 14 (70.00%) | |
| Age | 0.055 | |||
| ≥50 | 73 | 18 (24.66%) | 55 (75.34%) | |
| <50 | 27 | 12 (44.44%) | 15 (55.56%) | |
| AFP (ng/ml) | 0.239 | |||
| ≥20 | 51 | 18 (35.29%) | 33 (64.71%) | |
| <20 | 49 | 12 (24.49%) | 37 (75.51%) | |
| Vascular invasion | 0.752 | |||
| Yes | 22 | 6 (27.27%) | 16 (72.73%) | |
| No | 78 | 24 (30.77%) | 54 (69.23%) | |
| Lymph node metastasis | 0.399 | |||
| Yes | 22 | 5 (22.73%) | 17 (77.27%) | |
| No | 78 | 25 (32.05%) | 53 (67.95%) | |
| Distant metastasis | 0.040 | |||
| Yes | 9 | 0 (0.00%) | 9 (100.00%) | |
| No | 91 | 30 (32.97%) | 61 (67.03%) | |
| Number of tumors | 0.784 | |||
| Multiple | 51 | 13 (25.49%) | 37 (74.51%) | |
| Solitary | 49 | 17 (34.69%) | 33 (65.31%) | |
| Tumor size (cm) | 0.961 | |||
| >5 | 27 | 8 (29.63%) | 19 (70.37%) | |
| ≤5 | 73 | 22 (30.14%) | 51 (69.86%) | |
| TNM stage | 0.035 | |||
| Ι+ΙΙ | 61 | 23 (37.70%) | 38 (62.30%) | |
| ΙΙΙ+ΙV | 39 | 7 (17.95%) | 32 (82.05%) | |
| BCLC stage | 0.044 | |||
| A-C | 86 | 29 (33.72%) | 57 (66.28%) | |
| D | 14 | 1 (7.14%) | 13 (92.86%) |
MDM2: murine double minute-2; HCC: hepatocellular carcinoma; AFP: α-fetoprotein; TNM: tumor node metastasis; BCLC: Barcelona Clinic Liver Cancer.
Statistical analysis was performed using Chi-square test.
MDM2 promoter methylation is decreased in HBV-related HCC patients
The methylation frequency of MDM2 detected in PBMCs was 72.97% (27/37) in the CHB patients, 64.52% (20/31) in the LC patients, 30.00% (30/100) in the HBV-related HCC patients. The MDM2 methylation frequency in the HBV-related HCC patients was significantly lower than that in CHB patients [χ2=20.528, odds ratio (OR) = 6.300, 95% confidence interval (CI): 2.714-14.626, P=0.000] and LC (χ2=11.946, OR=4.242, 95%CI: 1.811-9.936, P=0.001) (Fig. 3A). However, no difference in the MDM2 methylation frequency was detected between the CHB and LC patients (χ2=0.563, P=0.452). The agarose gel electrophoresis results showing the MDM2 promoter methylation status are presented in Fig. 3B.
MDM2 promoter methylation is associated with tumor node metastasis (TNM) stage and Barcelona Clinic Liver Cancer (BCLC) stage
Next, we analyzed the correlation between MDM2 promoter methylation and clinicopathological characteristics in the HBV-related HCC patients (Table 3). The results revealed that MDM2 promoter methylation was significantly correlated with distant metastasis (P=0.040), TNM stage (P=0.035) and BCLC stage (P=0.044). There was no correlation between MDM2 promoter methylation and other clinicopathological characteristics, such as gender, age, AFP, vascular invasion, lymph node metastasis, number of tumors, tumor size.
Association between mRNA levels of MDM2 in PBMCs and clinicopathological characteristics
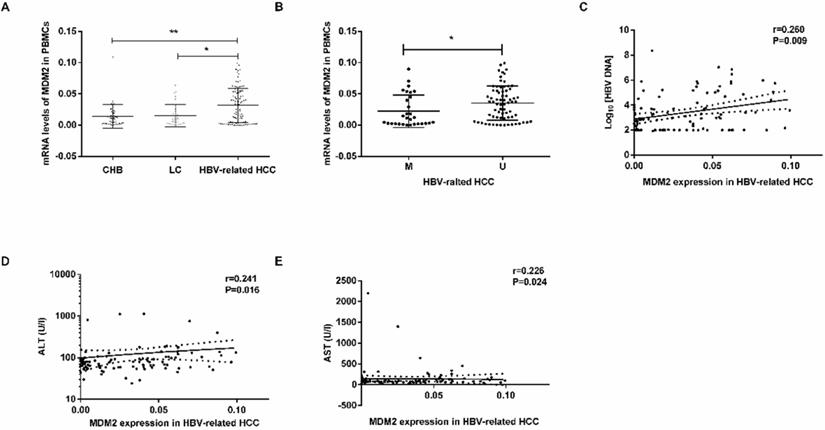
MDM2 mRNA levels were obviously higher in the HBV-related HCC patients than in the CHB patients (P=0.0034) LC patients (P=0.0102) (Fig. 4A). There were no significant differences in MDM2 mRNA levels between the CHB and LC patients (P>0.05). In the HBV-related HCC patients, the MDM2 mRNA levels in the unmethylated group were significantly higher than those in the methylated group (P=0.0236, Fig. 4B).
MDM2 mRNA level and its relationship with clinicopathological characteristics. (A) The mRNA levels of MDM2 among different samples from CHB, LC and HBV-related HCC. (B) MDM2 mRNA levels in HBV-related HCC patients in MDM2 M group and U group. (C-E) The linear correlation between MDM2 mRNA levels and clinicopathological characteristics in HBV-related HCC patients. CHB: chronic hepatitis B; LC: liver cirrhosis; HBV-related HCC: hepatitis B virus-related hepatocellular carcinoma; M: methylated group; U: unmethylated group; ALT: alanine aminotransferase; AST: aminotransferase aspartate. *P<0.05, **P<0.01.
Plasma levels of IL-6 and TNF-α in patients
| Cytokines (pg/ml) | CHB | LC | HBV-related HCC |
|---|---|---|---|
| IL-6 | 2.17 (0.89-3.62) | 2.25 (0.64-8.20) | 5.68 (2.13-10.43) |
| TNF-α | 7.76 (5.98-12.24) | 9.15 (5.37-13.26) | 13.90 (9.36-19.96) |
CHB: chronic hepatitis B; LC: liver cirrhosis; HBV-related HCC: hepatitis B virus-related hepatocellular carcinoma; IL-6: interleukin-6; TNF-α: tumor-necrosis factor-α.
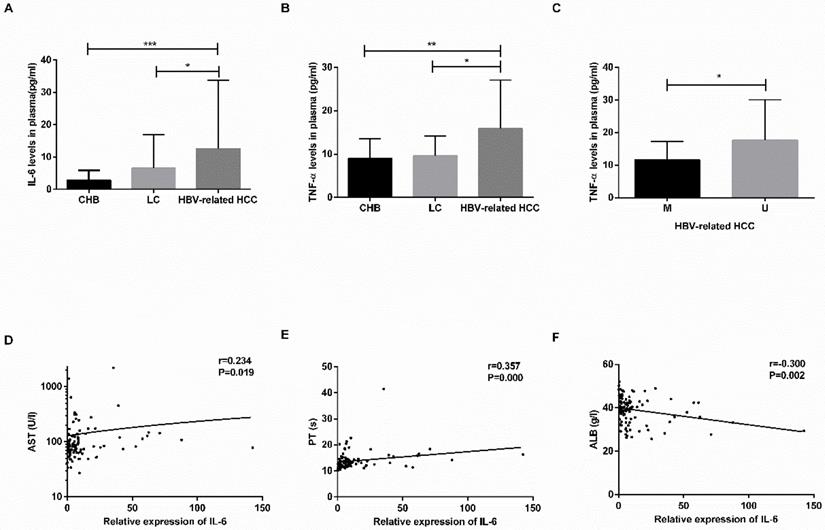
We next studied the correlation between MDM2 mRNA levels and clinicopathological characteristics in the HBV-related HCC patients. We found a positive correlation between MDM2 mRNA levels and HBV DNA (P=0.009), alanine aminotransferase (ALT) (P=0.016), aminotransferase aspartate (AST) (P=0.024) (Fig. 4C-E). However, there was no correlation between MDM2 mRNA levels and total bilirubin (TBIL), albumin (ALB) or prothrombin time (PT).
Levels of IL-6 and TNF-α are increased in plasma of HBV-related HCC patients
The plasma levels of IL-6 and TNF-α are shown in Table 4. The levels of IL-6 and TNF-α were increased in the HBV-related HCC patients compared with the CHB patients (IL-6: P=0.0001; TNF-α: P=0.0031) and LC patients (IL-6: P=0.0171; TNF-α: P=0.0176) (Fig. 5A-B). No significant difference was found between the CHB and LC patients (P>0.05).
In the HBV-related HCC patients, the TNF-α levels in the MDM2 unmethylated group were significantly increased in the MDM2 methylated group (P=0.0246) (Fig. 5C), the levels of IL-6 in the MDM2 unmethylated group were slightly higher than those in the MDM2 unmethylated group (P>0.05). The IL-6 levels were positively correlated with AST (P=0.019) and PT (P=0.000) in the HBV-related HCC patients (Fig. 5D-F) but were negatively correlated with ALB (P=0.002) (Fig. 5E). We did not find any possible relationship between TNF-α and clinicopathological features.
Combination of MDM2 promoter methylation and AFP can improve diagnostic efficiency in HBV-related HCC
The ROC curve was established, including AFP, methylation status of MDM2 and combined determination, to identify the HBV-related HCC, CHB and LC. We compared the diagnostic value of AFP (cut off point: 20 ng/ml), MDM2 methylation, and combined determination to distinguish HBV-related HCC from CHB. For the combination determination, the sensitivity was 89.00%, the specificity was 62.16%, the positive predictive value (PPV) was 87.25%, and the negative predictive value (NPV) was 62.07%, which was better than AFP (sensitivity: 52%, specificity: 62.16%, PPV: 78.79%, NPV: 32.39%) and MDM2 promoter methylation (sensitivity: 70.00%, specificity: 72.97%, PPV: 87.5%, NPV: 43.37%). Moreover, the area under curve (AUC) of the combined determination was 0.756 [standard error (SE)=0.0434, 95% CI=0.675-0.825)], which was significantly higher than AFP levels (AUC=0.639, SE=0.0471, 95% CI=0.553-0.720; P=0.0056) and slightly higher than MDM2 promoter methylation (AUC=0.715, SE=0.0436, 95% CI=0.632-0.789; P>0.05) (Fig. 6A).
The concentration of IL-6 and TNF-α in plasma and relationship with clinicopathological characteristics. (A-B) The plasma levels of IL-6 and TNF-α in CHB, LC, HBV-related HCC patients. (C) The difference in TNF-α levels in HBV-related HCC patients in the M group and U group. (D-F) The linear correlation between IL-6 levels and clinicopathological characteristics of HBV-related HCC patients. CHB: chronic hepatitis B; LC: liver cirrhosis; HBV-related HCC: hepatitis B virus-related hepatocellular carcinoma; IL-6: interleukin-6; TNF-α: tumor-necrosis factor-α; M: methylated group; U: unmethylated group; AST: aminotransferase aspartate; PT: prothrombin time; ALB: albumin. *P<0.05, ** P<0.01, *** P<0.001.
The receiver operating characteristic (ROC) curve of MDM2 promoter methylation and serum AFP in discriminating HBV-related HCC from CHB and LC patients. (A) Discriminating HBV-related HCC from CHB patients. (B) Discriminating HBV-related HCC from LC patients. AFP: alpha-fetoprotein; AUC: area under curve.
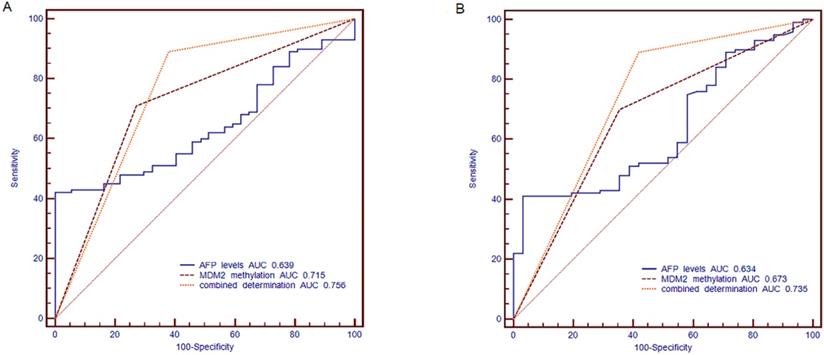
Meanwhile, AFP (cut off point: 20 ng/ml) combined with MDM2 methylation in distinguishing the HBV-related HCC patients from LC patients showed a sensitivity of 89.00%, specificity of 58.06%, PPV of 87.25%, and NPV of 62.07%, which were higher than the results of AFP (sensitivity: 52.00%, specificity: 58.06%, PPV: 80.00%, NPV: 27.27%) and MDM2 methylation (sensitivity: 70%, specificity: 64.52%, PPV: 86.42%, NPV: 40.00%). The combined determination AUC was 0.735 (SE=0.0477, 95% CI=0.651-0.809], which was higher than that of AFP alone (AUC=0.634, SE=0.0533, 95% CI=0.545-0.716; P=0.0128) and MDM2 methylation (AUC=0.673, SE=0.0494, 95% CI=0.585-0.752; P=0.0356) (Fig. 6B).
Discussion
Early-stage HCC is usually asymptomatic, and only 20% of cases of HCC are surgically removed [29]. Epigenetic alterations, particular DNA methylation, could be used for assessment in diagnosis [30]. In the present study, we first found that MDM2 promoter methylation existed in HBV-related HCC patients, LC patients and CHB patients. In addition, the MDM2 promoter methylation frequency in HBV-related HCC patients was lower than that in LC patients and CHB patients. MDM2 promoter methylation combined with AFP could be used as an effective diagnostic method for HBV-related HCC.
Oncogene activation, tumor suppressor function attenuation and inflammatory responses are involved in the pathogenesis of HCC. DNA methylation might be linked to the pathogenesis of HCC. MDM2 is an oncogene that can be expressed in PBMCs [31,32]. It is known that the products of MDM2 gene play an important role in HCC and promote tumor growth [33]. Previous study indicated that MDM2 gene promoter hypomethylation and overexpression occur in patients with pterygium [20]. We found that the methylation frequency of the MDM2 promoter in patients with HBV-related HCC was significantly decreased compared to that in LC and CHB patients. The decreased MDM2 promoter methylation showed a correlation with distant metastasis, TNM stage and BCLC stage in patients with HCC. These finds suggest that MDM2 gene promoter hypomethylation takes place in HBV-related HCC patients and is involved in HBV-related HCC progression. The exact mechanism of DNA methylation is not clear, several studies demonstrate that there is a correlation between oxidative stress and DNA methylation [34,35]. Oxidative stress could cause oncogene demethylation, which leads to oncogene overexpression and might play a role in the occurrence and development of HCC [36]. Our previous research has pointed out that IGFBP7 gene promoter methylation is related to oxidative stress in HBV-related HCC patients, while oxidative stress is involved in the development and progression of HCC [37]. We will make further study on the relationship between MDM2 gene promoter methylation and oxidative stress in the next research work.
MDM2 overexpression leads to loss of p53 function, resulting in the occurrence of tumors [38]. In humans, MDM2 overexpression is common in many different tumor types, such as soft tissue tumor, osteosarcoma and esophageal cancer [39]. Gene promoter methylation can lead to repressions of transcription. In this study, we showed that HBV-related HCC patients have increased MDM2 mRNA levels. This result is compatible with previous study by Zhang et al [40]. We also found that MDM2 mRNA levels were negatively correlated with MDM2 gene promoter methylation status in HBV-related HCC. This finding confirms a previous study that gene promoter methylation usually leads to transcriptional silencing [41]. However, it has already known that there is a mechanism of p53 degradation that is not mediated by MDM2 [42]. E3 ubiquitin ligases (E3s) can be divided into the really interesting new genes (RING)-finger protein family and the homologous to E6AP carboxyl terminus (HECT) family. MDM2 and Pirh2 belong to the single-subunit RING finger E3s, COP1 belongs to the subunit of multiunit RING finger E3s, as well as Arf-BP1 belongs to the HECT family. Previous studies reported elevated levels of MDM2, Pirh2, COP1 and Arf-BP1 in HCC and other tumors compared with those in normal tissues [43-46]. COP1 and Pirh2 are negative regulators of p53 by ubiquitination independent of MDM2 and could be the predictors for HCC survival [47,48]. Arf-BP1 is also an E3 ligase of the p53-independent and p53-dependent in vivo [49]. Although these E3 ligases are involved in MDM2-independent p53 degradation, the roles of these ligases in the regulation of p53 and the usefulness in the diagnosis of HCC remain to be explored.
MDM2 promoted the production of TNF-α and IL-6 through the MAPK and NF-κB pathways [16]. IL-6 and TNF-α play an important role in the occurrence and development of tumors, including HCC [50]. The occurrence and poor prognosis of HCC are associated with the higher serum levels of IL-6 [51,52]. Elevated TNF-α levels are found in several cancer tissues, such as ovarian and renal cancers [50,53]. Blocking MDM2 has anti-inflammatory and anti-cancer effects [54-56]. In this present study, we observed that plasma IL-6 and TNF-α levels were significantly higher in the HBV-related HCC patients. The TNF-α levels were lower in the methylated MDM2 promoter group than those without. These results indicate a link between MDM2 gene promoter methylation and inflammatory factors elevation in HBV-related HCC.
Serum AFP has been used in the diagnosis of HCC, however, serum AFP is often elevated in inflammation and other non-cancerous conditions; in addition, 40% of HCC patients do not manifest an elevation in serum AFP [57,58]. The promoter methylation of certain genes plays an important role in human cancers, these genes can be used as diagnostic biomarkers. In this study, we constructed the ROC curve to investigate the diagnostic value of combination MDM2 promoter methylation and AFP. The results indicated that the sensitivity and specificity of combining AFP and MDM2 methylation were higher compared to AFP alone. We also found that the decreased MDM2 promoter methylation correlates with the early stage of HBV-related HCC. These results suggested the method of diagnosing cancer that combined of AFP and MDM2 promoter methylation could be used to diagnose early HBV-related HCC. The present study first revealed the crucial value of the combination of MDM2 promoter methylation and serum AFP in the early diagnosis of HBV-related HCC. Sorafenib is a tyrosine kinase inhibitor and has a significant therapeutic effect on advanced HCC. It has been shown that circulating cell-free DNA may be potential biomarkers for determining the effect of HCC treatment [59]. The predictive value of MDM2 methylation for the treatment outcome of sorafenib will be performed in the future study.
There are some limitations to this research. First, our study was a retrospective single-center study. Further external validation with a large-sample, multiple-center is needed to verify the role of MDM2 promoter methylation as a biomarker. We should monitor the diagnostic value of MDM2 methylation. Second, there was a lack of liver tissues, therefore, we only performed this study in PBMCs. Third, we did not find alterations in MDM2 methylation in the CHB or LC patients from non-cancerous to cancerous diseases. However, the current report revealed the non-invasive early detection of HCC four years before conventional diagnosis using methylation biomarkers [60]. These results showed the great potential of methylation biomarker for predicting cancerous diseases in non-cancerous diseases. Therefore, whether unmethylated patients with CHB or LC are having risk should be verified in prospective cohort in the future. In addition, we only found that the MDM2 promoter methylation is a trend in HBV-related HCC, but the specificity of MDM2 promoter methylation for HBV-related HCC or non-HBV-related HCC should be noted and performed in the future study.
Conclusions
In conclusion, this study showed MDM2 promoter hypomethylation in HBV-related HCC patients. Moreover, the combination of MDM2 promoter methylation and serum AFP might improve the diagnostic efficiency of HBV-related HCC.
Abbreviations
MDM2: murine double minute-2; AFP: alpha-fetoprotein; CHB: chronic hepatitis B; LC: liver cirrhosis; HBV-related HCC: hepatitis B virus-related hepatocellular carcinoma; IL-6: interleukin-6; TNF-α: tumor-necrosis factor-α.
Acknowledgements
This work was funded by the Key Project of the Chinese Ministry of Science and Technology (2017ZX10202202, 2018ZX10302206-003-003, 2018ZX10302206-006-002), the National Natural Science Foundation of China (81970522), the Key Research and Development Project of Shandong Province (2019GSF108023).
Competing Interests
The authors have declared that no competing interest exists.
References
1. Llovet JM, Burroughs A, Bruix J. Hepatocellular carcinoma. Lancet (London, England). 2003;362:1907-17
2. Liu MX, Jin L, Sun SJ, Liu P, Feng X, Cheng ZL. et al. Metabolic reprogramming by PCK1 promotes TCA cataplerosis, oxidative stress and apoptosis in liver cancer cells and suppresses hepatocellular carcinoma. Oncogene. 2018;37:1637-53
3. El-Serag HB, Rudolph KL. Hepatocellular carcinoma: epidemiology and molecular carcinogenesis. Gastroenterology. 2007;132:2557-76
4. Zhang Z, Meng G, Wang L, Ma Y, Guan Z. The prognostic role and reduced expression of FOXJ2 in human hepatocellular carcinoma. Molecular medicine reports. 2016;14:254-62
5. El-Serag HB, Rudolph KL. Hepatocellular carcinoma: epidemiology and molecular carcinogenesis. Gastroenterology. 2007;132:2557-76
6. Oliner JD, Kinzler KW, Meltzer PS, George DL, Vogelstein B. Amplification of a gene encoding a p53-associated protein in human sarcomas. Nature. 1992;358:80-3
7. Weber JD, Taylor LJ, Roussel MF, Sherr CJ, Bar-Sagi D. Nucleolar Arf sequesters Mdm2 and activates p53. Nature cell biology. 1999;1:20-6
8. Haupt Y, Maya R, Kazaz A, Oren M. Mdm2 promotes the rapid degradation of p53. Nature. 1997;387:2 96-9
9. Simerzin A, Zorde-Khvalevsky E, Rivkin M, Adar R, Zucman-Rossi J, Couchy G. et al. The liver-specific microRNA-122*, the complementary strand of microRNA-122, acts as a tumor suppressor by modulating the p53/mouse double minute 2 homolog circuitry. Hepatology (Baltimore, Md). 2016;64:1623-36
10. Oliner JD, Saiki AY, Caenepeel S. The Role of MDM2 Amplification and Overexpression in Tumorigenesis. Cold Spring Harbor perspectives in medicine. 2016 6
11. Embade N, Fernandez-Ramos D, Varela-Rey M, Beraza N, Sini M, Gutierrez de Juan V. et al. Murine double minute 2 regulates Hu antigen R stability in human liver and colon cancer through NEDDylation. Hepatology (Baltimore, Md). 2012;55:1237-48
12. Feng C, Xian Q, Liu S. Micro RNA-518 inhibits gastric cancer cell growth by inducing apoptosis via targeting MDM2. Biomedicine & pharmacotherapy = Biomedecine & pharmacotherapie. 2018;97:1595-602
13. Pellegrino M, Mancini F, Lucà R, Coletti A, Giacchè N, Manni I. et al. Targeting the MDM2/MDM4 interaction interface as a promising approach for p53 reactivation therapy. Cancer Res. 2015;75:4560-72
14. Burgess A, Chia KM, Haupt S, Thomas D, Haupt Y, Lim E. Clinical Overview of MDM2/X-Targeted Therapies. Frontiers in oncology. 2016;6:7
15. Imanishi M, Yamamoto Y, Wang X, Sugaya A, Hirose M, Endo S. et al. Augmented antitumor activity of 5-fluorouracil by double knockdown of MDM4 and MDM2 in colon and gastric cancer cells. Cancer Science. 2019;110:639
16. Zhang L, Luo J, Wen H, Zhang T, Zuo X, Li X. MDM2 promotes rheumatoid arthritis via activation of MAPK and NF-kappaB. International immunopharmacology. 2016;30:69-73
17. Baylin SB, Ohm JE. Epigenetic gene silencing in cancer - a mechanism for early oncogenic pathway addiction? Nature reviews Cancer. 2006;6:107-16
18. Jones PA, Baylin SB. The epigenomics of cancer. Cell. 2007;128:683-92
19. Carvalho ATP, Gouveia L, Kanna CR, Wärmländer SKTS, Platts JA, Kamerlin SCL. Understanding the structural and dynamic consequences of DNA epigenetic modifications: computational insights into cytosine methylation and hydroxymethylation. Epigenetics. 2014;9:1604-12
20. Arish M, Kordi-Tamandani DM, Sangterash MH, Poyandeh R. Assessment of Promoter Hypermethylation and Expression Profile of P14ARF and MDM2 Genes in Patients With Pterygium. Eye Contact Lens. 2016;42:e4-7
21. Park JH, Park J, Choi JK, Lyu J, Bae MG, Lee YG. et al. Identification of DNA methylation changes associated with human gastric cancer. BMC medical genomics. 2011;4:82
22. Duruisseaux M, Esteller M. Lung cancer epigenetics: From knowledge to applications. Semin Cancer Biol. 2018;51:116-28
23. Tian MM, Fan YC, Zhao J, Gao S, Zhao ZH, Chen LY. et al. Hepatocellular carcinoma suppressor 1 promoter hypermethylation in serum. A diagnostic and prognostic study in hepatitis B. Clinics and research in hepatology and gastroenterology. 2017;41:171-80
24. Gao S, Sun FK, Fan YC, Shi CH, Zhang ZH, Wang LY. et al. Aberrant GSTP1 promoter methylation predicts short-term prognosis in acute-on-chronic hepatitis B liver failure. Alimentary pharmacology & therapeutics. 2015;42:319-29
25. Terrault NA, Lok ASF, McMahon BJ, Chang KM, Hwang JP, Jonas MM. et al. Update on prevention, diagnosis, and treatment of chronic hepatitis B: AASLD 2018 hepatitis B guidance. Hepatology (Baltimore, Md). 2018;67:1560-99
26. Fukui H, Saito H, Ueno Y, Uto H, Obara K, Sakaida I. et al. Evidence-based clinical practice guidelines for liver cirrhosis 2015. Journal of gastroenterology. 2016;51:629-50
27. Bruix J, Sherman M. Management of hepatocellular carcinoma: an update. Hepatology (Baltimore, Md). 2011;53:1020-2
28. Pasello G, Urso L, Mencoboni M, Grosso F, Ceresoli GL, Lunardi F. et al. MDM2 and HIF1alpha expression levels in different histologic subtypes of malignant pleural mesothelioma: correlation with pathological and clinical data. Oncotarget. 2015;6:42053-66
29. Zhu AX. Hepatocellular carcinoma: are we making progress? Cancer investigation. 2003;21:418-28
30. Weber M, Hellmann I, Stadler MB, Ramos L, Pääbo S, Rebhan M. et al. Distribution, silencing potential and evolutionary impact of promoter DNA methylation in the human genome. Nature genetics. 2007;39:457-66
31. Kondo I, Iida S, Takagi Y, Sugihara K. MDM2 mRNA expression in the p53 pathway may predict the potential of invasion and liver metastasis in colorectal cancer. Diseases of the colon and rectum. 2008;51:1395-402
32. Zhang CX, Chen J, Cai L, Wu J, Wang JY, Cao LF. et al. DNA induction of MDM2 promotes proliferation of human renal mesangial cells and alters peripheral B cells subsets in pediatric systemic lupus erythematosus. Molecular immunology. 2018;94:166-75
33. Meng X, Franklin DA, Dong J, Zhang Y. MDM2-p53 pathway in hepatocellular carcinoma. Cancer Res. 2014;74:7161-7
34. Wang JW, Wang JW, Zhang J, Wu CS, Fang Y, Su WW. et al. Decreased Methylation of IFNAR Gene Promoter from Peripheral Blood Mononuclear Cells Is Associated with Oxidative Stress in Chronic Hepatitis B. Journal of interferon & cytokine research: the official journal of the International Society for Interferon and Cytokine Research. 2018;38:480-90
35. Nishida N, Kudo M. Oxidative stress and epigenetic instability in human hepatocarcinogenesis. Dig Dis. 2013;31:447-53
36. Stefanska B, Huang J, Bhattacharyya B, Suderman M, Hallett M, Han Z-G. et al. Definition of the landscape of promoter DNA hypomethylation in liver cancer. Cancer Res. 2011;71:5891-903
37. Li F, Qiao CY, Gao S, Fan YC, Chen LY, Wang K. Circulating cell-free DNA of methylated insulin-like growth factor-binding protein 7 predicts a poor prognosis in hepatitis B virus-associated hepatocellular carcinoma after hepatectomy. Free radical research. 2018;52:455-64
38. Li M, Cong Y, Li Y, Zhong S, Wang R, Li H. et al. Insight Into the Binding Mechanism of p53/pDIQ-MDMX/MDM2 With the Interaction Entropy Method. Frontiers in chemistry. 2019;7:33
39. Momand J, Jung D, Wilczynski S, Niland J. The MDM2 gene amplification database. Nucleic acids research. 1998;26:3453-9
40. Zhang MF, Zhang ZY, Fu J, Yang YF, Yun JP. Correlation between expression of p53, p21/WAF1, and MDM2 proteins and their prognostic significance in primary hepatocellular carcinoma. Journal of translational medicine. 2009;7:110
41. Kim G-D, Ni J, Kelesoglu N, Roberts RJ, Pradhan S. Co-operation and communication between the human maintenance and de novo DNA (cytosine-5) methyltransferases. EMBO J. 2002;21:4183-95
42. Levine AJ, Oren M. The first 30 years of p53: growing ever more complex. Nat Rev Cancer. 2009;9:749-58
43. Embade N, Fernández-Ramos D, Varela-Rey M, Beraza N, Sini M, Gutiérrez de Juan V. et al. Murine double minute 2 regulates Hu antigen R stability in human liver and colon cancer through NEDDylation. Hepatology. 2012;55:1237-48
44. Huang X, Qian X, Cheng C, He S, Sun L, Ke Q. et al. Expression of Pirh2, a p27(Kip1) ubiquitin ligase, in hepatocellular carcinoma: correlation with p27(Kip1) and cell proliferation. Hum Pathol. 2011;42:507-15
45. Lee YH, Andersen JB, Song HT, Judge AD, Seo D, Ishikawa T. et al. Definition of ubiquitination modulator COP1 as a novel therapeutic target in human hepatocellular carcinoma. Cancer research. 2010;70:8264-9
46. Adhikary S, Marinoni F, Hock A, Hulleman E, Popov N, Beier R. et al. The ubiquitin ligase HectH9 regulates transcriptional activation by Myc and is essential for tumor cell proliferation. Cell. 2005;123:409-21
47. Dornan D, Wertz I, Shimizu H, Arnott D, Frantz GD, Dowd P. et al. The ubiquitin ligase COP1 is a critical negative regulator of p53. Nature. 2004;429:86-92
48. Wang XM, Yang LY, Guo L, Fan C, Wu F. p53-induced RING-H2 protein, a novel marker for poor survival in hepatocellular carcinoma after hepatic resection. Cancer. 2009;115:4554-63
49. Chen D, Kon N, Li M, Zhang W, Qin J, Gu W. ARF-BP1/Mule is a critical mediator of the ARF tumor suppressor. Cell. 2005;121:1071-83
50. Luo JL, Maeda S, Hsu LC, Yagita H, Karin M. Inhibition of NF-kappaB in cancer cells converts inflammation- induced tumor growth mediated by TNFalpha to TRAIL-mediated tumor regression. Cancer cell. 2004;6:297-305
51. Ohishi W, Cologne JB, Fujiwara S, Suzuki G, Hayashi T, Niwa Y. et al. Serum interleukin-6 associated with hepatocellular carcinoma risk: a nested case-control study. International journal of cancer. 2014;134:154-63
52. Chen L, Zhang Q, Chang W, Du Y, Zhang H, Cao G. Viral and host inflammation-related factors that can predict the prognosis of hepatocellular carcinoma. European journal of cancer (Oxford, England : 1990). 2012;48:1977-87
53. Harrison ML, Obermueller E, Maisey NR, Hoare S, Edmonds K, Li NF. et al. Tumor necrosis factor alpha as a new target for renal cell carcinoma: two sequential phase II trials of infliximab at standard and high dose. Journal of clinical oncology : official journal of the American Society of Clinical Oncology. 2007;25:4542-9
54. Thomasova D, Mulay SR, Bruns H, Anders HJ. p53-independent roles of MDM2 in NF-kappaB signaling: implications for cancer therapy, wound healing, and autoimmune diseases. Neoplasia (New York, NY). 2012;14:1097-101
55. Han NR, Oh HA, Nam SY, Moon PD, Kim DW, Kim HM. et al. TSLP induces mast cell development and aggravates allergic reactions through the activation of MDM2 and STAT6. The Journal of investigative dermatology. 2014;134:2521-30
56. Wang W, Qin JJ, Voruganti S, Nijampatnam B, Velu SE, Ruan KH. et al. Discovery and Characterization of Dual Inhibitors of MDM2 and NFAT1 for Pancreatic Cancer Therapy. Cancer research. 2018;78:5656-67
57. Lok AS, Sterling RK, Everhart JE, Wright EC, Hoefs JC, Di Bisceglie AM. et al. Des-gamma-carboxy prothrombin and alpha-fetoprotein as biomarkers for the early detection of hepatocellular carcinoma. Gastroenterology. 2010;138:493-502
58. Lee CW, Tsai HI, Lee WC, Huang SW, Lin CY, Hsieh YC. et al. Normal Alpha-Fetoprotein Hepatocellular Carcinoma: Are They Really Normal? J Clin Med. 2019 8
59. Oh CR, Kong SY, Im HS, Kim HJ, Kim MK, Yoon KA. et al. Genome-wide copy number alteration and VEGFA amplification of circulating cell-free DNA as a biomarker in advanced hepatocellular carcinoma patients treated with Sorafenib. BMC Cancer. 2019;19:292
60. Chen X, Gole J, Gore A, He Q, Lu M, Min J. et al. Non-invasive early detection of cancer four years before conventional diagnosis using a blood test. Nat Commun. 2020;11:3475
Author contact
![]() Corresponding author: Kai Wang, Professor, MD, Ph.D., Department of Hepatology, Qilu Hospital of Shandong University; Institute of Hepatology, Shandong University, Wenhuaxi Road 107#, Jinan 250012, Shandong, China. E-mail: wangdoc876com; Tel: +86-531-82169596; Fax: +86-531-8692754.
Corresponding author: Kai Wang, Professor, MD, Ph.D., Department of Hepatology, Qilu Hospital of Shandong University; Institute of Hepatology, Shandong University, Wenhuaxi Road 107#, Jinan 250012, Shandong, China. E-mail: wangdoc876com; Tel: +86-531-82169596; Fax: +86-531-8692754.

 Global reach, higher impact
Global reach, higher impact