Impact Factor
ISSN: 1449-1907
Int J Med Sci 2020; 17(18):3058-3064. doi:10.7150/ijms.50643 This issue Cite
Research Paper
BDNF Val66Met Polymorphism, the Allele-Specific Analysis by qRT-PCR - a Novel Protocol
1. Gdansk University of Physical Education and Sport, Faculty of Physical Education, Gdansk, Poland;
2. Mossakowski Medical Research Centre, Polish Academy of Sciences, Warsaw, Poland;
3. Department of Physical Therapy, Ariel University, Ariel, Israel.
4. International Institute of Molecular and Cell Biology in Warsaw, Poland.
Received 2020-7-13; Accepted 2020-9-28; Published 2020-10-18
Abstract
Background: Alteration in brain-derived neurotrophic factor (BDNF) production is a marker of neuropathological conditions, which has led to the investigation of Val66Met polymorphism occurring in the human BDNF gene (BDNF). Presently, there are no reported methods available for the analysis of Val66Met impact on human BDNF functioning.
Purpose: To develop a qRT-PCR protocol for the allele-specific expression evaluation of the Val66Met polymorphism in BDNF.
Methods: Using RNA extracted from muscle samples of 9 healthy volunteers (32.9 ± 10.3 y) at rest and following a maximal effort aerobic capacity exercise test, a protocol was developed for the detection of Val66/Met66 allele-specific BDNF expression in Real-Time Quantitative Reverse Transcription PCR (qRT-PCR) - relative to housekeeping genes - and validated by absolute quantification in Droplet Digital Polymerase Chain Reaction (ddPCR).
Results: Differences in the relative values of BDNF mRNA were confirmed by ddPCR analysis. HPRT1 and B2M were the most stable genes expressed in muscle tissue among different metabolic conditions, while GAPDH revealed to be metabolic responsive.
Conclusion: Our qRT-PCR protocol successfully determines the allele-specific detection and changes in BDNF expression regarding the Val66Met polymorphism.
Keywords: Brain-derived Neurotrophic Factor, Allele-specific detection, ddRT-PCR, Gene expression, Exercise metabolism, Reference gene
Introduction
Brain-derived neurotrophic factor (BDNF) is a bioactive protein from the family of neurotrophins, neurotrophic growth factors, with the most abundant expression in the neural system development and maturity throughout the mammalian lifespan [1,2]. With compulsory roles in neurogenesis, synaptogenesis, neuronal repair and protection, alteration in BDNF dynamics may result in various neurological disorders and neurodegenerative diseases [3,4,5,6].
Predominantly expressed in neural and skeletal muscle cells as a protein signaler [7], BDNF synthesis results in the formation of several precursor isoforms in a multistage process involving cleavage and storage [2,8]. The BDNF gene (BDNF) transcript encodes a precursor protein (pre-pro-BDNF isoform) which loses its secretory signal peptide (pre-region) and dimer into the pro-BDNF isoform. Pro-BDNF is subsequently cleaved by proprotein convertases in either the Golgi apparatus or in secretory vesicles to form mature BDNF [2,9]. Released BDNF acts locally via both autocrine and paracrine mechanisms [10,11].
A single nucleotide polymorphism (SNP) has been identified in the human BDNF gene, resulting in a valine (Val) for methionine (Met) substitution in the 66th position of pre-pro-BDNF (Val66Met). This SNP in the BDNF gene is thought to have important implications for the gene and protein functioning. The BDNF Val66Met has been suggested to impair activity dependent release of BDNF [12], play a role in BDNF/pro-BDNF secretion ratios [13], and is also associated with increased susceptibility to cognitive deficits and neuropsychiatric disorders [14,15]. Implications for the Met66-allele presence have been evidenced in experimental models [16;17] in which the complexity of the post-translational mechanisms was unable to determine impairments in the neurotrophin's function [18]. In a clinical perspective, circulating BDNF concentration is likely to be associated with a greater acute response of cortisol to stress, especially in individuals with a psychiatric disorder [19]. Although circulating blood concentrations of BDNF may provide some indication of BDNF production [20], it provides little understanding of the BDNF Val66Met polymorphism expression.
The differential expression of BDNF Val66Met polymorphic alleles to physical exercise was first detected in a recent study [21]. Most clinical studies have reported changes in circulating BDNF concentrations during aerobic activities [22], but inconsistent in regards to resistance training [23]. Increases in BDNF concentrations appear to be associated with the intensity of aerobic exercise [24,25], but may decrease during the recovery period if the exercise stimulus created a significant elevation in oxidative stress [20]. Considering the increasing interest in the Val66Met polymorphism [26,27], and its possible implications in a variety of neuropathological processes, the purpose of this study was to determine the validity of assessing human BDNF expression in vivo by developing a Real-Time Quantitative Reverse Transcription Polymerase Chain Reaction (qRT-PCR) protocol using a standard maximum aerobic capacity test. Based upon the more consistent response of BDNF to higher intensity endurance exercise protocols, we believed that this protocol would best provide for the differential analysis of BDNF expression regarding the Val66Met polymorphism validated by absolute quantification in QX200™ Droplet Digital™ Polymerase Chain Reaction (ddPCR).
Materials and methods
Participants
Nine apparently healthy and physically active volunteers participating in medium and long distance Polish race circuits (10 and 21 km), with known genotypes for the BDNF gene agreed to participate in this study. Descriptive characteristics for all participants can be observed in Table 1. After an explanation of all procedures, risks, and benefits, each participant completed a Par-Q questionnaire [28] and provided his or her written informed consent to participate in the study. This study was conducted in accordance to Regulation 2016/679 of the European Union Parliament Council of 27 April 2016 on the protection of individuals with regard to the processing of personal data and on the free movement of such data, and repealing Directive 95/46 / EC (general data protection regulations) approved by the Ethical Committee of the Central Clinic Hospital of the MSWiA in Warsaw, Poland.
Characteristic of Participants in ddPCR™ and qRT-PCR analyses
| Subject | Age (years) | BMI (kg/m2) | VO2max (ml·kg/min) | Genotype |
|---|---|---|---|---|
| Female | 36 | 19.84 | 44.00 | VAL/VAL |
| Male | 24 | 21.97 | 60.00 | VAL/VAL |
| Male | 46 | 22.35 | 62.00 | VAL/VAL |
| Male | 28 | 21.97 | 59.38 | VAL/MET |
| Male | 43 | 26.31 | 41.40 | VAL/VAL |
| Male | 26 | 23.10 | 45.62 | VAL/MET |
| Male | 22 | 22.74 | 51.38 | VAL/VAL |
| Male | 46 | 23.70 | 56.06 | VAL/VAL |
| Male | 28 | 23.89 | 61.40 | VAL/VAL |
| Male | 36 | 20.67 | 54.24 | VAL/VAL |
| Means ± SD | 32.9 ± 10.3 | 23.25 ± 1.4 | 54.7 ± 7.7 |
*VO2max, maximal oxygen intake; BMI, body mass index.
Experimental Design
All study participants reported to the Human Performance Laboratory in the Mossakowski Medical Research Centre, Polish Academy of Science, Warsaw, Poland. Prior to the testing session, participants reported to the laboratory 2-hours post-prandial and were instructed not to consume any caffeine or alcohol; they were also instructed not to perform any lower body physical activity within 48 hours of testing. During the visit, all participants were provided with a brief medical exam that included anthropometric measurements. In addition, two muscle biopsies were obtained from participants prior to, and following performance of a maximal aerobic capacity test.
Muscle Biopsies
Participants were required to rest in a supine position for 30 min. A muscle biopsy (50 mg wet weight) from the middle portion of the Vastus lateralis was obtained by a medical doctor, using local anesthesia and a semi-automatic 14-gauge biopsy needle (Guillotine needle double shoot DSGBL 14/10, Tsunami, Italy). Following the first sampling, participants were required to perform a maximal aerobic capacity test. Following the conclusion (within 1- 2 min) of the aerobic capacity test, a second muscle sample was obtained. Muscle samples were immediately placed in dry ice before storage at -80° C. All tests were performed between 3 p.m. and 5 p.m. (GMT+2).
Maximal Aerobic Capacity Test
The maximal aerobic capacity test was conducted using the standard Bruce Protocol [29]. The treadmill protocol began with each participant performing a 3-min walk at 2.7 km·h-1 and at a 10% incline. A progressive increase in both speed and inclination occurred every 3-min (stages), until participant's volitional fatigue. VO2max (maximum rate of oxygen consumption) was determined to be the highest 30-s VO2 value recorded during the test and coincided with at least two of the following three criteria: (a) 90% of age-predicted maximum heart rate; (b) respiratory exchange ratio > 1.1; and/or (c) a plateau of oxygen uptake (less than 150 mL/min increase in VO2 during the last 60 s of the test). Ventilation parameters were monitored and recorded from rest and throughout the protocol. Heart rate (bpm) and respiratory gases were assessed using Vmax29-Sensor Medics (CareFusion, USA) gas analyzer, and CardioSys software, version 4.1 (Marquette Hellige GmbH, Germany) (Data are presented in Table 1).
Muscle RNA extraction
The muscle samples (50 mg) were manually homogenized in Eppendorf tubes with 0.5 ml of TRIzol reagent (T9424, Sigma-Aldrich, USA) and TRIzol-chlorophorm RNA extraction was performed according to the manufacturer's protocol. The total RNA was dissolved in 0.03 ml of RNAse-free water and concentrations were determined by NanoDrop™ 2000 Spectrophotometer (Thermo Scientific, USA). RNA samples were stored at -80°C. One µg of RNA was used for cDNA synthesis in 0.02 ml of reaction mixture using the iScript™ Reverse Transcription kit (BioRad, USA) according to the manufacturer's protocol. Samples of 1 and 3 μL of this cDNA solution was used as a template for qRT-PCR and ddPCR analyses.
Reference Gene Establishment
To quantify BDNF mRNA expression in muscle we first established a gene of reference - which is expressed in a constant and stable rate both during homeostasis and stressed (e.g. exercise) conditions - i.e., housekeeping gene. To select the most applicable reference gene we used the muscle samples of three participants from the set, at both resting and post-exercise conditions, and performed a qRT-PCR analysis using the BioRad Prime PCR Pathway assay and Reference gene panel, and SsoAdvanced™ Universal SYBR® Green Supermix. Participant samples from both sexes with a maximal discrepancy in age were used for the reference gene establishment in order to address the highest heterogeneity.
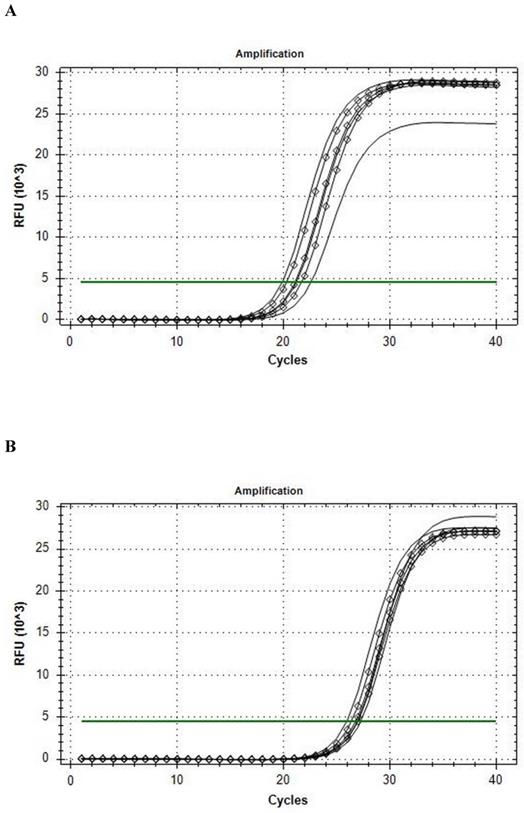
The Reference gene panel included the known candidate genes frequently recommended for muscle tissue analysis [30], and the internal PCR performance, reverse transcription, RNA quality, and genomic DNA (gDNA) admixture controls. Fourteen eligible housekeeping genes were tested for comparison in qRT-PCR analysis: ACTB, B2M, G6PD, GAPDH, GUSB, HMBS, HPRT1, PGK1, RPL13A, RPLPO, RPS18, TBP, TFRC, YWHAZ. CFX Connect Real-Time PCR System analysis revealed that the B2M was the most stable gene enabled for further usage in ddPCR, and that HPRT1 was the most stable gene during the conditions of our intervention (see Fig. 1). On the base of stability in gene candidates' expression, B2M was included into the qRT-PCR and ddPCR analysis of the BDNF as a reference gene.
qRT-PCR BDNF polymorphism specific assay
One µl of cDNA solution from the same participant's samples, excluding the female participant, were used in a qRT-PCR with allele-specific BDNF assay containing hydrolyzed probes (TaqMan) with both HEX (hexachloro-fluorescein) and FAM (6-carboxyfluorescein) systems of detection for the Val66- and Met66-coding BDNF allele's products, respectively (ddPCR Mutation Assay: rs6265, dHsaMDS320493890, BioRad, USA). The BDNF allele-specific primers were designed according to Sheikh and colleagues [31]. A HEX-based B2M Expression Probe Assay (qHsaCPE5053101, BioRad, USA) with the same SsoAdvanced Universal Probes Supermix (BioRad, USA) was ran in parallel and the melting curves were used as a product specificity control. To count the genomic DNA admixture contribution in the mRNA, samples of the participant's total RNA solution were used as a template in concentrations equalized to the residual in cDNA samples. All the reactions were performed in triplicate and the mean was used in subsequent calculations.
Unlike the absolute quantities achieved by ddPCR, reports of FAM and HEX probe detection corresponding to Met66- and Val66-coding BDNF mRNA levels from qRT-PCR could not be compared directly by threshold cycles (C(t)s), probably according to difference in FAM and HEX fluorescence detection sensitivity. Therefore, BDNF allele content in genome DNA admixture of RNA samples was used for the mathematical conversion of FAM to HEX detection, then Met66-coding allele's expression was standardized to HEX detection.
PCR program used was: 95°C - 10 min and 40 cycles of 94°C - 30 sec, 55°C - 60 sec with fluorescence detection. The C(t)s - number of cycles that takes to reach threshold - were determined on the HEX and FAM channels by CFX Connect Real-Time PCR System (BioRad, USA). BDNF expression levels were calculated by 'delta-delta C(t)' method [32]. BDNF C(t)s of each sample was corrected according to the contribution of gDNA admixture using C(t) meanings achieved for the corresponding RNA samples (C(t)RNA) by the following formula:
C(t)correctBDNF = C(t)BDNF + log2((1 + 2(C(t)BDNF - C(t)RNA) / 1)
C(t) for samples where RNA did not give a registered signal were accepted without correction. Corrections of C(t) were performed for FAM and HEX detection independently.
Values of BDNF Val66- and Met66-coding alleles present in RNA samples (the alleles' ratio in genome DNA admixture = 1:1) were used as controls for equalizing HEX and FAM detection. The C(t)s of the Met-coding allele were unified to HEX detection afterward in the following formula:
C(t)unifMetBDNF = C(t)FAMMetBDNF + (C(t)HEXRNA - C(t)FAMRNA)
Amplification curves of in rest (line) and post-exercise (diamond) conditions. A, B2M; B, HPRT1.
Total BDNF expression in heterozygous subjects [C(t) - C(t)sumBDNF] was calculated with the formula:
C(t)sumBDNF = C(t)HEXBDNF - log2((1 + 2(C(t)HEXBDNF - C(t)unifMetBDNF) / 1)
Statistics
Normality of values was determined by a Kolmogorov-Smirnov test. A paired Student's t-test was used to compare condition/changes description and Spearman's rank test for ddPCR to qRT-PCR correlation coefficient in SPSS. An α level of p ≤ 0.05 was used to determine statistical significance. All data are reported as means ± SD.
Results
Among tested gene candidates as a reference the best stability in the metabolic stressed conditions in muscle was detected for B2M (less than 2.5 C(t) range inter- and intra-individually) (Fig. 1a) and HPRT1 (less than 2 C(t) range inter- and intra-individually) (Fig. 1b) while the least stable for qRT-PCR analysis of stress conditions in the muscle was found to be GAPDH, (more than 4 C(t) range inter- and intra-individually). The other analyzed genes also showed low range of variability (less than 3.5 C(t) range inter- and intra-individually) and could possibly be used as a reference gene as well. The results of qRT-PCR Val66Met allele-specific BDNF expression analysis with the all mathematical proceeding can be observed in Table 2.
Real-time qPCR analysis detected the relative levels of BDNF expression with specificity for the Val66Met polymorphism, according to the absolute quantification performed by ddPCR (see Table 3). The BDNF expression showed small variability - often less than 1 C(t) in both the inter-individual differences (K-S(8) = 0.172, p > 0.05) and in regard to testing condition (Rest mean = 1.84 ± 0.79; Stress mean = 0.95 ± 0.34, t(8) = 4.64, p = 0.002) and genotype. Values detected by ddPCR were reported by qRT-PCR with a moderate negative correlation (ρ = - 0.456), suggesting the higher BDNF mRNA copy number, the lower C(t).
qRT-PCR BDNF analysis. Threshold cycles, C(t)s, calculations.
| Genotype | Fluorescense detection | B2Mpre mean | B2Mpost mean | BDNFpre mean | BDNFpost mean | mRNApre mean | mRNApost mean | BDNFpre HEX/FAM correction | BDNFpost HEX/FAM correction | TotalBDNFpre expression in heterozygous | TotalBDNFpost expression in heterozygous |
|---|---|---|---|---|---|---|---|---|---|---|---|
| V/V | HEX | 24.55 | 23.26 | 35.43 | 34.44 | 40.11 | 36.95 | 35.49 | 34.67 | ||
| FAM | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | |||
| V/V | HEX | 21.34 | 22.46 | 32.33 | 34.10 | 38.22 | 41.00 | 32.35 | 34.11 | ||
| FAM | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | |||
| V/M | HEX | 23.91 | 24.05 | 34.21 | 34.68 | 36.46 | 40.30 | 34.48 | 34.7 | 33.86 | 34.01 |
| FAM | 0 | 0 | 29.98 | 30.83 | 31.50 | 35.78 | 35.37 | 35.39 | |||
| V/V | HEX | 26.12 | 23.60 | 35.7 | 33.34 | 38.62 | 36.02 | 35.88 | 33.54 | ||
| FAM | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | |||
| V/M | HEX | 23.92 | 23.8 | 34.42 | 34.98 | 36.30 | 36.56 | 34.76 | 35.39 | 33.96 | 34.66 |
| FAM | 0 | 0 | 30.09 | 31.01 | 31.62 | 32.13 | 35.19 | 35.99 | |||
| V/V | HEX | 23.21 | 23.17 | 32.29 | 33.37 | 35.29 | 35.63 | 32.45 | 33.64 | ||
| FAM | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | |||
| V/V | HEX | 24.37 | 24.61 | 34.04 | 34.23 | 36.94 | 37.71 | 34.22 | 34.35 | ||
| FAM | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | |||
| V/V | HEX | 24.6 | 22.95 | 33.19 | 32.19 | 36.65 | 36.65 | 33.32 | 32.25 | ||
| FAM | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
Pre, Analyses from the rest condition; Post, Analyses from after effort test
qRT-PCR results validation by ddPCR. Condition- and allele-specific BDNF expression differences.
| Genotype | qRT-PCR | ddPCR | qRT-PCR | ddPCR | ||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Tpre BDNF-B2M | Tpost BDNF-B2M | T BDNF post /pre | Tpre BDNF /1000B2M | Tpost BDNF /1000B2M | T BDNF post /pre | Pre MetBDNF /ValBDNF | Post MetBDNF /ValBDNF | Pre MetBDNF /ValBDNF | Post MetBDNF /ValBDNF | |
| V/V | 10.94 | 11.41 | 0.721 | 1.444 | 0.703 | 0.487 | ||||
| V/V | 11.01 | 11.66 | 0.641 | 1.792 | 1.17 | 0.653 | ||||
| V/M | 9.950 | 9.960 | 0.996 | 1.427 | 0.926 | 0.649 | 0.541 | 0.621 | 0.707 | 0.614 |
| V/V | 9.760 | 9.950 | 0.877 | 0.793 | 0.498 | 0.628 | ||||
| V/M | 10.04 | 10.86 | 0.566 | 1.524 | 0.910 | 0.598 | 0.741 | 0.662 | 0.660 | 0.968 |
| V/V | 9.250 | 10.48 | 0.427 | 2.319 | 0.745 | 0.321 | ||||
| V/V | 9.850 | 9.740 | 1.077 | 2.045 | 1.110 | 0.543 | ||||
| V/V | 8.720 | 9.300 | 0.665 | 3.460 | 1.607 | 0.464 | ||||
| Means ± SD | T BDNF post/pre, 0.736 ± 0.205 | T BDNF post/pre, 0.543 ± 0.115 | MetBDNF/ValBDNF, 0.641 ± 0.084 | MetBDNF/ValBDNF, 0.737 ± 0.158 | ||||||
Pre, Analyses from the rest condition; Post, Analyses from exercise protocol; Tpre and Tpost - total (both alleles) BDNF expression, ValBDNF, MetBDNF - allele-specific BDNF expression.
Discussion
BDNF and its gene polymorphism Val66Met have taken on a greater focus of attention regarding its crucial role in neural system development and functioning. Whereas alteration in BDNF production has been suggested to be a marker of neuropathological conditions [3,11,19,33]. Investigations examining the effect of exercise on changes in circulating BDNF concentrations have gained much interest as a possible target for the therapy and/or prevention of stress and neurodegeneration-related disorders [22,34,35]. Therefore, a precise and accessible method proposed for the laboratory analysis of the allele-specific expression of BDNF is a valuable tool for a broader assessment of the BDNF Val66Met polymorphism in a clinical and research scenario. By assessing relatively small, but detectable by absolute quantification changes in mRNA levels, we were able to design a protocol for the identification of BDNF expression in human tissue regarding to the allele specificity and the metabolic condition by qRT-PCR.
Our results showed that amongst the genes expressing constant patterns (stability) in the human muscle regardless of sex and physiological conditions, the most stable one is the gene of hypoxanthine phosphoribosyl transferase (HPRT1). The establishment of a tissue-specific reference gene can help discriminate both distinctive characteristics, and genetic-based changes in cell functioning among different physiological conditions [36,37,38,39;40]. Precision in the determination of a gene of reference for comparative analysis of the expression of specific proteins is crucial to enable the assessment of factors that are expressed in low quantities in tissue. These results contribute to the establishment of the HPRT1 as the most stable gene expressed in muscle tissue, and can be used as reference for comparative analyses of gene expression in humans, regardless of sex and metabolic conditions.
Furthermore, our results indicate that the GAPDH (Glyceraldehyde-3-Phosphate Dehydrogenase) gene shows unacceptable parameters (the lowest among the genes tested) as a reference gene for comparisons in different metabolic conditions. Although initially considered a housekeeping gene and widely used as an internal control in experiments on proteins, mRNA, and DNA [41], recent evidence has shown that GAPDH, besides having a role in energy metabolism, participates is a more complex regulatory mechanism [42]. Nonetheless, the changes in expression patterns of this gene noted during the exercise protocol suggest that GAPDH should not be considered as a reference gene when testing different metabolic conditions.
Using the absolute quantification of ddPCR analysis as a reference, we have demonstrated that the identification of the allele-specific expression of BDNF by qRT-PCR is feasible. The differential detection of BDNF alleles' expression enables the objective analysis of the Val66Met polymorphism role on BDNF functioning, supporting the previous speculations [43,44,45]. Moreover, this methodology has proven to be applicable even in small samples, in spite of the small variability showed for the human BDNF expression. The qRT-PCR analysis was able to report the results of the ddPCR analysis of BDNF expression both in allele- and condition-specific manner (Table 3).
In conclusion, our results indicated that the qRT-PCR protocol was successful in determining the measurement of BDNF expression levels in muscle tissue, as well as metabolic stress-delivered and genotype-associated changes in these levels. This protocol appears to be useful for the detection of the Val66Met polymorphism specific BDNF expression and can be used in heterogeneous populations.
Acknowledgements
Authors are appreciated to the support of Professor Jacek Kuznicki and Doctor Lukasz Majewski from the International Institute of Molecular and Cell Biology in Warsaw, and the estimated Professors Paweł Cięszczyk and Andrzej Ziemba from the Gdansk University of Physical Education and the Mossakowski Medical Research Centre respectively.
Author Contribution
Conceptualization, G.G.A. and E.V.G.; Sample Collection and Analysis, G.G.A. and E.V.G.; Resources, E.V.G.; Data Curation, E.V.G.; Writing, G.G.A., J.R.H. and E.V.G.; Project Administration, G.G.A., Funding, G.G.A. and E.V.G.
Competing Interests
The authors have declared that no competing interest exists.
References
1. Lamb YN, Mckay NS, Thompson CS. et al. Brain-derived neurotrophic factor Val66Met polymorphism, human memory, and synaptic neuroplasticity. Wiley Interdiscip Rev Cogn Sci. 2015;6:97-108 https://doi.org/10.1002/wcs.1334
2. Rafieva LM, Gasanov EV. Neurotrophin Propeptides: Biological Functions and Molecular Mechanisms. Curr Protein Pept Sci. 2016;17:298-305 https://doi.org/10.2174/1389203716666150623104145
3. Ventriglia M, Zanardini R, Bonomini C. et al. Serum brain-derived neurotrophic factor levels in different neurological diseases. Biomed Res Int. 2013;2013:901082. https://doi.org/10.1155/2013/901082
4. Gonzalez A, Moya-Alvarado G, Gonzalez-Billaut C, Bronfman FC. Cellular and molecular mechanisms regulating neuronal growth by brain-derived neurotrophic factor. Cytoskeleton. 2016;73:612-28 https://doi.org/10.1002/cm.21312
5. Schmidt RH, Nickerson JM, Boatright JH. Exercise as Gene Therapy: BDNF and DNA Damage Repair. Asia-Pacific J Ophthalmol. 2016;5:309-11 https://doi.org/10.1097/APO.0000000000000226
6. Zhao H, Alam A, San C-YY. et al. Molecular mechanisms of brain-derived neurotrophic factor in neuro-protection: Recent developments. Brain Res. 2017;1665:1-21 https://doi.org/10.1016/j.brainres.2017.03.029
7. Matthews VB, Aström M-BB, Chan MHS. et al. Brain-derived neurotrophic factor is produced by skeletal muscle cells in response to contraction and enhances fat oxidation via activation of AMP-activated protein kinase. Diabetologia. 2009;52:1409-18 https://doi.org/10.1007/s00125-009-1364-1
8. Kowiański P, Lietzau G, Czuba E. et al. BDNF: A Key Factor with Multipotent Impact on Brain Signaling and Synaptic Plasticity. Cell Mol Neurobiol. 2018;38:579-93 https://doi.org/10.1007/s10571-017-0510-4
9. Chiaruttini C, Vicario A, Li Z. et al. Dendritic trafficking of BDNF mRNA is mediated by translin and blocked by the G196A (Val66Met) mutation. Proc Natl Acad Sci. 2009;106:16481-6 https://doi.org/10.1073/pnas.0902833106
10. Pardridge WM, Kang YS, Buciak JL. Transport of human recombinant brain-derived neurotrophic factor (BDNF) through the rat blood-brain barrier in vivo using vector-mediated peptide drug delivery. Pharm Res. 1994;11:738-46
11. Zhang Y, Pardridge WM. Conjugation of brain-derived neurotrophic factor to a blood-brain barrier drug targeting system enables neuroprotection in regional brain ischemia following intravenous injection of the neurotrophin. Brain Res. 2001;889:49-56 https://doi.org/10.1016/S0006-8993(00)03108-5
12. Ieraci A, Madaio AI, Mallei A. et al. Brain-Derived Neurotrophic Factor Val66Met Human Polymorphism Impairs the Beneficial Exercise-Induced Neurobiological Changes in Mice. Neuropsychopharmacology. 2016;41(13):3070-9 doi: 10.1038/npp.2016.120
13. Lemos JR, Alves CR, de Souza SBC. et al. Peripheral vascular reactivity and serum BDNF responses to aerobic training are impaired by the BDNF Val66Met polymorphism. Physiol Genomics. 2016;48:116-23 https://doi.org/10.1152/physiolgenomics.00086.2015
14. Chen ZY, Jing D, Bath KG. et al. Genetic variant BDNF (Val66Met) polymorphism alters anxiety-related behavior. Science. 2006;314(5796):140-3 doi: 10.1126/science.1129663
15. Ward DD, Summers MJ, Saunders NL. et al. The BDNF Val66Met polymorphism moderates the relationship between cognitive reserve and executive function. Transl Psychiatry. 2015;5(6):e590. doi: 10.1038/tp.2015.82
16. Egan MF, Kojima M, Callicott JH. et al. The BDNF val66met polymorphism affects activity-dependent secretion of BDNF and human memory and hippocampal function. Cell. 2003;112:257-69 https://doi.org/10.1016/S0092-8674(03)00035-7
17. Chen ZY, Patel PD, Sant G. et al. Variant Brain-Derived Neurotrophic Factor (BDNF) (Met66) Alters the Intracellular Trafficking and Activity-Dependent Secretion of Wild-Type BDNF in Neurosecretory Cells and Cortical Neurons. J Neurosci. 2004;24:4401-11 https://doi.org/10.1523/JNEUROSCI.0348-04.2004
18. Jiang X, Zhou J, Mash DC. et al. Human BDNF isoforms are differentially expressed in cocaine addicts and are sorted to the regulated secretory pathway independent of the Met66 substitution. NeuroMolecular Med. 2009;11:1-12 https://doi.org/10.1007/s12017-008-8051-0
19. de Assis GG, Gasanov EV. BDNF and Cortisol integrative system - Plasticity vs. degeneration: Implications of the Val66Met polymorphism. Front Neuroendocrinol. 2019;55:100784. https://doi.org/10.1016/j.yfrne.2019.100784
20. Brunelli A, Dimauro I, Sgrò P. et al. Acute exercise modulates BDNF and pro-BDNF protein content in immune cells. Med Sci Sports Exerc. 2012;44:1871-80 https://doi.org/10.1249/MSS.0b013e31825ab69b
21. de Assis GG. et al. Val66Met allele-specific BDNF expression in human skeletal muscle in vivo and the effect of maximal effort on it. Sci Rep. In press
22. de Assis GG, Gasanov EV, de Sousa MBC. et al. Brain derived neutrophic factor, a link of aerobic metabolism to neuroplasticity. J Physiol Pharmacol. 2018;69:351-9 https://doi.org/10.26402/jpp.2018.3.12
23. Dinoff A, Herrmann N, Swardfager W. et al. The Effect of exercise training on resting concentrations of peripheral brain-derived neurotrophic factor (BDNF): A meta-analysis. PLoS One. 2016;11:e0163037. https://doi.org/10.1371/journal.pone.0163037
24. Ferris LT, Williams JS, Shen CL. The effect of acute exercise on serum brain-derived neurotrophic factor levels and cognitive function. Med Sci Sports Exerc. 2007;39:728-34 https://doi.org/10.1249/mss.0b013e31802f04c7
25. Schmidt-Kassow M, Schädle S, Otterbein S. et al. Kinetics of serum brain-derived neurotrophic factor following low-intensity versus high-intensity exercise in men and women. Neuroreport. 2012;23:889-93 https://doi.org/10.1097/WNR.0b013e32835946ca
26. Harrisberger F, Smieskova R, Schmidt A. et al. BDNF Val66Met polymorphism and hippocampal volume in neuropsychiatric disorders: A systematic review and meta-analysis. Neurosci Biobehav Rev. 2015;55:107-18 https://doi.org/10.1016/j.neubiorev.2015.04.017
27. Lim YY, Rainey-Smith S, Lim Y. et al. BDNF Val66Met in preclinical Alzheimer's disease is associated with short-term changes in episodic memory and hippocampal volume but not serum mBDNF. Int Psychogeriatrics. 2017;29:1825-34 https://doi.org/10.1017/S1041610217001284
28. Warburton DER, Gledhill N, Jamnik VK. et al. Evidence-based risk assessment and recommendations for physical activity clearance: Consensus Document 2011. Appl Physiol Nutr Metab. 2011;36:266-98 https://doi.org/10.1139/H11-062
29. Bruce RA, Blackmon JR, Jones JW, Strait G. Exercising testing in adult normal subjects and cardiac patients. Ann Noninvasive Electrocardiol. 2004;9:291-303 https://doi.org/10.1111/j.1542-474X.2004.93003.x
30. Kuang J, Yan X, Genders AJ. et al. An overview of technical considerations when using quantitative real-time PCR analysis of gene expression in human exercise research. PLoS One. 2018;13(5):e0196438. https://doi.org/10.1371/journal.pone.0196438
31. Sheikh HI, Hayden EP, Kryski KR. et al. Genotyping the BDNF rs6265 (val66met) polymorphism by one-step amplified refractory mutation system PCR. Psychiatric Genetics. 2010;20(3):109-12 https://doi.org/10.1097/YPG.0b013e32833a2038
32. Schmittgen TD, Livak KJ. Analyzing real-time PCR data by the comparative C(T) method. Nat Protoc. 2008;3(6):1101-8 doi: 10.1038/nprot.2008.73. PMID: 18546601
33. Shen T, You Y, Joseph C. et al. BDNF polymorphism: A review of its diagnostic and clinical relevance in neurodegenerative disorders. Aging Dis. 2018;9:523-36 https://doi.org/10.14336/AD.2017.0717
34. de Assis GG, de Almondes KM. Exercise-dependent BDNF as a modulatory factor for the executive processing of individuals in course of cognitive decline. A systematic review. Front Psychol. 2017;8:1-8 https://doi.org/10.3389/fpsyg.2017.00584
35. de Assis GG. BDNF and Neurodegeneration: The Rise of the Exercise as a Preventive Care. Biomed J Sci Tech Res. 2020;25:19502-4 https://doi.org/10.26717/BJSTR.2020.25.004265
36. Wang H, Wang J, Jiang J. et al. Reference genes for normalizing transcription in diploid and tetraploid arabidopsis. Sci Rep. 2014;4:6781. https://doi.org/10.1038/srep06781
37. Lee WJ, Jang SJ, Lee SC. et al. Selection of reference genes for quantitative real-time polymerase chain reaction in porcine embryos. Reprod Fertil Dev. 2015;29(2):357-67 https://doi.org/10.1071/RD14393
38. Li J, Yoshikawa A, Brennan MD. et al. Genetic predictors of antipsychotic response to lurasidone identified in a genome wide association study and by schizophrenia risk genes. Schizophr Res. 2018;192:194-204 https://doi.org/10.1016/j.schres.2017.04.009
39. Yokoyama T, Omotehara T, Hirano T. et al. Identification of reference genes for quantitative PCR analyses in developing mouse gonads. J Vet Med Sci. 2018;80(10):1534-9 https://doi.org/10.1292/jvms.18-0417
40. Sainz J, Prieto C, Crespo-Facorro B. Sex differences in gene expression related to antipsychotic induced weight gain. PLoS One. 2019;14(4):e0215477. https://doi.org/10.1371/journal.pone.0215477
41. Kozera B, Rapacz M. Reference genes in real-time PCR. J Appl Genet. 2013;54:391-406 https://doi.org/10.1007/s13353-013-0173-x
42. Zhang JY, Zhang F, Hong CQ. et al. Critical protein GAPDH and its regulatory mechanisms in cancer cells. Cancer Biol Med. 2015;12(1):10-22 https://doi.org/10.7497/j.issn.2095-3941.2014.0019
43. Sim MS, Mohamed Z, Hatim A, Rajagopal VL, Habil MH. Association of brain-derived neurotrophic factor (Val66Met) genetic polymorphism with methamphetamine dependence in a Malaysian population. Brain Res. 2010;1357:91-96 https://doi.org/10.1016/j.brainres.2010.08.053
44. Sutherland HG, Maher BH, Rodriguez-Acevedo AJ. et al. Investigation of brain-derived neurotrophic factor (BDNF) gene variants in migraine. Headache. 2014;54:1184-93 https://doi.org/10.1111/head.12351
45. Wang DD, Tian T, Dong Q. et al. Transcriptome profiling analysis of the mechanisms underlying the BDNF Val66Met polymorphism induced dysfunctions of the central nervous system. Hippocampus. 2014;24:65-78 https://doi.org/10.1002/hipo.22204
Author contact
![]() Corresponding author: Eugene V. Gasanov: egasanovgov.pl; Phone: +48 733 298 421
Corresponding author: Eugene V. Gasanov: egasanovgov.pl; Phone: +48 733 298 421

 Global reach, higher impact
Global reach, higher impact