3.2
Impact Factor
ISSN: 1449-1907
Int J Med Sci 2020; 17(17):2718-2727. doi:10.7150/ijms.47224 This issue Cite
Research Paper
Construction and Validation of a Protein Prognostic Model for Lung Squamous Cell Carcinoma
1. Department of Medical Oncology, Guangzhou First People's Hospital, School of Medicine, South China University of Technology, Guangzhou, Guangdong, China, 510180
2. Department of Medical Oncology, Guangzhou First People's Hospital, Guangzhou Medical University, Guangzhou, Guangdong, China, 510180
#These authors contributed equally to this work.
Received 2020-4-20; Accepted 2020-9-14; Published 2020-9-23
Abstract
Lung squamous cell carcinoma (LUSCC), as the major type of lung cancer, has high morbidity and mortality rates. The prognostic markers for LUSCC are much fewer than lung adenocarcinoma. Besides, protein biomarkers have advantages of economy, accuracy and stability. The aim of this study was to construct a protein prognostic model for LUSCC. The protein expression data of LUSCC were downloaded from The Cancer Protein Atlas (TCPA) database. Clinical data of LUSCC patients were downloaded from The Cancer Genome Atlas (TCGA) database. A total of 237 proteins were identified from 325 cases of LUSCC patients based on the TCPA and TCGA database. According to Kaplan-Meier survival analysis, univariate and multivariate Cox analysis, a prognostic prediction model was established which was consisted of 6 proteins (CHK1_pS345, CHK2, IRS1, PAXILLIN, BRCA2 and BRAF_pS445). After calculating the risk values of each patient according to the coefficient of each protein in the risk model, the LUSCC patients were divided into high risk group and low risk group. The survival analysis demonstrated that there was significant difference between these two groups (p= 4.877e-05). The area under the curve (AUC) value of the receiver operating characteristic (ROC) curve was 0.699, which suggesting that the prognostic risk model could effectively predict the survival of LUSCC patients. Univariate and multivariate analysis indicated that this prognostic model could be used as independent prognosis factors for LUSCC patients. Proteins co-expression analysis showed that there were 21 proteins co-expressed with the proteins in the risk model. In conclusion, our study constructed a protein prognostic model, which could effectively predict the prognosis of LUSCC patients.
Keywords: Lung squamous cell carcinoma, The Cancer Protein Atlas, The Cancer Genome Atlas, Protein prognostic risk model, Overall survival
Introduction
Lung cancer is the most common and the leading cause of cancer-related malignancy in the world [1]. According to the Cancer statistics 2018, there were about 2.09 million new cases and 1.76 million deaths per year [2]. Despite advancements in chemotherapy, radiotherapy, targeted therapy, biotherapy and immunotherapy, the 5-year survival rate of LUSCC is only 18% [3-5]. And no individualized treatment strategy is recommended to LUSCC due to the lack of clear histological features [6]. Besides, the economic impact of LUSCC is significant and is increasing yearly. Therefore, it is urgent and necessary to find more specific and reliable diagnostic and prognostic biomarkers for LUSCC.
Several studies have suggested prognostic factors for LUSCC, including proteins, mRNAs, lncRNAs and miRNAs. Tumor protein biomarkers demonstrated important roles in cancer detection, clinical outcome prediction or effective therapy selection [7-9]. Of note, protein biomarkers are economical, reliable and easily measurable. For instance, carcinoembryonic antigen (CEA), as the only FDA-approved biomarker for colorectal cancer (CRC), is a selective biomarker for detecting disease recurrence and monitoring response to routine treatment [10, 11]. Besides, proteins, such as tripartite motif-containing protein59 (TRIM59), minichromosome maintenance protein 2 (MCM2) and CD271 were highly expressed, while the expression of interferon-induced proteins with tetratricopeptide repeats 2 (IFIT2) was decreased in LUSCC cell lines or tissues [12]. The overall survival (OS) of patients with high TRIM59, MCM2, CD271 expression or low IFIT2 expression were significantly better than the opposite group [12-16]. However, the sensitivity and specificity was not satisfactory using one biomarker. Prognostic model using multiple protein biomarkers would demonstrate great potential in the diagnosis and prognosis of LUSCC patients.
In this study, we presented a prognostic risk model which was consisted of 6 proteins by identifying 237 proteins from TCPA database and TCGA database. In addition, we determined that the risk model was an independent prognostic model for LUSCC patients. This novel prognostic risk model could provide evidence for the diagnosis and prognosis of LUSCC patients.
Material and methods
Protein data download and data processing
Protein expression data of LUSCC patients were downloaded from The Cancer Proteome Atlas (TCPA) database up to January 2020. The “impute” package in the R software was used to fill up the missing data. This study complied with the publication guidelines for TCPA. And no additional ethical consent was required.
Clinical data download and data consolidation
Clinical data of LUSCC patients were downloaded from the TCGA database up to January 2020. Perl software was used to extract survival information and merge it with the protein expression data. This study complied with the publication guidelines of TCGA. And no additional ethical consent was required.
Kaplan-Meier survival analysis and univariate Cox analysis
The correlations between protein expression and overall survival of LUSCC patients were analyzed using Kaplan-Meier survival analysis and univariate Cox analysis via “survival” package in R software. A P value < 0.05 was used as the criterion for screening proteins with significant difference. The "ggplot2" and "ggrepel" packages were used to draw the volcano map of differently expressed proteins.
Construction of a prognostic risk model
The proteins with statistical difference were analyzed by multivariate Cox analysis to build a prognostic risk model. The coefficients of each protein in the model and the risk values of all samples were obtained at the same time. According to the risk values, patients were divided into high risk group and low risk group.
Assessment of the performance of the prognostic risk model
The "survminer” and “survival” package in R software were used to analyze the relationships between component proteins, risk values and survival status, as well as the survival curve. Using the "pheatmap" package in the R software, we built a risk curve for risk value, survival status and protein expression. The survival time, survival state, risk value, age, gender, staging, T, M and N states of the samples were combined with Perl software, followed by univariate and multivariate COX analysis to observe the correlations between the above clinical states, risk value and survival state. The ROC curve with the AUC value were assessed by the "survivalROC" package in R software.
Co-expressed proteins analysis
Other proteins co-expressed with the proteins in the risk model were screened by R software with the correlation coefficient was set by > 0.5 and the P value was set by < 0.001. The packages of "ggalluvial, ggplot2 and dplyr" in R software were used to construct the sankyl diagram for the related co-expressed proteins.
Statistical analysis
The retrieval of protein data and clinical information from TCPA and TCGA dataset were performed in R software (R 3.6.2) or Perl software Strawberry Perl (64-bit). All statistical analyses were assessed by R software. A P-value < 0.05 was statistically significant.
Results
Protein and clinical data download and data processing
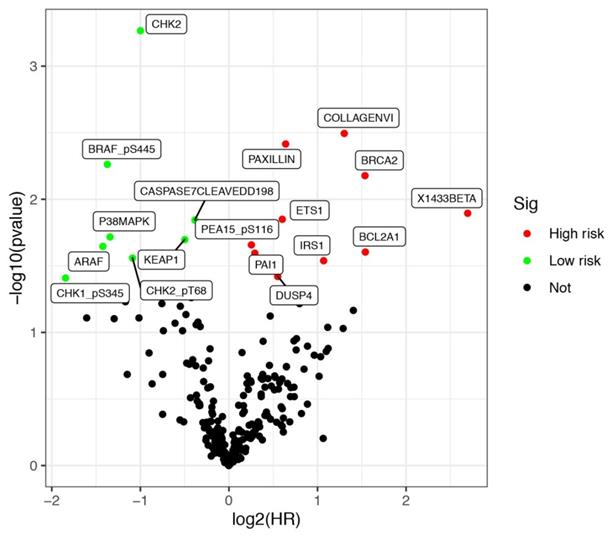
Protein expression data of LUSCC patients were downloaded from the TCPA database up to January 2020. The database contains 8167 tumor samples in total. And it is mainly consisted of TCGA tumor tissue sample sets, which including more than 32 cancer types in TCGA, and about 500 samples from an independent patient cohort. Bisides, The database also focuses on reverse-phase protein arrays (RPPA) data for cancer cell lines, containing >665 individual cell lines across 19 lineages (https://tcpaportal.org/tcpa/index.html). A total of 237 proteins were identified from 325 cases of LUSCC patients. Meanwhile, clinical data of 504 cases of LUSCC patients were downloaded from TCGA database. After screening 320 cases with both survival information and protein expression data, a total of 17 proteins were identified significantly associated with the survival of LUSCC patients. All the differentially expressed proteins was demonstrated by volcano plots (Figure 1). Among them, COLLAGENVI, PAXILLIN, BRCA2, X1433BETA, ETS1, PEA15_pS116, BCL2A1, PAI1, IRS1 and DUSP4 were regarded as high risk factors, while BRAF_pS445, CASPASE7CLEAVEDD198, P38MAPK, KEAP1, ARAF, CHK2_pT68 and CHK1_pS345 were viewed as low risk factors.
Kaplan-Meier survival analysis and univariate Cox analysis
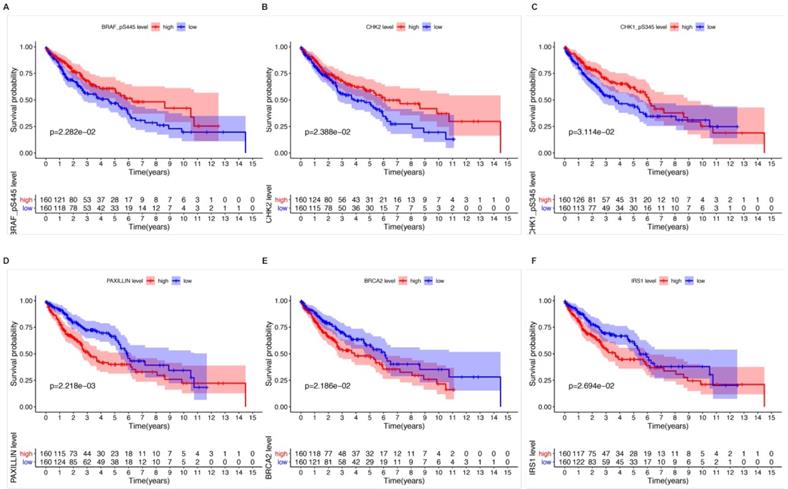
Kaplan-Meier survival analysis and univariate Cox analysis demonstrated that 6 proteins were significantly correlated with OS and could be viewed as independent prognostic factors for LUSCC patients at a criteria of P value < 0.05 (Table 1). High expression of CHK1_pS345 (P=3.114e-02, Figure 2A), CHK2 (P=2.388e-02, Figure 2B), BRAF_pS445 (P=2.282e-02, Figure 2C) were positively correlated with better overall survival. While high expression of PAXILLIN (P=2.218e-03, Figure 2D), BRCA2 (P=2.186e-02, Figure 2E) and IRS1 (P=2.694e-02, Figure 2F) indicated poorer OS for LUSCC patients.
Significant proteins from Kaplan-Meier and univariate COX analysis (P < 0.05)
| protein | Kaplan-Meier | HR | HR.95L | HR.95H | P value |
|---|---|---|---|---|---|
| CHK1_pS345 | 0.031 | 0.278 | 0.0826 | 0.937 | 0.039 |
| CHK2 | 0.024 | 0.500 | 0.338 | 0.741 | 0.001 |
| IRS1 | 0.027 | 2.100 | 1.079 | 4.088 | 0.029 |
| PAXILLIN | 0.002 | 1.560 | 1.154 | 2.108 | 0.004 |
| BRCA2 | 0.022 | 2.905 | 1.345 | 6.273 | 0.007 |
| BRAF_pS445 | 0.023 | 0.387 | 0.198 | 0.756 | 0.005 |
Differentially expressed proteins in lung squamous cell carcinoma (LUSCC). Data were retrieved from TCPA and TCGA database. A total of 320 cases of LUSCC patients with both protein expression data and clinical parameters were included.
The relationships between six independent prognostic proteins and overall survival (OS) of LUSCC patients. High expression of CHK1_pS345 (A), CHK2 (B), BRAF_pS445 (C) were positively correlated with better OS. High expression of PAXILLIN (D), BRCA2 (E) and IRS1(F) indicated poor OS.
Construction of a prognostic risk model
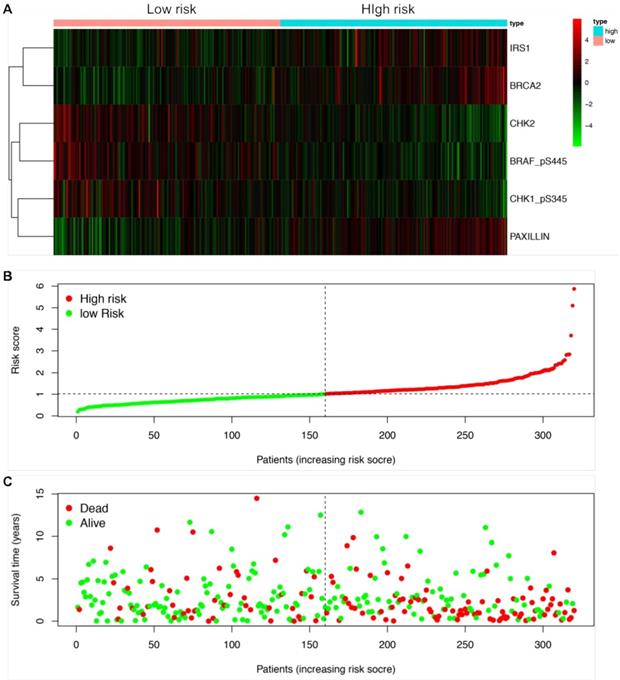
We next established a prognostic risk model using multivariate Cox regression analysis. According to the multivariate Cox analysis, a total of 6 proteins were included in the prognostic risk model, which could be viewed as independent prognostic factors for LUSCC patients (Table 2). The coefficients of each protein were demonstrated in Table 2. After calculating the risk value of each patient according to the coefficient of each protein in the risk model, the LUSCC patients were divided into high risk group and low risk group. Each group contained 160 cases. The heatmap shows the protein expression profiles of the prognostic risk model based on the risk scores (Figure 3A). Figure 3B shows the risk score distribution of the LUSCC patients. Figure 3C shows the survival status of the LUSCC patients based on the risk scoring model.
Significant proteins from multivariate Cox analysis
| protein | coef | HR | HR.95L | HR.95H | P value |
|---|---|---|---|---|---|
| CHK1_pS345 | -0.982 | 0.375 | 0.093 | 1.503 | 0.166 |
| CHK2 | -0.324 | 0.723 | 0.454 | 1.153 | 0.173 |
| IRS1 | -0.077 | 0.926 | 0.399 | 2.147 | 0.858 |
| PAXILLIN | 0.443 | 1.557 | 1.132 | 2.140 | 0.006 |
| BRCA2 | 0.563 | 1.755 | 0.600 | 5.135 | 0.304 |
| BRAF_pS445 | -0.706 | 0.494 | 0.249 | 0.979 | 0.043 |
Assessment of the performance of the prognostic risk model
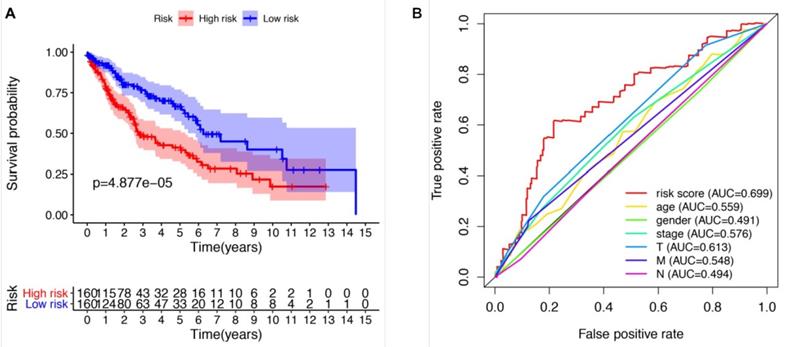
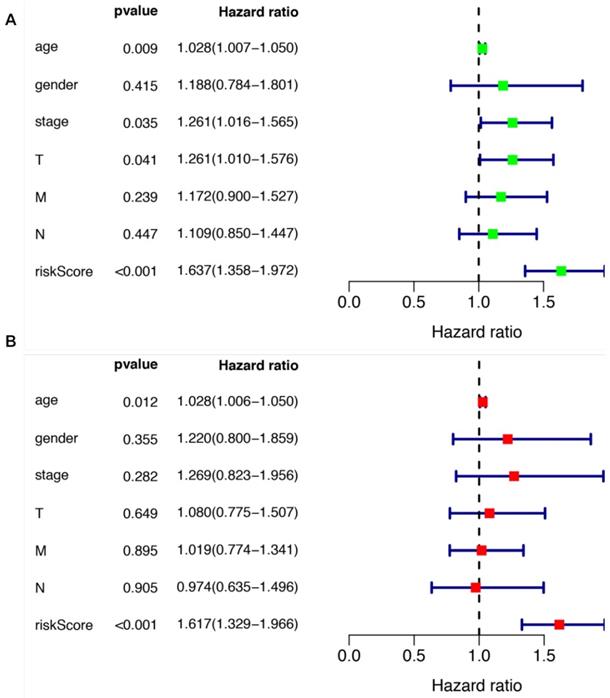
We further evaluated the performance of the prognostic risk model. After being divided into two groups based on the risk scores, there was significant difference between high risk group and low risk group in the OS (P = 4.877e-05, Figure 4A). As shown in Figure 4B, the area under the curve (AUC) value was 0.699 in the receiver operating characteristic (ROC) curve of the risk model, which suggesting that the prognostic model can effectively predict the survival of LUSCC patients. Moreover, combining the risk value with survival time, survival state, age, gender, stage, T, M and N states with Perl software, univariate Cox analysis demonstrated that the risk value (HR 1.673(1.358-1.972), P<0.001), age (HR 1.028(1.007-1.050), P=0.009), stage (HR 1.261(1.016-1.565), P=0.035), T states (HR 1.261(1.010-1.576), P=0.041) were statistical significance (Figure 5A). Further multivariate Cox analysis revealed that the risk value (HR 1.617, 95%CI 1.329-1.966, P=1.51e-06) and age (HR 1.028, 95%CI 1.006-1.050, P=0.012) could be viewed as independent prognostic factors for LUSCC patients (Figure 5B).
Co-expressed protein analysis
Other proteins co-expressed with the proteins included in the prognostic model were screened by R software with the correlation coefficient was set by > 0.5 and the P value was set by < 0.001. AKT, BETACATENIN, KU80, MRE11, NCADHERIN, TUBERIN, NRAS, PKCPANBETAII_pS660, RBM15, TAZ, X1433BETA, ACVRL1, MSH2, BCL2A1, CHK1_pS296, IGF1R_pY1135Y1136 and CASPASE9 were found to be co-expressed with BRCA2. And proteins co-expressed with CHK2 were CYCLINB1, MSH2, MSH6 and BRD4. The sankyl diagram for the related co-expressed proteins was constructed by packages of “ggalluvial, ggplot2 and dplyr” in R software (Figure 6).
Discussion
With the development of next-generation sequencing (NGS) technologies, there were more and more risk factors being identified for LUSCC patients, including miRNAs, lncRNAs and mRNAs [17-20]. However, some disadvantages of NGS, such as cost, reproducibility, and data analysis, had widely limited its application. Moreover, tumor protein biomarker was economical, reliable and clinical feasible [7, 21]. Besides, it would be more accessible in health-providing centers. Some studies had successfully validated protein prognostic model in malignancies, such as bladder cancer, lung adenocarcinoma and diffuse large B-cell lymphoma [22-24]. The protein models demonstrated vital role for predicting prognosis, which including bladder cancer, esophageal squamous cell carcinoma and pancreatic ductal adenocarcinoma [25-27]. Abnormal expression of various protein biomarkers in cancer cells or inflammatory cells demonstrated prediction potential in cancers [21, 28, 29]. And a prediction model would demonstrate much better specificity and sensitivity than one biomarker.
Construction of a protein prognostic risk model in LUSCC. The patients were divided into high-risk group and low-risk group according to the risk values. (A) The heatmap demonstrated the expression of the six proteins between high risk group and low risk group. Upregulated expression of CHK1_pS345, CHK2 and BRAF_pS445 were detected in low-risk group, while upregulated expression of PAXILLIN, BRCA2 and IRS1 were detected in high-risk group. (B) Scatter diagram shows the distributions of risk scores of LUSCC patients. (C) Scatter diagram shows the survival status of the patients based on this prognostic risk model.
The prognostic risk model could effectively predict the survival of LUSCC patients. (A) LUSCC patients in high-risk group demonstrated poor OS than that in the low-risk group. (B) Receiver operating characteristic (ROC) curve revealed the performance of the prognostic risk model in LUSCC.
The prognostic significances of clinicopathological parameters and the risk model. (A) Univariate Cox analysis was performed to assess the prognostic values of various clinicopathological factors and risk score. (B) Multivariate Cox analysis revealed the independent prognostic values of various clinicopathological factors and risk score in the survival of LUSCC patients.
Sankyl diagram shows other proteins co-expressed the six proteins included in the prognostic risk model.

In this study, a total of 320 cases of LUSCC patients with both clinical information and protein expression data were identified from the TCPA and TCGA database. We further established a prognostic risk model that including 6 proteins (CHK1_pS345, CHK2, IRS1, PAXILLIN, BRCA2 and BRAF_pS445) using multivariate Cox regression analysis. These six proteins were significantly correlated with OS of LUSCC patients. In addition, the performance of the prognostic risk model was evaluated according to Kaplan-Meier survival analysis and ROC curve. Moreover, we screened other proteins co-expressed with the proteins in the risk model to find more related proteins. Our study validated a prognostic risk model which was with good sensitivity and specificity, and could effectively predict the survival of LUSCC patients.
Some of the proteins in our prognostic risk model have been reported to be involved in the tumorigenesis and prognosis of malignancies. Check-point kinase (Chk) is an enzyme that plays multiple roles in cancer development, including (1) blocking cell cycle and DNA damage, (2) mediating the detection and repair of damaged DNA, (3) guaranteeing the quality of DNA replication and distribution of chromosome[30]. Chk1 and Chk2 were the most common enzymes in ChK family, both played extremely important roles in DNA damage repair[31]. The deletion or overexpression of Chk1 or Chk2 may cause the occurrence and development of some tumors and affect the prognosis of cancer patients [32-34]. Insulin Receptor Substrate (IRS) proteins are the main cytoplasmic adaptor molecules involving in transducing extracellular signals from receptors to downstream proteins. IRS family is consisted of four proteins (IRS1-IRS4). IRS1 is located on human chromosome 2q34-37 and encoded by single exon. It is widely expressed in human tissues [35]. IRS1 expression was over-expressed in laryngeal squamous cell carcinoma and colorectal cancer [36, 37]. Using single-cell analysis, Eyler et al. found that glioma cells could activate the AKT signaling pathway through IRS1 to acquire drug resistance capability [38]. Furthermore, IRS1 Gly972Arg variations may be associated with an increased susceptibility to develop gastric cancer [39].The important cellular adhesion factor Paxillin (PXN) is a hallmark protein in focal adhesion (FA) site. It acted as a signal binding protein of tumorigenic tyrosine kinases and was involved in various physiological processes, such as cell proliferation, tissue reconstruction, cell adhesion and migration [40]. High expression of PXN was correlated with worse prognosis in liver and colorectal cancer [41-43]. Various researches have suggested that breast cancer susceptibility gene 2 (BRCA2) was widely involved in gene transcription and cell apoptosis [44, 45]. Mutation of BRCA2 genes led to function loss of BRCA2 protein, which resulted in tumorigenesis, progression and poor prognosis [46-48]. BRAF pS445 was an essential kinase in the mitogen-activated protein kinase (MAPK) pathway. Mutation of BRAF gene led to uncontrollable cell growth and ultimately developed to cancer cell [49]. Besides, application of BRAF inhibitors could improve outcomes of melanoma, colorectal cancer and non-small cell lung cancer patients [50-52].
In addition, some studies have revealed the expression and biological functions of these proteins in lung cancer. CHK1 was upregulated in lung cancer cells. Inhibition of CHK1 could seriously damage the cell cycle characteristics of lung cancer cells [53]. Activation of Chk1 was the earliest and most important event in NSCLC stem cells receiving chemotherapy. Differentiated NSCLC cells were usually accompanied with weak Chk1 activation and more sensitive to chemotherapeutic agents than undifferentiated tumor cells. In vitro, chemotherapy combined with Chk1 inhibitor can significantly reduce the survival rate of NSCLC stem cell [54]. Targeting Chk2 reduced phosphorylation and increased the efficacy of cisplatin [55]. Researches showed that genomic changes of IRS1 could lead to development of lung cancer [56]. Silencing IRS1 caused lung cancer cell proliferation and induced phosphorylation of anaplastic lymphoma kinase(AKT) [57]. Importantly, Paxillin played a crucial role in the progression of lung cancer [58]. Studies by Jagadeeswaran et al. [59] identified a total of 21 unique paxillin mutations in lung cancer specimens and cell lines. BRCA2 gene has been proved to carry a high risk of hereditary in breast and ovarian cancer. A stop-gain mutation of this gene, K3326* (rs11571833), was significantly associated with small cell lung cancer [60]. Besides, activating BRCA2 by MIR-1245 suppressed the proliferation and invasion of lung cancer cells [61]. The polymorphism rs144848 in gene BRCA2 often suggested a lower incidence of lung cancer in Chinese women [62]. BRAF mutations were detected in 3% of NSCLC, in which 58% was BRAF V600E mutation and the mutations were always associated with smoking [63, 64]. Recent reports revealed that it acted as resistance pathways in 3-10% of osimertinib-treated patients [65-68]. To targeting BRAF mutations, vemurafenib was an option[69]. Another therapeutic approach such as ulixertinib worked by blocking downstream signaling in the MAP kinase pathway[70]. However, there are lack of studies concerning these protein biomarkers in LUSCC. Thus, the expression and biological functions of these proteins in LUSCC are needed to be further elucidated in our future work.
In conclusion, we have established and validated a protein-based prognostic risk model for LUSCC patients. This model demonstrated good sensitively and specificity. Integrated analysis of proteins would provide economic, accuracy and stability pattern in research of diagnosis and prognosis biomarkers for LUSCC patients.
Abbreviations
LUSCC, Lung squamous cell carcinoma; TCPA, The Cancer Protein Atlas; TCGA, The Cancer Genome Atlas; ROC, receiver operating characteristic; AUC, area under the curve; CEA, carcinoembryonic antigen; CRC, colorectal cancer; TRIM59, tripartite motif-containing protein59; MCM2, minichromosome maintenance protein 2; IFIT2, interferon-induced proteins with tetratricopeptide repeats 2; OS, overall survival; NGS, next-generation sequencing; Chk, Check-point kinase; IRS, Insulin receptor substrate; PXN, Paxillin; FA, focal adhesion; MAPK, mitogen-activated protein kinase; AKT, Anaplastic lymphoma kinase.
Acknowledgements
Funding
This research was funded by the National Natural Science Foundation of China (No. 81672900 and No. 81502563), the Natural Science Foundation of Guangdong Province, China (No. 2016A030310109), Guangzhou Science and Technology Program (No. 201904010427 and No.201707010262) and Guangzhou General Science and technology project of Health and Family Planning (grant number 20201A011007).
Availability of data and material
All date generated or analyzed during this study are included within the article. The datasets used and analyzed during the current study are available from the corresponding author on reasonable request.
Author Contributions
Conception and design: Xisheng Fang and Guolong Liu; Collection and assembly of data: Xia Liu, Yong Wu and Haibo Mao; Data analysis and interpretation: Lin Lu, Baoxiu Li, Chengyin Weng, and Mingmei Guan; Manuscript writing: Lin Lu and Xisheng Fang; Manuscript revising: Guolong Liu; Final approval of manuscript: All authors.
Competing Interests
The authors have declared that no competing interest exists.
References
1. Acosta AM, Kadkol SS. Mitogen-Activated Protein Kinase Signaling Pathway in Cutaneous Melanoma: An Updated Review. Arch Pathol Lab Med. 2016;140:1290-6
2. Ferlay J, Colombet M, Soerjomataram I, Dyba T, Randi G, Bettio M. et al. Cancer incidence and mortality patterns in Europe: Estimates for 40 countries and 25 major cancers in 2018. Eur J Cancer. 2018;103:356-87
3. Derman BA, Mileham KF, Bonomi PD, Batus M, Fidler MJ. Treatment of advanced squamous cell carcinoma of the lung: a review. Transl Lung Cancer Res. 2015;4:524-32
4. Masters GA, Johnson DH, Temin S. Systemic Therapy for Stage IV Non-Small-Cell Lung Cancer: American Society of Clinical Oncology Clinical Practice Guideline Update. J Oncol Pract. 2016;12:90-3
5. Theelen W, Peulen HMU, Lalezari F, van der Noort V, de Vries JF, Aerts J. et al. Effect of Pembrolizumab After Stereotactic Body Radiotherapy vs Pembrolizumab Alone on Tumor Response in Patients With Advanced Non-Small Cell Lung Cancer: Results of the PEMBRO-RT Phase 2 Randomized Clinical Trial. JAMA Oncol. 2019
6. Coudray N, Ocampo PS, Sakellaropoulos T, Narula N, Snuderl M, Fenyo D. et al. Classification and mutation prediction from non-small cell lung cancer histopathology images using deep learning. Nat Med. 2018;24:1559-67
7. Sun N, Sun S, Gao Y, Li Y, Lu Z, Yuan Z. et al. Utility of IDH1 as a Serum Protein Biomarker for the Early Detection of NSCLC: A Multicenter In Vitro Diagnostic Clinical Trial. Cancer Sci. 2020
8. Sun R, Hunter C, Chen C, Ge W, Morrice N, Liang S. et al. Accelerated Protein Biomarker Discovery from FFPE Tissue Samples Using Single-Shot, Short Gradient Microflow SWATH MS. J Proteome Res. 2020
9. Neubert H, Shuford CM, Olah TV, Garofolo F, Schultz GA, Jones BR. et al. Protein Biomarker Quantification by Immunoaffinity Liquid Chromatography-Tandem Mass Spectrometry: Current State and Future Vision. Clin Chem. 2020;66:282-301
10. Locker GY, Hamilton S, Harris J, Jessup JM, Kemeny N, Macdonald JS. et al. ASCO 2006 update of recommendations for the use of tumor markers in gastrointestinal cancer. J Clin Oncol. 2006;24:5313-27
11. Duffy MJ, van Dalen A, Haglund C, Hansson L, Holinski-Feder E, Klapdor R. et al. Tumour markers in colorectal cancer: European Group on Tumour Markers (EGTM) guidelines for clinical use. Eur J Cancer. 2007;43:1348-60
12. Su W, Xiao W, Chen L, Zhou Q, Zheng X, Ju J. et al. Decreased IFIT2 Expression In Human Non-Small-Cell Lung Cancer Tissues Is Associated With Cancer Progression And Poor Survival Of The Patients. Onco Targets Ther. 2019;12:8139-49
13. Mochizuki M, Nakamura M, Sibuya R, Okazaki T, Abe J, Nakagawa T. et al. CD271 is a negative prognostic factor and essential for cell proliferation in lung squamous cell carcinoma. Lab Invest. 2019;99:1349-62
14. Wu W, Wang X, Shan C, Li Y, Li F. Minichromosome maintenance protein 2 correlates with the malignant status and regulates proliferation and cell cycle in lung squamous cell carcinoma. Onco Targets Ther. 2018;11:5025-34
15. Ma G, Ji D, Qu X, Liu S, Yang X, Wang G. et al. Mining and validating the expression pattern and prognostic value of acetylcholine receptors in non-small cell lung cancer. Medicine (Baltimore). 2019;98:e15555
16. Lou M, Gao Z, Zhu T, Mao X, Wang Y, Yuan K. et al. TRIM59 as a novel molecular biomarker to predict the prognosis of patients with NSCLC. Oncol Lett. 2020;19:1400-8
17. Sousa D, Matthiesen R, Lima RT, Vasconcelos MH. Deep Sequencing Analysis Reveals Distinctive Non-Coding RNAs When Comparing Tumor Multidrug-Resistant Cells and Extracellular Vesicles with Drug-Sensitive Counterparts. Cancers (Basel). 2020 12
18. Tang RX, Chen WJ, He RQ, Zeng JH, Liang L, Li SK. et al. Identification of a RNA-Seq based prognostic signature with five lncRNAs for lung squamous cell carcinoma. Oncotarget. 2017;8:50761-73
19. Zheng R, Shen Q, Mardekian S, Solomides C, Wang ZX, Evans NR 3rd. Molecular profiling of key driver genes improves staging accuracy in multifocal non-small cell lung cancer. J Thorac Cardiovasc Surg. 2019
20. Paik PK, Kim RK, Ahn L, Plodkowski AJ, Ni A, Donoghue MTA. et al. A Phase II Trial of Albumin-Bound Paclitaxel and Gemcitabine in Patients with Newly Diagnosed Stage IV Squamous Cell Lung Cancers. Clin Cancer Res. 2020
21. Russell MR, Graham C, D'Amato A, Gentry-Maharaj A, Ryan A, Kalsi JK. et al. Diagnosis of epithelial ovarian cancer using a combined protein biomarker panel. Br J Cancer. 2019;121:483-9
22. Bosch M, Akhter A, Chen BE, Mansoor A, Lebrun D, Good D. et al. A bioclinical prognostic model using MYC and BCL2 predicts outcome in relapsed/refractory diffuse large B-cell lymphoma. Haematologica. 2018;103:288-96
23. Li W, Gao LN, Song PP, You CG. Development and validation of a RNA binding protein-associated prognostic model for lung adenocarcinoma. Aging (Albany NY). 2020;12:3558-73
24. Semeniuk-Wojtas A, Lubas A, Cierniak S, Brzoskowska U, Syrylo T, Zielinski H. et al. Selected protein expression in a new prognostic model for patients with non-muscle-invasive bladder cancer. J Cancer Res Clin Oncol. 2020
25. Burki T. Protein biomarker for pancreatic ductal adenocarcinoma. Lancet Oncol. 2017;18:e511
26. Tsai CH, Chen YT, Chang YH, Hsueh C, Liu CY, Chang YS. et al. Systematic verification of bladder cancer-associated tissue protein biomarker candidates in clinical urine specimens. Oncotarget. 2018;9:30731-47
27. Wang M, Smith JS, Wei WQ. Tissue protein biomarker candidates to predict progression of esophageal squamous cell carcinoma and precancerous lesions. Ann N Y Acad Sci. 2018;1434:59-69
28. Duangkumpha K, Stoll T, Phetcharaburanin J, Yongvanit P, Thanan R, Techasen A. et al. Discovery and Qualification of Serum Protein Biomarker Candidates for Cholangiocarcinoma Diagnosis. J Proteome Res. 2019;18:3305-16
29. Enroth S, Berggrund M, Lycke M, Broberg J, Lundberg M, Assarsson E. et al. High throughput proteomics identifies a high-accuracy 11 plasma protein biomarker signature for ovarian cancer. Commun Biol. 2019;2:221
30. HM B, A M, LS C, D M. Therapeutic potential of investigational CHK-1 inhibitors for the treatment of solid tumors. Expert opinion on investigational drugs. 2017;26:1063-72
31. G A, YC L, B C, DSS L, NLY N, AC C. et al. Csk-homologous kinase (Chk) is an efficient inhibitor of Src-family kinases but a poor catalyst of phosphorylation of their C-terminal regulatory tyrosine. Cell communication and signaling: CCS. 2017;15:29
32. Grabauskiene S, Bergeron EJ, Chen G, Thomas DG, Giordano TJ, Beer DG. et al. Checkpoint kinase 1 protein expression indicates sensitization to therapy by checkpoint kinase 1 inhibition in non-small cell lung cancer. J Surg Res. 2014;187:6-13
33. Wang WT, Catto JW, Meuth M. Differential response of normal and malignant urothelial cells to CHK1 and ATM inhibitors. Oncogene. 2015;34:2887-96
34. Zhang Y, Hunter T. Roles of Chk1 in cell biology and cancer therapy. Int J Cancer. 2014;134:1013-23
35. Gorgisen G, Gulacar I, Ozes O. The role of insulin receptor substrate (IRS) proteins in oncogenic transformation. Cellular and molecular biology (Noisy-le-Grand, France). 2017;63:1-5
36. Cui C, Li Y, Cui X, Wu X. lncRNA XIST promotes the progression of laryngeal squamous cell carcinoma by sponging miR-144 to regulate IRS1 expression. Oncology reports. 2020;43:525-35
37. Liu Y, Li Y, Hou R, Shu Z. Knockdown GREM1 suppresses cell growth, angiogenesis, and epithelial-mesenchymal transition in colon cancer. Journal of cellular biochemistry. 2019;120:5583-96
38. Wang S, Brooks A, Poole A, Simoes-Barbosa A. Determinants of translation efficiency in the evolutionarily-divergent protist Trichomonas vaginalis. BMC molecular and cell biology. 2020;21:54
39. Gorgisen G, Karatas U, Ates C, Oksuz M, Gulacar I. Association of IRS1 Gly972Arg and IRS2 Gly1057Asp polymorphisms with gastric cancer in Turkish subjects. Oncology letters. 2020;20:2016-20
40. Pasquin S, Sharma M, Gauchat JF. Cytokines of the LIF/CNTF family and metabolism. Cytokine. 2016;82:122-4
41. Mathlouthi A, Pennacchietti E, De Biase D. Effect of Temperature, pH and Plasmids on In Vitro Biofilm Formation in Escherichia coli. Acta Naturae. 2018;10:129-32
42. Sutherland L, Ruhe M, Gattegno-Ho D, Mann K, Greaves J, Koscielniak M. et al. LIF-dependent survival of embryonic stem cells is regulated by a novel palmitoylated Gab1 signalling protein. J Cell Sci. 2018 131
43. Humer D, Spadiut O. Wanted: more monitoring and control during inclusion body processing. World J Microbiol Biotechnol. 2018;34:158
44. Soenderstrup IMH, Laenkholm AV, Jensen MB, Eriksen JO, Gerdes AM, Hansen TVO. et al. Clinical and molecular characterization of BRCA-associated breast cancer: results from the DBCG. Acta Oncol. 2018;57:95-101
45. Martinez-Useros J, Garcia-Foncillas J. The Role of BRCA2 Mutation Status as Diagnostic, Predictive, and Prognosis Biomarker for Pancreatic Cancer. Biomed Res Int. 2016;2016:1869304
46. Cui M, Gao XS, Gu X, Guo W, Li X, Ma M. et al. BRCA2 mutations should be screened early and routinely as markers of poor prognosis: evidence from 8,988 patients with prostate cancer. Oncotarget. 2017;8:40222-32
47. Le Page C, Rahimi K, Rodrigues M, Heinzelmann-Schwarz V, Recio N, Tommasi S. et al. Clinicopathological features of women with epithelial ovarian cancer and double heterozygosity for BRCA1 and BRCA2: A systematic review and case report analysis. Gynecol Oncol. 2020;156:377-86
48. Kuang S, Li H, Feng J, Xu S, Le Y. Correlation of BRCA2 gene mutation and prognosis as well as variant genes in invasive urothelial carcinoma of the bladder. Cancer Biomark. 2019;25:203-12
49. Amaral T, Sinnberg T, Meier F, Krepler C, Levesque M, Niessner H. et al. The mitogen-activated protein kinase pathway in melanoma part I - Activation and primary resistance mechanisms to BRAF inhibition. Eur J Cancer. 2017;73:85-92
50. Post JB, Roodhart JML, Snippert HJG. Colorectal Cancer Modeling with Organoids: Discriminating between Oncogenic RAS and BRAF Variants. Trends Cancer. 2020;6:111-29
51. Patel H, Yacoub N, Mishra R, White A, Long Y, Alanazi S. et al. Current Advances in the Treatment of BRAF-Mutant Melanoma. Cancers (Basel). 2020 12
52. O'Leary CG, Andelkovic V, Ladwa R, Pavlakis N, Zhou C, Hirsch F. et al. Targeting BRAF mutations in non-small cell lung cancer. Transl Lung Cancer Res. 2019;8:1119-24
53. Sheng X, Jin J, He Z, Huang Y, Zhou A, Wang J. et al. Pazopanib versus sunitinib in Chinese patients with locally advanced or metastatic renal cell carcinoma: pooled subgroup analysis from the randomized, COMPARZ studies. BMC cancer. 2020;20:219
54. Bartucci M, Svensson S, Romania P, Dattilo R, Patrizii M, Signore M. et al. Therapeutic targeting of Chk1 in NSCLC stem cells during chemotherapy. Cell death and differentiation. 2012;19:768-78
55. Zhang H, Shao F, Guo W, Gao Y, He J. Knockdown of KLF5 promotes cisplatin-induced cell apoptosis via regulating DNA damage checkpoint proteins in non-small cell lung cancer. Thoracic cancer. 2019;10:1069-77
56. Houghton A, Rzymkiewicz D, Ji H, Gregory A, Egea E, Metz H. et al. Neutrophil elastase-mediated degradation of IRS-1 accelerates lung tumor growth. Nature medicine. 2010;16:219-23
57. Gorgisen G, Hapil F, Yilmaz O, Cetin Z, Pehlivanoglu S, Ozbudak I. et al. Identification of novel mutations of Insulin Receptor Substrate 1 (IRS1) in tumor samples of non-small cell lung cancer (NSCLC): Implications for aberrant insulin signaling in development of cancer. Genetics and molecular biology. 2019;42:15-25
58. Kanteti R, Batra S, Lennon F, Salgia R. FAK and paxillin, two potential targets in pancreatic cancer. Oncotarget. 2016;7:31586-601
59. Jagadeeswaran R, Surawska H, Krishnaswamy S, Janamanchi V, Mackinnon A, Seiwert T. et al. Paxillin is a target for somatic mutations in lung cancer: implications for cell growth and invasion. Cancer research. 2008;68:132-42
60. Rafnar T, Sigurjonsdottir G, Stacey S, Halldorsson G, Sulem P, Pardo L. et al. Association of BRCA2 K3326* With Small Cell Lung Cancer and Squamous Cell Cancer of the Skin. Journal of the National Cancer Institute. 2018;110:967-74
61. Yang L, Wang J, Fan Y, Yu K, Jiao B, Su X. Hsa_circ_0046264 up-regulated BRCA2 to suppress lung cancer through targeting hsa-miR-1245. Respiratory research. 2018;19:115
62. Lin Y, He F, Zhang X, Yu T, Liu Z, Cai L. Polymorphism rs144848 in BRCA2 may reduce lung cancer risk in women: a case-control study in southeast China. Tumori. 2016;102:150-5
63. Friedlaender A, Banna G, Patel S, Addeo A. Diagnosis and Treatment of ALK Aberrations in Metastatic NSCLC. Current treatment options in oncology. 2019;20:79
64. Friedlaender A, Drilon A, Weiss G, Banna G, Addeo A. KRAS as a druggable target in NSCLC: Rising like a phoenix after decades of development failures. Cancer treatment reviews. 2020;85:101978
65. Bearz A, De Carlo E, Doliana R, Schiappacassi M. Acquired BRAF V600E Mutation as Resistant Mechanism after Treatment with Third-Generation EGFR Tyrosine Kinase Inhibitor. Journal of thoracic oncology: official publication of the International Association for the Study of Lung Cancer. 2017;12:e181-e2
66. Ding P, Becker T, Bray V, Chua W, Ma Y, Xu B. et al. Plasma next generation sequencing and droplet digital PCR-based detection of epidermal growth factor receptor (EGFR) mutations in patients with advanced lung cancer treated with subsequent-line osimertinib. Thoracic cancer. 2019;10:1879-84
67. Del Re M, Crucitta S, Gianfilippo G, Passaro A, Petrini I, Restante G. et al. EGFRUnderstanding the Mechanisms of Resistance in -Positive NSCLC: From Tissue to Liquid Biopsy to Guide Treatment Strategy. International journal of molecular sciences. 2019 20
68. Mehlman C, Cadranel J, Rousseau-Bussac G, Lacave R, Pujals A, Girard N. et al. Resistance mechanisms to osimertinib in EGFR-mutated advanced non-small-cell lung cancer: A multicentric retrospective French study. Lung cancer (Amsterdam, Netherlands). 2019;137:149-56
69. Mazieres J, Cropet C, Montané L, Barlesi F, Souquet P, Quantin X. et al. Vemurafenib in non-small-cell lung cancer patients with BRAF and BRAF mutations. Annals of oncology: official journal of the European Society for Medical Oncology. 2020;31:289-94
70. Sullivan R, Infante J, Janku F, Wong D, Sosman J, Keedy V. et al. First-in-Class ERK1/2 Inhibitor Ulixertinib (BVD-523) in Patients with MAPK Mutant Advanced Solid Tumors: Results of a Phase I Dose-Escalation and Expansion Study. Cancer discovery. 2018;8:184-95
Author contact
![]() Corresponding author: Dr. Guolong Liu, Guangzhou First People's Hospital, South China University of Technology, Guangzhou, Guangdong, China, 510180. Tel: +86-20-81045125, Fax: +86-20-81045125, E-mail: eyglliuedu.cn or Dr. Lin Lu, Tel: +86-20-81045230, Fax: +86-20-81045230, E-mail: eylinlvedu.cn
Corresponding author: Dr. Guolong Liu, Guangzhou First People's Hospital, South China University of Technology, Guangzhou, Guangdong, China, 510180. Tel: +86-20-81045125, Fax: +86-20-81045125, E-mail: eyglliuedu.cn or Dr. Lin Lu, Tel: +86-20-81045230, Fax: +86-20-81045230, E-mail: eylinlvedu.cn

 Global reach, higher impact
Global reach, higher impact