3.2
Impact Factor
ISSN: 1449-1907
Int J Med Sci 2020; 17(12):1763-1772. doi:10.7150/ijms.48162 This issue Cite
Research Paper
FLAD1 is up-regulated in Gastric Cancer and is a potential prediction of prognosis
1. Breast Cancer Center, the Third Affiliated Hospital of Sun Yat-sen University, Guangzhou, 510000, P.R. China.
2. Department of Pathology, the Third Affiliated Hospital of Sun Yat-sen University, Guangzhou510000, P. R. China.
3. Department of Urologic Oncology, Chongqing University Cancer Hospital & Chongqing Cancer Institute & Chongqing Cancer Hospital, Chongqing, 400030, P.R. China.
4. Joint Surgery/Orthopedic Trauma Department, the Third Affiliated Hospital of Sun Yat-sen University, Guangzhou 510000, P. R. China.
5. General Surgery Department, the Third Affiliated Hospital of Sun Yat-sen University, Guangzhou, 510000, P.R. China.
*These authors contributed equally to this work.
Received 2020-5-13; Accepted 2020-6-23; Published 2020-7-6
Abstract
Background: Gastric cancer (GC) is a common malignancy throughout the world. Biomarkers for prognosis and risk evaluation of GC are rapidly discovered. We investigated the prognostic role of FLAD1, an important protein-coding gene that affects cell cycle and survival.
Methods: The expression of FLAD1 at mRNA levels in GC tumor tissues and normal tissues was mined and analyzed in Oncomine database and verified in 10 pairs of GS tissues and their adjacent normal tissues in our center by RT qPCR. The FLAD1 protein expression were detected in 106 paraffin-embedded GC tissues by immunohistochemistry (IHC). Statistical analyses were applied to evaluate the clinical significance of FLAD1. The prognostic value of FLAD1 mRNA expression was also analyzed using the Kaplan-Meier plotter (
Results: Statistics obtained from online database suggested FLAD1 mRNA was overexpressed in GC tissues. The results were further validated in 10 pairs of GS tissues and adjacent normal tissues in our center (p=0.021). IHC and survival analysis of GC samples from 106 patients showed FLAD1 was overexpressed in 63/106 (59.4%) patients and was associated to higher TNM stage (p=0.026). Multivariate analysis revealed FLAD1 was an independent prognostic factor for GC (p < 0.001). Furthermore, FLAD1 mRNA was associated to unfavorable overall survival (OS), first progression (FP), and post-progression survival (PPS) of GC (p<0.001).
Conclusion: FLAD1 in GC is overexpressed at both mRNA and protein level and could be a potential biomarker for GC prognosis.
Keywords: FLAD1, biomarker, overexpression, gastric cancer, prognosis
Introduction
Gastric cancer (GC) is the fifth most common and the third most mortal malignancy worldwide, and the second most common cause of morbidity and mortality in China[1, 2]. Most GCs are diagnosed at late stages when patients are complaining of abdominal pain, anorexia, and Cachexia. Consequently, the five-year survival rate of GC is dismal, despite great advances in surgery and adjuvant treatments [3]. Hence, illuminating the molecular mechanisms of GC occurance and development is of great importance for improving clinical outcomes [4]. Efforts are made to identify prognostic factors for better cost-effectiveness of GC management, such as coding and non-coding RNAs, which have been found to impact proliferation, apoptosis, and invasion of GC tumor cell [5, 6].
FLAD1 is a protein-coding gene for flavin adenine dinucleotide synthetase (FADS) ubiquitously expressed in various human tissue, with the highest level in lymph node [7]. FADS is a key enzyme in the FAD biosynthesis process [8], which contains an N-terminal molybdopterin -binding (MPTb) domain and a C-terminal domain (FADS domain). MPTb domain has FAD hydrolase activity, and FADS domain catalyzes FAD synthesis [9]. FADS is closely related to oxidation-reduction chain of cell. It participates in FMN biosynthesis, phosphor-adenosine phosphosulfate metabolism, and the oxidative-reduction process.
Human FLAD1 is ubiquitously expressed in lymph node, thyroid, and 25 other tissues [7]. FLAD1 gene is located on the long arm of chromosome 1q21.3, the deficiency can cause multiple acyl-CoA dehydrogenase deficiencies (MADDs), which is a severe metabolic disorder with mitochondrial respiratory-chain deficiency [8, 10]. FADS functions are critical to tumor cell as well. Due to its function in the oxidation-reduction chain, FLAD1 is strongly associated with the survival of malignant tumor cells and thus the prognosis of malignant tumor. Eeles et al. have found FLAD1, together with other 22 genes, was associated with prostate cancer [11]. Target-identification phenotypic screening and competitive affinity-based proteome profiling have targeted FLAD1 as a potential target for treatment [12]. Mitra et al. reported the relationship between FLAD1 expression and non-small cell lung cancer, especially in recurrent tumors, suggesting FLAD1 might be a biomarker for tumor relapse [13]. Unfortunately, there is no study up to date investigating the relationship between FLAD1 and GC; whether FLAD1 can serve as a prognostic biomarker is unknown. To fill this gap, we mined the online database and employed our own sample library to validate the results. By investigating FLAD1 expression level in GC and its correlation to prognosis, we found FLAD1 was a valuable biomarker for the prognosis of GC.
Methods and Materials
Oncomine 4.5
To obtain the gene expression profile of FLAD1 in GC tissue and normal tissue, we searched Oncomine (www.oncomine.org), an open-access online microarray database The data sets covered major types of cancer, including GC, and provided gene expression profiles based on more than 700 studies [14, 15]. All data in our analysis were extracted in January 2020. The differences in FLAD1 expression between GC tissues, and normal gastric tissues were analyzed by Chi‑square test. The threshold value was determined as 2.0-fold change of expression level, p < 0.05, and top 10% gene rank. The details of 10 involved studies are as follows: 1. Diffuse Gastric Adenocarcinoma vs. Normal; p = 0.049, fold change = 1.537,5722 samples. Chen Gastric, Mol Biol Cell, 2003. 2. Gastric Intestinal Type Adenocarcinoma vs. Normal; p = 2.94E-11, fold change = 2.373, 343 samples. Chen Gastric, Mol Biol Cell, 2003. 3. Gastric Mixed Adenocarcinoma vs. Normal; p = 1.70E-4, fold change = 2.175, 971 samples. Chen Gastric, Mol Biol Cell, 2003. 4. Diffuse Gastric Adenocarcinoma vs. Normal; p = 0.001, fold change = 1.309,1088 samples. Cho Gastric, Clin Cancer Res, 2011. 5. Gastric Adenocarcinoma vs. Normal; p = 0.064, fold change = 1.440, 3202 samples. Cho Gastric, Clin Cancer Res, 2011. 6. Gastric Intestinal Type Adenocarcinoma vs. Normal; p = 0.056, fold change = 1.213, 5353 samples Cho Gastric, Clin Cancer Res, 2011. 7. Gastric Mixed Adenocarcinoma vs. Normal; p = 0.063, old change = 1.092, 5821 samples, Cho Gastric, Clin Cancer Res, 2011. 8. Gastric Cancer vs. Normal; p = 0.384, fold change = 1.047, 7615 samples. Cui Gastric, Nucleic Acids Res, 2011. 9. Gastric Intestinal Type Adenocarcinoma vs. Normal; p =3.92E-8, fold change = 1.440, 3202 samples. DErrico Gastric, Eur J Cancer, 2009. 10. Gastric Cancer vs. Normal; p = 0.003, fold change = 1.454, 865 samples.Wang Gastric, Med Oncol, 2010.
Patients and Specimens
106 patients who were diagnosed with GC at the Third Affiliated Hospital of Sun Yat-sen University from Aug 2001 to Nov 2004 were included in the study. Among them 39 were male and 67 were female. The mean patient age at diagnosis was 57 (IQR: 43-68). The post-operative pathologic diagnoses confirmed gastric adenocarcinoma. None of the patients received neo-adjuvant chemotherapy. The clinicopathologic characteristics were evaluated according to the AJCC recommendation [16, 17] (Table 1). Follow-up time was defined from diagnosis to death or the lasted census date. Overall survival (OS) was defined from the date of first diagnosis to the date of death for any reason or to the last follow-up. The follow-up time of the GC cohort ranged from 1 to 118 months (median 21 months). We also collect of 10 pairs of GC tissues and their corresponding adjacent normal tissues in our center to investigate the different RNA and protein expression level of FLAD1 in GC and noncancerous tissues. The fresh tissue samples for Real-time PCR (RT-PCR) analysis were immersed into RNAlater (Sigma-Aldrich R0901, St. Louis., MO, USA) immediately during surgery and stored at 4˚C overnight, and then preserved at‑80˚C. The fresh tissue samples for western boltting analysis were preserved at ‑80 °C. Informed consent was obtained from all patients and the study procedure is approved by the ethical committee of the Third Affiliated Hospital of Sun Yat-sen University; Institutional Review Board (IRB) number, [2019] 02-071-01.
Association between FLAD1 expression level and clinicopathological characteristics
| Patient characteristics | FLAD1 expression | Total (%) | P-value | |
|---|---|---|---|---|
| Low (%) | High (%) | |||
| Sex | 0.629 | |||
| Male | 17 (16.0) | 22 (20.8) | 39 (36.8) | |
| Female | 26 (24.5) | 41 (38.7) | 67 (63.2) | |
| Age | 0.024 | |||
| < 60 years | 30 (28.3) | 30 (28.3) | 60 (56.6) | |
| ≥60 years | 13 (12.3) | 33 (31.1) | 46 (43.4) | |
| Size | 0.083 | |||
| < 5cm | 17 (16.0) | 15 (14.2) | 32 (30.2) | |
| ≥5cm | 26 (24.5) | 48 (44.0) | 74 (69.8) | |
| pT | < 0.001 | |||
| 1 | 9 (8.5) | 1 (0.9) | 10 (9.4) | |
| 2 | 9 (8.5) | 1 (0.9) | 10 (9.4) | |
| 3 | 24 (22.6) | 60 (56.6) | 84 (79.2) | |
| 4 | 1 (0.9) | 1 (0.9) | 2 (1.9) | |
| pN | < 0.001 | |||
| 0 | 16 (15.1) | 5 (4.7) | 21 (19.8) | |
| 1 | 22 (20.8) | 16 (15.1) | 38 (35.8) | |
| ≥2 | 5 (4.7) | 42 (39.6) | 47 (44.3) | |
| Metastatic disease | 1.000 | |||
| Yes | 3 (2.8) | 4 (3.8) | 7 (6.6) | |
| No | 40 (37.7) | 59 (55.7) | 99 (93.4) | |
| TNM stage | 0.026 | |||
| I | 7 (6.6) | 6 (5.7) | 13 (12.3) | |
| II | 11 (10.4) | 7 (6.6) | 18 (17.0) | |
| III | 25 (23.6) | 43 (40.6) | 68 (64.2) | |
| IV | 0 (0) | 7 (6.6) | 7 (6.6) | |
| Pathologic grade | 0.321 | |||
| Low | 14 (13.2) | 15 (14.2) | 29 (27.4) | |
| High | 29 (27.4) | 48 (45.3) | 77 (72.6) | |
| Infiltration | 0.646 | |||
| Yes | 3 (2.8) | 2 (1.9) | 5 (4.7) | |
| No | 40 (37.7) | 61 (57.5) | 101 (95.3) | |
| Ki67 expression | ||||
| Negative | 16 (40.0) | 24 (60.0) | 40 (37.8) | 0.926 |
| Positive | 27 (40.9) | 39 (59.1) | 66 (62.2) | |
Real-time PCR (RT-PCR) analysis
Total RNA samples were extracted from 10 pairs of GC tissues and their corresponding adjacent normal tissues using TRIzol reagent (Invitrogen, CA, USA). The extracted RNA was pretreated with RNase-free DNase, and 2 μg was used for cDNA synthesis. An initial amplification using FLAD1 specific primers was performed for the PCR amplification of FLAD1 cDNA, with denaturation at 95 °C for 10 min followed by 28 cycles of denaturation at 95 °C for 60 s, primer annealing at 58 °C for 30 s, and primer extension at 72 °C for 30 s. A final extension at 72 °C for 5 min was performed to complete of the cycles. The reaction mixture was then stored at 4 °C. Real-time PCR was performed to investigate the fold increase of FLAD1 mRNA in each pairs of GC and normal gastric tissue using the following primer sequences designed by Primer Express v 2.0 software (Applied Biosystems): FLAD1 fragments, 5'-TGACCCCTACTCCTGTAGCC-3' (forward) and 5'-AGCTGACGCAGAAAATCCCA-3' (reverse); and GAPDH, 5'-TGTTGCCATCAATGACCCC-3' (forward), 5'-CTCCACGACGTACTCAGC-3' (reverse). Glyceraldehyde-3-phosphate dehydrogenase (GAPDH) was used as an internal control; the relative expression level of FLAD 1 was calculated using the 2-ΔΔCT method. All experiments were performed in triplicate.
Western blotting analysis
Cells at 70‑80% confluency were lysed by radioimmunoprecipitation assay buffer (RIPA; Cell Signaling Technology, Inc., Danvers, MA) with complete protease inhibitor cocktail (Roche Applied Science, Mannheim, Germany) on ice. Fresh 6 pairs of GC tissues and their corresponding adjacent normal tissues were ground into powder in liquid nitrogen and then lysed using SDS-PAGE sample buffer. A total of 20 μg samples were separated on 10.5% SDS polyacrylamide gels and transferred onto PVDF membranes (Immobilon P, Millipore, MA). PVDF membranes were blocked with 5% fat-free milk in Tris-buffered saline with 0.1% Tween-20 at room temperature for 1 h, and then incubated with anti-FLAD1 antibody (Abnova Corp., TW, catalog Number PAB22183) at 1:500 dilution overnight at 4°C, and then incubated with horseradish peroxidase-conjugated goat anti-rabbit IgG (Santa Cruz Biotechnology, SC-2004). FLAD1 expression was detected by ECL Western blotting detection reagent (Amersham) according to the manufacturer. GAPDH was used as loading control.
IHC Staining and Analysis
The paraffin-embedded samples of 106 human gastric cancer in the Third Affiliated Hospital of Sun Yat-sen University from Aug 2001 to Nov 2004 were obtained to perform IHC staining. Each 4-μm-thick paraffin slide was treated with xylene and rehydrated with alcohol solution in a descending concentration. The slides were then treated with EDTA antigenic retrieval buffer in microwave at 650W for 3min and then 350W twice more. Hydrogen peroxide (3%, in methanol) was used to suppress the endogenous peroxidase activity. The samples were incubated with 1% BSA at room temperature for 60min to block unspecific binding, and then incubated with rabbit polyclonal antibody raised against recombinant FLAD1 (Abnova Corp., TW, catalog Number PAB22183), 4 °C at 1:500 dilution level overnight. Common goat serum (Santa Cruz Biotechnology, Inc.) was used as the negative control. After 3 washes in PBS, the samples were incubated with anti-rabbit secondary antibody at room temperature for 30min, and further incubated with a streptavidin-horseradish peroxide complex (1:1500, Abcam, Cambridge, UK) at room temperature for 30min. After incubation, the slides were stained with 3-amino-9-ethyl carbazole for 3 min at room temperature and then counterstained with 10% Mayer's hematoxylin for 30s. The samples were then dehydrated for analysis.
The immunostaining was analyzed by two pathologists blinded to patient-related information. The extent of IHC staining was categorized into 0-no staining (no visible difference from control group), 1-weak staining (light yellow), 2-moderate staining (yellow), and 3-strong staining (dark yellow). The proportion of tumor cell in the sample was scored as 0-no positive cells, 1-positive cells consists 1~25% of the sample, 2-positive cells consists 26~50% of the sample, 3-positive cells consists 51~75% of the sample, and 4-positive cells consists 76~100% of the sample. The FLAD1 expression was evaluated by the extent score multiplied by the proportion score. Total score 0: negative (-); total score 1~4: weakly positive (+); total score 6~8: positive (++); and total score 9~12: strongly positive (+++).The chosen of cut-off values for FLAD1 was based on the heterogeneity using log-rank test concerning overall survival. A staining score of ≥8 was used to define high FLAD1 expression, and <8 was used to define low FLAD1 expression.
Statistical Analyses
The IHC results and corresponding clinicopathologic features were analyzed by SPSS Statistics 23 (IBM Corp., NY, US). The correlation between FLAD1 expression and other clinical features (age, sex, grade, TNM stage, and tumor infiltration) was analyzed using the χ2 test and Fisher's exact test while n < 40. Cox-regression model was used to analyze the impact of FLAD1 and covariates on survival. The impact of FLAD1 expression overall survival was validated by our 106 cases using the Kaplan-Meier plot, and evaluated by log-rank p-value and HR.
The Kaplan-Meier plotter
The Kaplan-Meier plotter (www.kmplot.com) is an online graphic tool to explore the relationship between gene expression and prognosis. Using the website's built-in data, the Kaplan-Meier plotter can provide survival curves according to different gene expression level. It also provides utilities including subgroup analyses based on several clinicopathological parameters (clinical staging, receptor status, etc.) [18]. The cutoff value of gene expression level was determined automatically by selecting the “auto select best cutoff” option. The survival curves of the two cohorts were plotted to evaluate OS, FP, and PPS. To quantify the impact of FLAD1 overexpression on prognosis, the number at risk, log-rank p-value and hazard ratio (HR) with 95% confidence interval (CI) were calculated and annotated for each plot.
Results
FLAD1 was overexpressed in GC
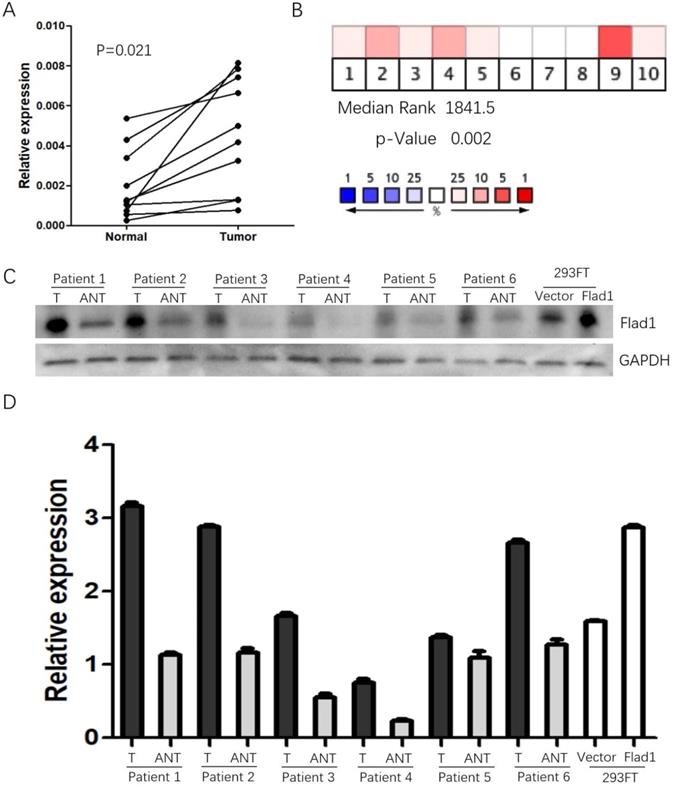
The data obtained from Oncomine suggested elevated FLAD1 expression in GC compared to normal tissues (median rank: 1841.5.0, p =0.002, Figure 1A). As a validation of the results, the qPCR analysis of 10 paired (normal vs GC) samples from our patient cohort was performed (Figure 1B). Consistent with these data, FLAD1 protein was also found to be upregulated in 6 fresh GC tissues compared with non-cancerous tissues (Figure 1C, D).
FLAD1 overexpression was associated with poor outcome of GC
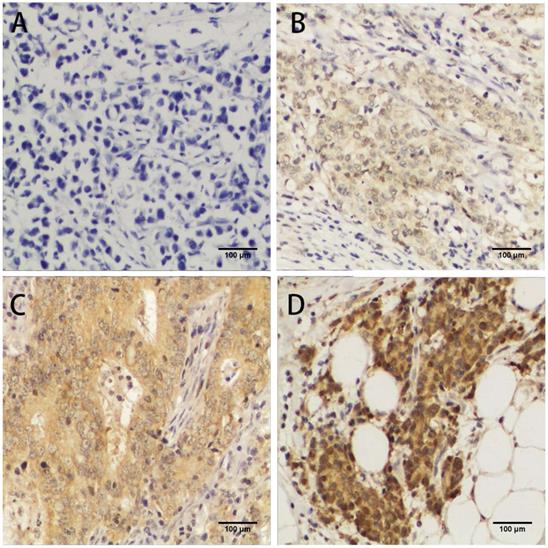
To investigate the correlation between FLAD1 overexpression at protein level and prognosis of GC, we performed IHC analysis based on the 106 patient samples. The extent of staining was scored into five levels compared to non-cancerous tissue (Figure 2). The staining for FLAD1 was positive for 63/106 (59.4%) patients. The sex, tumor size, tumor grade, metastasis, infiltration status and Ki-67 expression level were not of significant difference between the FLAD1-positive and the FLAD1-negative group (Table 1), though the majority of patients had pT3 disease (79.2%). FLAD1 overexpression was correlated to patient age ≥ 60 years (Pearson χ2 = 5.014, p = 0.024), higher pathologic T stage (Pearson χ2 = 25.358, p < 0.001), higher pathologic N stage (Pearson χ2 = 33.247, p < 0.001), and higher TNM stage (Pearson χ2 = 9.288, p = 0.026). The correlation was further supported by non-parametric testing (Table 2).
Pearson χ2 and Spearman rank-sum correlation coefficient of clinicopathologic characteristics associated with FLAD1 overexpression
| Clinicopathologic features | Pearson χ2 | Correlation coefficient |
|---|---|---|
| Age | 5.104 | 0.219 |
| pT | 25.358 | 0.449 |
| pN | 33.247 | 0.555 |
| TNM stage | 9.288 | 0.270 |
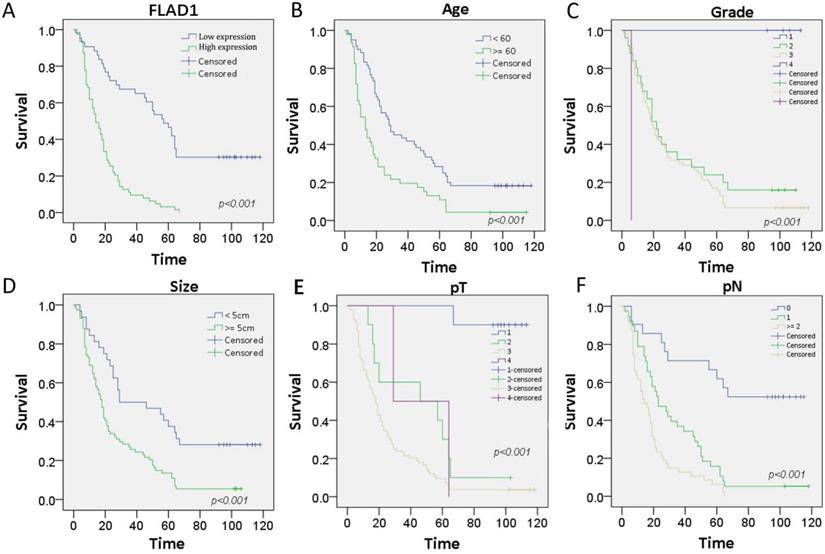
The prognostic role of FLAD1 was further validated by the patient cohorts in our center using Cox regression model. The median follow-up time for this cohort is 21 months. On univariate analysis, we confirmed FLAD1-positive individuals had higher risk of death (HR = 4.388 p < 0.001). Meanwhile, patient age ≥ 60 years, tumor size ≥ 5cm, and higher TNM stage were also death-related risk factors. On multivariate analysis, our results supported FLAD1 (HR = 2.937, p < 0.001) was an independent risk factor for OS (Table 3). The Kaplan-Meier plots revealed FLAD1-positive GC patients had worse OS compared to FLAD1-negative ones (Log-rank χ2 = 41.978, p < 0.001, Figure 3A). Elder age, higher grade, greater tumor size, higher pT and pN stage also indicated poor outcomes (Figure 3B-F).
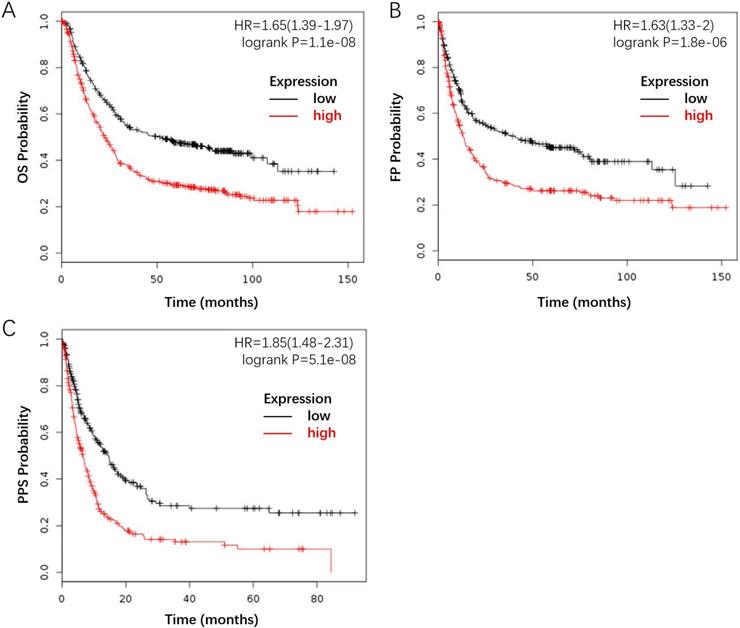
Furthermore, we investigated the correlation between FLAD1 overexpression at mRNA level and prognosis of GC by plotting and comparing the OS, FP, and PPS of GC patients to healthy individuals through Kaplan-Meier plotter (www.kmplot.com). FLAD1 overexpression was associated with worse OS (HR = 1.65, 95% CI: 1.39-1.97, p < 0.001), FP (HR = 1.63, 95% CI: 1.33-2.00, p < 0.001), and PPS (HR = 1.85, 95% CI: 1.48-2.31, p < 0.001, Figure 4A-C). Taken together, these findings supported that FLAD1 was a satisfactory prognostic factor for GC patients.
Expression of FLAD1 at mRNA level in GC and non-cancerous tissue. A. Heat map revealed over-expression of FLAD1 in GC across 10 studies. B. Expression levels of FLAD1 in 10 paired (normal vs tumor) GC samples were detected by real-time PCR. C. FLAD1 protein expression levels in six paired gastric carcinoma tissues and over expression of FLAD1 in 293FT cells by Western blotting. D. Quantitative analysis of FLAD1 protein in C. T, Gastric carcinoma tissues, ANT, matched adjacent non-tumor gastric tissues.
Immunochemistry analyses of FLAD1 expression in GC tissue samples. FLAD1 expression was mainly localized in cytoplasm. Representative images of A. Negative staining of FLAD1, B. weakly positive staining (+) of FLAD1, C. positive staining (++) of FLAD1, and D. strongly positive staining (+++) of FLAD1. The magnification was 400×.
Cox-regression analysis of various prognostic parameters in GC patients
| Risk factor | Univariate | Multivariate | ||
|---|---|---|---|---|
| HR (95% CI) | P-value | HR (95% CI) | P-value | |
| Age ≥60 | 2.077 (1.375-3.139) | 0.001 | 1.533 (0.978-2.404) | 0.063 |
| Size ≥5 cm | 2.277 (1.410-3.678) | 0.001 | 1.176 (0.690-2.001) | 0.551 |
| pT | 0.001 | 0.005 | ||
| 1 | Referent | Referent | ||
| 2 | 17.539 (2.207‑139.398) | 0.007 | 16.415 (1.870‑144.062) | 0.012 |
| 3 | 36.233 (4.970‑264.173) | < 0.001 | 23.874 (2.760-206.479) | 0.004 |
| 4 | 16.855 (1.516‑188.064) | 0.022 | 13.411 (0.973‑184.922) | 0.052 |
| pN | < 0.001 | 0.577 | ||
| 0 | Referent | Referent | ||
| 1 | 4.022 (1.955‑8.274) | < 0.001 | 1.212 (0.525‑2.798) | 0.652 |
| ≥2 | 7.015 (3.421‑14.386) | < 0.001 | 1.511 (0.641-3.514) | 0.346 |
| TNM stage | 0.013 | 0.051 | ||
| I | Referent | |||
| II | 2.015 (0.866-4.688) | 0.104 | 2.891 (1.121-7.457) | 0.028 |
| III | 2.257 (1.076-4.736) | 0.031 | 3.910 (1.397-7.287) | 0.006 |
| IV | 2.665 (0.963-7.380) | 0.059 | 3.194 (1.007-9.470) | 0.036 |
| FLAD1 expression | 4.388 (2.715-7.092) | < 0.001 | 2.937 (1.522-5.760) | 0.001 |
Discussion
GC is the second leading cause of cancer-related death worldwide [19]. Current treatment for GC includes surgery, chemotherapy, adjuvant/neoadjuvant chemotherapy, and radiotherapy [20-23]. The alternatives of treatment necessitate accurate prediction of tumor-related risks. The TNM-stage-based classification proposed by the AJCC was the most adopted evaluation for GC [24]; meanwhile, histopathologic grading has been reported to be an independent prognostic factor, with several related grading system under construction [25]. Clinicopathologic characteristics such as cellular dysmorphism, tumor location, comorbidity, and complications are also involved in some monograms [26]. However, these prognostic factors have some limitations. First, despite satisfactory accuracy and prognostic power, the traditional TNM staging system is largely based on surgery, which is not regular in non-operable patients. Secondly, the TNM staging system cannot well adapt to the fact that the anatomic location of the primary tumor is influential to prognosis [24]. Thirdly, the prognostic power of clinical features is often refined to specific stages; besides, there is no unanimous agreement on the threshold value and criteria for these factors. As a response to these challenges, tumor biomarkers are increasingly evaluated for the prognosis of GC [27-29].
FLAD1 is an essential gene in flavin metabolism and oxidative-reduction chain. It is expressed across more than 200 species. In the human genome, FLAD1 is located on chromosome 1 (1q21.3) and most highly expressed in lymph nodes, although ubiquitously expressed in all tissue types [7]. FLAD1's protein product, FADS, possesses basic yet indispensable oxidative-reduction bioactivity. For this reason, it is widely distributed in the cytoplasm and membranous organelles such as mitochondrion [30]. More importantly, due to the significant role in electron transportation, it is closely correlated to cell metabolism. FLAD1 has already been identified to correlate with susceptibility and outcome of hepatocellular carcinoma, prostate cancer and lung cancer [11, 13, 31]. To our knowledge, there is no current study investigating the prognostic role of FLAD1 in GC. Our study first confirmed a correlation between FLAD1 overexpression and poor outcome of GC.
The present study verified that FLAD1 mRNA was overexpressed in GC tissues compared with normal matched tissues staining which provided novel evidence that the up-regulation of FLAD1 was closely associated with poor survival rates in GC patients. Multivariate analysis revealed FLAD1 was an independent prognostic factor for OS in patients with GC. To investigate the clinical significance of FLAD1 expression at mRNA level, we analyzed its relationship with prognosis of GC by plotting and comparing the OS, FP, and PPS of GC patients to healthy individuals by using The Kaplan-Meier plotter (www.kmplot.com) and found that FLAD1 mRNA was associated to unfavorable OS, FP, and PPS of GC (p<0.001). Taken together, these findings clearly demonstrated that FLAD1 was a satisfactory prognostic factor for GC patients. It may serve as a biomarker for GC, which may aid early diagnosis and precise therapy. However, at present, the precise functions of FLAD1in malignant cancer remain obscure. Probing the precise mechanism underlying FLAD1 in GC requires further investigation.
Kaplan-Meier survival curves with univariate analysis (log-rank test) of the 106-patient cohorts confirmed FLAD1-positive individuals had worse OS. Patient age, tumor grade, tumor size, and TNM stage also had impact on the OS of GC. A.OS rate for GC patients with high FLAD1 expression compares to those with low FLAD1 expression. B. OS rate for GC patients with age over 60 years compares to those with age under 60 years. C.OS rate for GC patients with grade 1, grade 2, grade 3, and grade 4. D. OS rate for GC patients with tumor size over 5cm and under 5cm. E. OS rate for GC patients with pT1(Tumor invades lamina propria, muscularis mucosae, or submucosa), pT2 (Tumor invades muscularis propria), pT3(Tumor penetrates subserosal connective tissue without invasion of visceral peritoneum or adjacent structures), and pT4(Tumor invades serosa (visceral peritoneum) or adjacent structures). F. OS rate for GC patients with pN0 (No regional lymph node metastasis), pN1(Metastasis in 1-2 regional lymph nodes), and pN2 (Metastasis in 3-6 regional lymph nodes).
FLAD1-overexpressing individuals had a worse outcome compared to the baseline. The survival plot was generated by the Kaplan-Meier Plotter using the website's built-in dataset. A. OS rate for GC patients with high FLAD1 expression (480 patients) compares to those with low FLAD1 expression (396 patients). B. FP rate for GC patients with high FLAD1 expression (327 patients) compares to those with low FLAD1 expression (314 patients). C.PPS rate for GC patients with high FLAD1 expression (248 patients) compares to those with low FLAD1 expression (251 patients).
We additionally investigated the association between FLAD1 expression and other clinical features of patients with GC. FLAD1 overexpression was correlated to patient age ≥60 years, higher pathologic T stage, higher pathologic N stage, and higher TNM stage, which revealed that FLAD1 may serve as a molecular biomarker for a subpopulation of patients with more aggressive disease. However, the sex, tumor size, tumor grade, metastasis, and infiltration status were not of significant difference between the FLAD1-positive and the FLAD1-negative group.
To testify whether FLAD1 is a satisfactory prognostic factor for GC, we used the Kaplan-Meier plotter to analyze the correlation between survival and FLAD1 expression level. The OS, FP, and PPS were all reduced in FLAD1-overexpressing groups, after a follow-up time of 150 months (OS/FP) and 80 months (PPS), respectively.
The survival analyses for different clinical stages, lymph node metastasis status, and HER2 status were also conducted. Recently researchers have started to investigate the association between FLAD1 and cancer. However, the exact role of FLAD 1 in human cancers does not appear to be clearly identified. Another study of our team in breast cancer has found the expression of FLAD 1 is closely associated with genes regulating DNA replication, microtubule, mitosis, cytoskeleton, cell cycle, cell division, p53 signaling pathway, nucleotide excision repair, and mismatch repair (data not showed). Based on these relationships, it could be hypothesized that FLAD1 may participated in cancer cell proliferation, migration and DNA damage repair. One limitation of this study was short of research on molecular mechanism. It remains ambiguous how the FLAD1 exerts unique impact on cancer development. Nevertheless, FLAD1 expression still offers a path forward towards a future GC risk assessment system.
In conclusion, to the best of our knowledge, we first reported FLAD1 was overexpressed and associated with poor prognosis of GC. Current understanding of the mechanism underlying our findings is lacking; the paucity of research on the pathway involved in FLAD1 overexpression also limits further development of its therapeutic application. Future researches are needed to investigate the causality and molecular mechanism of FLAD1 overexpression. Nevertheless, our study still highlights FLAD1 as a potential prognostic factor for GC in the clinical context. Its expression status may serve for the early diagnosis and stratification of patients and create a new therapeutic selection for GC patients.
Acknowledgements
This study was supported by grants from the National Natural Science Foundation of China (81902416) and the Natural Science Foundation of Guangdong Province, China (2017A030313858, 2018A0303130324).
Compliance with ethical standards
Ethical approval
The study was approved by Committee on the Ethics of the Third Affiliated Hospital of Sun Yat-sen University. All procedures performed in studies involving human participants were in accordance with the ethical standards of the institutional or national research committee and with the 1964 Helsinki declaration and its later amendments or comparable ethical standards.
Informed consent
Informed consent was obtained from all individual participants included in the study. And each patient had given a written informed consent about the use of the samples for medical research.
Competing Interests
The authors have declared that no competing interest exists.
References
1. Fitzmaurice C, Abate D, Abbasi N, Abbastabar H, Abd-Allah F, Abdel-Rahman O. et al. Global, Regional, and National Cancer Incidence, Mortality, Years of Life Lost, Years Lived With Disability, and Disability-Adjusted Life-Years for 29 Cancer Groups, 1990 to 2017: A Systematic Analysis for the Global Burden of Disease Study. JAMA Oncol. 2019
2. Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68:394-424
3. Biagioni A, Skalamera I, Peri S, Schiavone N, Cianchi F, Giommoni E. et al. Update on gastric cancer treatments and gene therapies. Cancer Metastasis Rev. 2019;38:537-48
4. Zhang JX, Chen ZH, Chen DL, Tian XP, Wang CY, Zhou ZW. et al. LINC01410-miR-532-NCF2-NF-kB feedback loop promotes gastric cancer angiogenesis and metastasis. Oncogene. 2018;37:2660-75
5. Yang Q, Zhang RW, Sui PC, He HT, Ding L. Dysregulation of non-coding RNAs in gastric cancer. World journal of gastroenterology. 2015;21:10956-81
6. Zhang Z, Li Z, Li Y, Zang A. MicroRNA and signaling pathways in gastric cancer. Cancer gene therapy. 2014;21:305-16
7. Fagerberg L, Hallstrom BM, Oksvold P, Kampf C, Djureinovic D, Odeberg J. et al. Analysis of the human tissue-specific expression by genome-wide integration of transcriptomics and antibody-based proteomics. Molecular & cellular proteomics: MCP. 2014;13:397-406
8. Barile M, Giancaspero TA, Leone P, Galluccio M, Indiveri C. Riboflavin transport and metabolism in humans. J Inherit Metab Dis. 2016;39:545-57
9. Giancaspero TA, Galluccio M, Miccolis A, Leone P, Eberini I, Iametti S. et al. Human FAD synthase is a bi-functional enzyme with a FAD hydrolase activity in the molybdopterin binding domain. Biochem Biophys Res Commun. 2015;465:443-9
10. Auranen M, Paetau A, Piirila P, Pohju A, Salmi T, Lamminen A. et al. Patient with multiple acyl-CoA dehydrogenation deficiency disease and FLAD1 mutations benefits from riboflavin therapy. Neuromuscular disorders: NMD. 2017;27:581-4
11. Eeles RA, Olama AA, Benlloch S, Saunders EJ, Leongamornlert DA, Tymrakiewicz M. et al. Identification of 23 new prostate cancer susceptibility loci using the iCOGS custom genotyping array. Nature genetics. 2013;45:385-91 91e1-2
12. Cheng K, Lee JS, Hao P, Yao SQ, Ding K, Li Z. Tetrazole-Based Probes for Integrated Phenotypic Screening, Affinity-Based Proteome Profiling, and Sensitive Detection of a Cancer Biomarker. Angewandte Chemie (International ed in English). 2017;56:15044-8
13. Mitra R, Lee J, Jo J, Milani M, McClintick JN, Edenberg HJ. et al. Prediction of postoperative recurrence-free survival in non-small cell lung cancer by using an internationally validated gene expression model. Clin Cancer Res. 2011;17:2934-46
14. Rhodes DR, Yu J, Shanker K, Deshpande N, Varambally R, Ghosh D. et al. ONCOMINE: a cancer microarray database and integrated data-mining platform. Neoplasia (New York, NY). 2004;6:1-6
15. Rhodes DR, Kalyana-Sundaram S, Mahavisno V, Varambally R, Yu J, Briggs BB. et al. Oncomine 3.0: genes, pathways, and networks in a collection of 18,000 cancer gene expression profiles. Neoplasia. 2007;9:166-80
16. Union for International Cancer Control. TNM History, Evolution, and Milestones. 2017. http://wwwuiccorg/sites/main/files/private/History.Evolution_Milestones
17. American Joint Committee on Cancer. Desk References.2016. https://cancerstagingorg/references-tools/deskreferences/pages/default
18. Nagy A, Lanczky A, Menyhart O, Gyorffy B. Validation of miRNA prognostic power in hepatocellular carcinoma using expression data of independent datasets. Sci Rep. 2018;8:9227
19. Torre LA, Bray F, Siegel RL, Ferlay J, Lortet-Tieulent J, Jemal A. Global cancer statistics, 2012. CA: a cancer journal for clinicians. 2015;65:87-108
20. Macdonald JS, Smalley SR, Benedetti J, Hundahl SA, Estes NC, Stemmermann GN. et al. Chemoradiotherapy after surgery compared with surgery alone for adenocarcinoma of the stomach or gastroesophageal junction. The New England journal of medicine. 2001;345:725-30
21. Sasako M, Sakuramoto S, Katai H, Kinoshita T, Furukawa H, Yamaguchi T. et al. Five-year outcomes of a randomized phase III trial comparing adjuvant chemotherapy with S-1 versus surgery alone in stage II or III gastric cancer. Journal of clinical oncology: official journal of the American Society of Clinical Oncology. 2011;29:4387-93
22. Shinohara S, Korenaga D, Edagawa A, Koushi K, Itoh S, Kawanaka H. et al. Significant prognostic factors in patients with Stage IV gastric cancer with special reference to the curability of surgery. Surgery today. 2013;43:40-7
23. Toyokawa T, Ohira M, Sakurai K, Kubo N, Tanaka H, Muguruma K. et al. The Role of Adjuvant Chemotherapy for Patients with Stage IB Gastric Cancer. Anticancer research. 2015;35:4091-7
24. Washington K. 7th edition of the AJCC cancer staging manual: stomach. Annals of surgical oncology. 2010;17:3077-9
25. Zhu Z, Sun X, Wang J, Sun Z, Wang Z, Zheng X. et al. Histopathology-based prognostic score is independent prognostic factor of gastric carcinoma. BMC cancer. 2014;14:663
26. Aoyama T, Yoshikawa T, Fujikawa H, Hayashi T, Ogata T, Cho H. et al. Prognostic factors in stage IB gastric cancer. World journal of gastroenterology. 2014;20:6580-5
27. Liu HS, Xiao HS. MicroRNAs as potential biomarkers for gastric cancer. World journal of gastroenterology. 2014;20:12007-17
28. Wang DS, Ren C, Qiu MZ, Luo HY, Wang ZQ, Zhang DS. et al. Comparison of the prognostic value of various preoperative inflammation-based factors in patients with stage III gastric cancer. Tumour biology: the journal of the International Society for Oncodevelopmental Biology and Medicine. 2012;33:749-56
29. Hamakawa T, Kukita Y, Kurokawa Y, Miyazaki Y, Takahashi T, Yamasaki M. et al. Monitoring gastric cancer progression with circulating tumour DNA. British journal of cancer. 2015;112:352-6
30. Galluccio M, Brizio C, Torchetti EM, Ferranti P, Gianazza E, Indiveri C. et al. Over-expression in Escherichia coli, purification and characterization of isoform 2 of human FAD synthetase. Protein expression and purification. 2007;52:175-81
31. Ye C, Zhang X, Chen X, Cao Q, Zhou Y, Li W. et al. Multiple novel hepatocellular carcinoma signature genes are commonly controlled by the master pluripotency factor OCT4. Cell Oncol (Dordr). 2019
Author contact
![]() Corresponding authors: Dr. Nana Zhang, Department of Pathology, the Third Affiliated Hospital of Sun Yat-sen University. 600 Tianhe Road, Guangzhou, Guangdong Province, 510230, P.R. China. Tel: +86-020-85253359; E-mail: zhangnn7sysu.edu.cn and Dr. Jiani Wang, Breast Cancer Center, the Third Affiliated Hospital of Sun Yat-sen University. 600 Tianhe Road, Guangzhou, Guangdong Province, 510230, P.R. China. Tel: +86-020-85252154; E-mail: wangjni2sysu.edu.cn.
Corresponding authors: Dr. Nana Zhang, Department of Pathology, the Third Affiliated Hospital of Sun Yat-sen University. 600 Tianhe Road, Guangzhou, Guangdong Province, 510230, P.R. China. Tel: +86-020-85253359; E-mail: zhangnn7sysu.edu.cn and Dr. Jiani Wang, Breast Cancer Center, the Third Affiliated Hospital of Sun Yat-sen University. 600 Tianhe Road, Guangzhou, Guangdong Province, 510230, P.R. China. Tel: +86-020-85252154; E-mail: wangjni2sysu.edu.cn.

 Global reach, higher impact
Global reach, higher impact