3.2
Impact Factor
ISSN: 1449-1907
Int J Med Sci 2020; 17(11):1639-1651. doi:10.7150/ijms.46125 This issue Cite
Research Paper
PODXL2 maintains cellular stemness and promotes breast cancer development through the Rac1/Akt pathway
1. Department of Biochemistry and Molecular Biology, National Cheng Kung University, Tainan 70101, Taiwan
2. Institute of Basic Medical Sciences, College of Medicine, National Cheng Kung University, Tainan 70101, Taiwan
3. PhD Program for Cancer Molecular Biology and Drug Discovery, College of Medical Science and Technology, Taipei Medical University, Taipei 11031, Taiwan
4. Graduate Institute of Cancer Biology and Drug Discovery, College of Medical Science and Technology, Taipei Medical University, Taipei 11031, Taiwan
5. NTT Institute of Hi-Technology, Nguyen Tat Thanh University, Ho Chi Minh City, Vietnam
6. School of Chinese Medicine for Post-Baccalaureate, I-Shou University, Kaohsiung 82445, Taiwan
7. Department of Radiology and Biomedical Imaging, University of California, San Francisco, CA 94143, USA
8. Department of Biotechnology, Chia Nan University of Pharmacy and Science, Tainan 71710, Taiwan
9. Department of Senior Citizen Service Management, Chia Nan University of Pharmacy and Science, Tainan 71710, Taiwan
10. Department of Emergency Medicine, Kaohsiung Medical University Hospital, Kaohsiung Medical University, Kaohsiung 80708, Taiwan
11. Department of Surgery, National Cheng Kung University Hospital, College of Medicine, National Cheng Kung University, Tainan 70101, Taiwan
12. Department of Biostatistics, Vanderbilt University Medical Center, Nashville, TN 37232, Tennessee, USA
*Equal contributors.
Received 2020-3-18; Accepted 2020-5-21; Published 2020-6-29
Abstract
The cluster of differentiation 34 (CD34) family, which includes CD34, podocalyxin-like protein 1 (PODXL), and PODXL2, are type-I transmembrane sialomucins and markers of hematopoietic stem cells (HSCs) and vascular-associated tissues. CD34 family proteins are expressed by endothelial cells and hematopoietic precursors. PODXL is well known to be associated with invadopodia formation and to promote the epithelial-mesenchymal transition, tumor migration and invasion. PODXL expression was correlated with poor survival of cancer patients. However, the role of PODXL2 in cancer has been less fully explored. To reveal the novel role of PODXL2 in breast cancer, the present study evaluated PODXL2 levels in relation to clinical outcomes of cancer patients by performing a bioinformatics analysis using the Oncomine database, Kaplan-Meier plots, and the CCLE database. Empirical validation of bioinformatics predictions was conducted utilizing the short hairpin (sh)-RNA silencing method for PODXL2 in the BT474 invasive ductal breast carcinoma cell line. The bioinformatics analysis revealed that PODXL2 overexpression was correlated with poor survival of breast cancer patients, suggesting an oncogenic role of PODXL2 in breast carcinoma. In a validation experiment, knockdown of PODXL2 in BT474 cells slightly influenced cell proliferation, suppressed migration, and inhibited expressions of downstream molecules, including Ras-related C3 botulinum toxin substrate 1 (Rac1), phosphorylated (p)-Akt (S473), and p-paxillin (Y31) proteins. In addition, knockdown of PODXL2 reduced expression levels of cancer stem cell (CSC) markers, including Oct-4 and Nanog, and the breast CSC marker aldehyde dehydrogenase 1 (ALDH1). Collectively, our present study demonstrated that PODXL2 plays a crucial role in cancer development and could serve as a potential prognostic biomarker in breast cancer patients.
Keywords: PODXL2, Breast cancer, Bioinformatics, Cancer stem cells.
Introduction
Cancer stem cells (SCs; CSCs) promote tumor progression, and cancers cause serious mortality worldwide [1]. The main cause of death is usually a delayed diagnosis and distant metastasis to other organs [2]. Another cause of death is the development of resistance after standard therapy [3]. CSCs, also known as tumor-initiating cells (TICs), are located inside the tumor. Theoretically, CSCs establish a reservoir of self-sustaining cells. CSCs have the ability to self-renew and produce heterogeneous cancer lineages [4]. CSCs were reported in many malignancies, including leukemia, brain, breast, lung, colorectal cancers, etc. Moreover, CSCs are also considered to cause distant metastasis, cancer recurrence, and chemoresistance through different signaling pathways [5-7]. Different CSCs exhibit different surface antigens. Cluster of differentiation 34 (CD34) is one of the surface antigens which distinguish leukemic SCs [8]. CD34 and its family of proteins are considered hematopoietic SC (HSC) markers involved in cancer progression in previous studies [9, 10]. CD34 family proteins contain CD34, podocalyxin-like protein 1 (PODXL, podocalyxin, or PCLP1), and podocalyxin-like protein 2 (PODXL2, endoglycan, or PCLP2) [11, 12]. These molecules are classified in the same family because their structures of protein domains and genomic organization of introns/exons are similar [12]. PODXL is overexpressed in several malignancies, such as breast [13], ovarian [14], lung [15], prostate [16], liver [17], colon [18], and pancreatic cancers [19]. High expression of PODXL is strongly associated with a poor prognosis of cancer patients. PODXL also promotes cancer cell migration and invasion, the epithelial-mesenchymal transition (EMT), and immune suppression [20, 21]. Moreover, PODXL is one of the CSC markers in leukemia, and breast, pancreatic, and small-cell lung cancers [22]. PODXL influences the CSC signature and chemoresistance by regulating the transcriptional co-activator with the PDZ domain-binding motif (TAZ) and the hippo-YAP signaling cascade [23].
PODXL2 is the third and newest family member of the CD34 family. PODXL2 was identified through homological presentation in cytoplasmic tails of CD34 and PODXL using a GenBank expressed sequence tag [24]. PODXL2 functions as an L-selectin (SELL) ligand like the other two members [25, 26]. PODXL2 is expressed by T cells, podocytes, and neurons [11]. Although CD34 family proteins have similar domain structures, PODXL2 has an extra non-glycosylated and glutamic acid-rich region in the extracellular domain. Moreover, the cysteine-rich domain of PODXL2 contains more unpaired cysteines, which are related to homodimerization [12]. Intracellular binding partners of PODXL2 may be casein kinase II and protein kinase C because the cytoplasmic domain of PODXL2 consists of several potential phosphorylation sites. However, the correlation between PODXL2 and cancer is not yet well studied. PODXL2 was implied to have a crucial role in breast cancer because it is a type-I transmembrane sialomucin. Mucins stimulate the EMT by disrupting cell-cell junctions and promoting cell growth [29], and aberrant overexpression of mucins was detected in breast [27], gastric [28], and colon cancers [29]. PODXL2 shares the PDZ domain-binding motif (aspartate-threonine-histidine-leucine, DTHL) in the cytosolic tail with PODXL. The PDZ domain interacts with the intracellular binding proteins, ezrin and Na+/H+ exchanger regulatory factor 1/2 (NHERF1/2), to induce downstream phosphoinositide 3-kinase (PI3K) and G protein-coupled receptor signaling [30, 31]. In the present study, we explored the function of PODXL2 in cancer and analyzed the influence of expression of the PODXL2 protein on clinical outcomes of cancer patients. We hypothesized that PODXL2 plays a similar role as PODXL in breast cancer, and increased PODXL2 expression in breast cancer promotes tumor progression.
Materials and Methods
Bioinformatics Analysis
The Oncomine online database provides 715 datasets and 86,733 samples for DNA and messenger (m)RNA expression levels from analysis of high-throughput databases of cancer-related genes in 20 common tumors [32]. We set up search criteria with a p value <10-4, a fold change of PODXL2 gene >1.5, and a gene rank in the top 10%. Coexpression profiles of PODXL2 in Oncomine were used to check a set of genes with similar expression patterns, and these were illustrated in a heat map format.
The Human Protein Atlas is a well-known database with millions of publicly available high-resolution immunohistochemical (IHC) images [33]. We collected IHC images from normal breast and breast cancer tissues for comparison and tested our hypothesis.
The Kaplan-Meier plotter is an online database that provides a method to evaluate impacts of 54,675 genes on patient outcomes. In total, 13,316 cancer samples were collected, including 6234 breast, 3452 lung, 2190 ovarian and 1440 gastric cancer patients. We analyzed PODXL2 (Affymetrix ID: 219152_at) with the JetSet best probe set [34] and automatically selected the best cutoff. Overall survival (OS), recurrence-free survival (RFS), and distant metastasis-free survival (DMFS) of breast cancer patients were analyzed.
The cBioPortal database allows users to download, analyze, and visualize publicly available cancer genomics datasets [35]. We collected data on coexpressed genes of PODLX2 mRNA expression in three breast cancer datasets: Molecular Taxonomy of Breast Cancer International Consortium (METABRIC) [36], The Cancer Genome Atlas (TCGA) Nature [37], and Cell [38]. We individually selected the top 10% of coexpressed genes and obtained processing networks after analysis with the METACORE database. We also downloaded Cancer Cell Line Encyclopedia (CCLE) data [39] for in vitro experiments. The CCLE project has assembled DNA copy numbers, mRNA expressions, and mutation information from 1000 human cancer cell lines. We obtained raw data of PODXL2 mRNA levels. After categorizing the data, we used GENE-E software to present visual data by a heat map as we previously described [40-42]. Red represents higher PODXL2 mRNA expression, while blue represents lower expression.
The STRING database includes about 9.6 million protein data points in many organisms to predict protein-protein interactions (PPIs). We keyed in PODLX2 proteins to establish signaling pathways. An interaction network among PODXL2 and the top 50 genes from coexpressing gene lists of the TCGA and METABRIC datasets were used as input to ClueGO and CluePedia for a gene ontology (GO) analysis. The networks were built on molecular structures and biological functions of uploaded genes. All GO terms and pathways were updated to 12 January 2020, including biological processes, cellular components, immune processes, molecular functions, and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathways. ClueGO version 2.5.5 and CluePedia version 1.5.5 are plug-in packages that we used with Cytoscape V3.7.2 for analyses under academic license.
Cell Culture, Short Hairpin (sh)RNA Transfection, and Western Blotting
The BT-474 human cell line of ductal carcinoma of the breast was obtained from ATCC (Manassas, VA, USA). shRNAs targeting PODXL2 or luciferase genes were purchased from the National RNAi Core Facility (Academia Sinica, Taiwan; http://rnai.genmed.sinica.edu.tw). Accession number TRCN0000245516 was the target sequence 5'-CCA TCT GCA TCA TCA TCA TTG-3', and TRCN0000245515 interacted with the sequence 5'-GGG ACC CAC TGC AGA TTA TGT-3'. A construct (pLKO.1 containing a non-silencing shRNA against luciferase) was purchased as an expression control. Stable clones of BT-474 cells with knockdown of the PODXL2 gene were established by LipofetamineTM3000 and P3000TM. Cell lysates were collected, and proteins were extracted. Western blotting was performed to confirm the suppressive efficacy of PODXL2-shRNA with an anti-PODXL2 polyclonal antibody (GeneTex, Irvine, CA, USA). Downstream molecules were studied with an anti-glyceraldehyde-3-phosphate dehydrogenase (GAPDH) polyclonal antibody (pAb), anti-aldehyde dehydrogenase 1 (ALDH1) pAb (GeneTex), anti-Ras-related C3 botulinum toxin substrate 1 (Rac1) pAb, anti-AKT serine/threonine kinase 1 (Akt) (C67E7) monoclonal antibody (mAb), anti-phospho-Akt (pS473) (D9E) mAb (Cell Signaling, Danvers, MA, USA), anti-paxillin mAb, anti-phosphorylated (p)-paxillin (pY31) mAb, anti-Nanog monoclonal antibody, anti-octamer-binding transcription factor 4 (Oct-4) mAb, and anti-SRY-related HMG-box2 (SOX2) mAb (Epitomics, Burlingame, CA, USA).
RNA Extraction and Real-Time Polymerase Chain Reaction (PCR)
RNA from stable clones of BT474 cells was extracted with the TRIzol reagent (MD Biosciences, Zürich, Switzerland). An iScript™ cDNA Synthesis Kit (Bio-Red, Hercules, CA, USA) was used for complementary (c)DNA generation as a template in a real-time quantitative (q)PCR method. Three replicates of cDNA were analyzed using StepOne™ (Applied Biosystems, Life Technologies, CA, USA). The Universal Probe Library (Roche Applied Science, Penzberg, Germany) was used for primer design. A qPCR was used for the following primer set: PODXL2-sense: 5'-TGC CTT CAG TCA CCC CAA CTA-3' and PODXL2-antisense: 5'-AGC CTC GAA CTC TAC CCC AAG-3'; NHERF1-sense: 5'-AGG GAA ACT GAC GAG TTC TT-3' and NHERF1-antisense: 5'-TTC ACG ACT GTT CTC CTT CT-3'; and GAPDH-sense: 5'-AGC CAC ATC GCT CAG ACA C-3' and GAPDH -antisense: 5'-GCC CAA TAC GAC CAA ATC C-3'. Appropriate Taqman probes, TaqMan PCR Master Mix Kit, and an ABI StepOne™ sequence detector (Applied Biosystems, Foster City, CA, USA) were used for the qPCRs. The ΔΔCt value was used to determine mRNA levels of each gene. GAPDH was used as the housekeeping gene.
MTT Assay, Colony-Forming Assay, and Wound-Healing Assay
Cell proliferation was assessed by a colorimetric assay with 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT) dye (Millipore, Burlington, MA, USA). The final reactivity was measured as the OD570 value. Anchorage-independent growth was evaluated by a colony-formation assay. Cells were fixed with 100% methanol and stained with 2% methylene blue, and then the number of colonies was counted. The ability of cells to migrate was examined by scratching to form a wound and conducting a wound-healing assay. A scratch between cell layers in culture plates was made with a pipette tip to simulate physical separation. To compare the wounded areas before and after 48 h of incubation, we used a light microscope to capture images from the culture plate. The distance cells moved was calculated by subtracting the distance at 0 h from that at 48 h.
Statistical Analysis
Comparisons between groups were made using a one-way analysis of variance (ANOVA) and Tukey's multiple-comparison test. p values of <0.05 were considered a statistically significant. GraphPad Prism 5.0 (GraphPad Software, San Diego, CA) was used for these analyses.
Results
Expression of PODXL2 in Different Types of Cancer
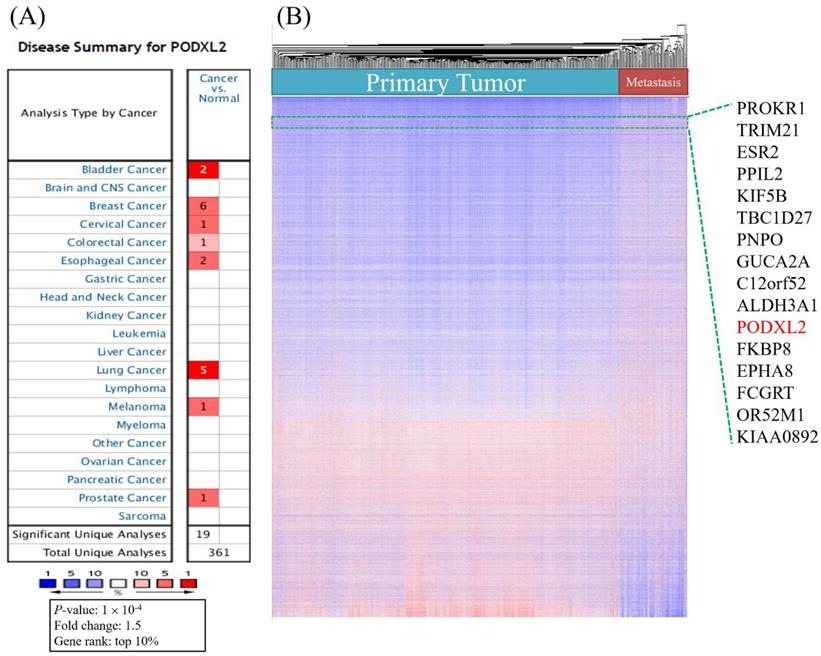
We analyzed mRNA expression levels of PODXL2 in 20 common cancers from the Oncomine database (Table 1). The analyses showed that the mRNA level of PODXL2 was overexpressed in 8 of 20 types of cancers compared to normal tissues (Figure 1A). It was enhanced in bladder cancer (2 of 10 studies), breast cancer (6 of 43 studies), cervical cancer (1 of 10 studies), colorectal cancer (1 of 32 studies), esophageal cancer (2 of 7 studies), lung cancer (4 of 20 studies), melanomas (1 of 5 studies), and prostate cancer (1 of 9 studies) (Table 1). We found 6 of 43 analyses of breast cancer meet our threshold. In other 39 analyses, four analyses showed fold change of PODXL2 less than 0 without any statistical significance. All other 35 analyses showed fold change of PODXL2 larger than 0, which meant cancer tissue have higher expression of PODXL2 than normal tissue (Supplementary Table 1). No studies detected the downregulation of PODXL2 mRNA with statistical significance in the present analysis. Upregulated genes in public datasets are listed in Figure 1B. Gene ranks are displayed in a heat map and were ordered from highest to lowest values of raw expression in primary and metastatic tumors of breast cancer. PODXL2 expression was higher in primary and metastatic breast cancers.
Correlation of PODXL2 mRNA and Protein Levels with Poor Prognoses of Breast Cancer Patients
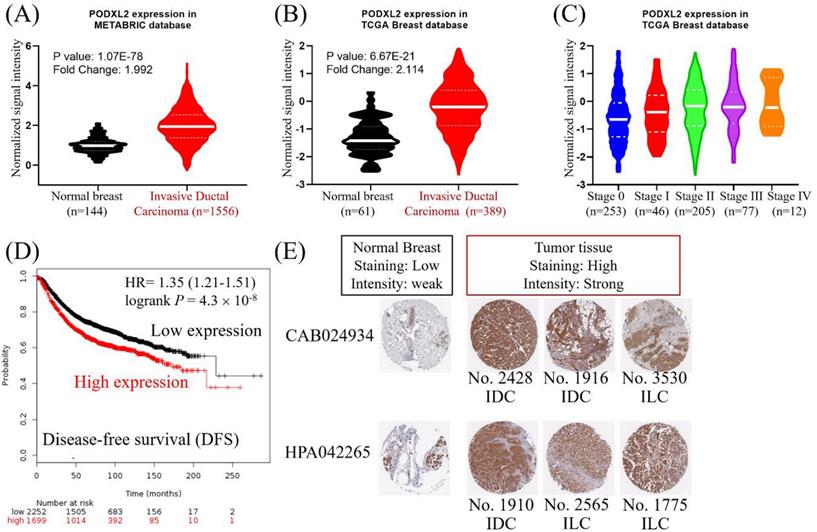
PODXL2 mRNA expression in breast cancers was upregulated in the METABRIC database (Figure 2A). The value from normal breast (n = 144, mean =0.986) was compared with the value from invasive ductal breast carcinoma (n = 1556, mean = 1.980). The mean difference and standard deviation (mean ± SD of difference) of PODXL2 between cancer and normal was 0.994 ± 3.010 with p-value 2.826E-40. PODXL2 also had high expression in breast cancers from the TCGA dataset (Figure 2B). The value from normal breast (n = 61, mean = -1.314) was compared with the value from invasive ductal breast carcinoma (n = 389, mean = -0.2347). The mean ± SD of difference of PODXL2 between cancer and normal was 1.080 ± 2.524 with p-value 3.148E-18. PODXL2 mRNA expression increased from stage 0 to stage IV breast cancer (Figure 2C) and the value from stage 0 (n = 253, mean = -0.600 ± 0.895), stage II (n = 205, mean = -0.251 ± 0.907), stage III (n = 77, mean = -0.159 ± 0.882), elevated till stage IV (n = 12, mean = -0.079 ± 0.853). Comparison between different cancer stage showed significant difference between stage II vs. stage 0, stage III vs. stage 0, and stage IV vs. stage 0. High expression of PODXL2 mRNA was correlated with poor disease-free survival of breast cancer patients in the Kaplan-Meier Plotter (Figure 2D). PODXL2 protein expression was investigated using the online database of The Human Protein Atlas. Low expression of PODXL2 with weak intensity was detected in normal breast tissues. Breast cancer cells showed high expression with strong intensity (Figure 2E). These results were consistent with two kinds of primary antibodies (CAB024934 or HPA042265). Our results demonstrated that PODXL2 mRNA and protein were highly expressed in breast cancer.
Participation of PODXL2-Coexpressed Genes in the Cell Cycle and EMT
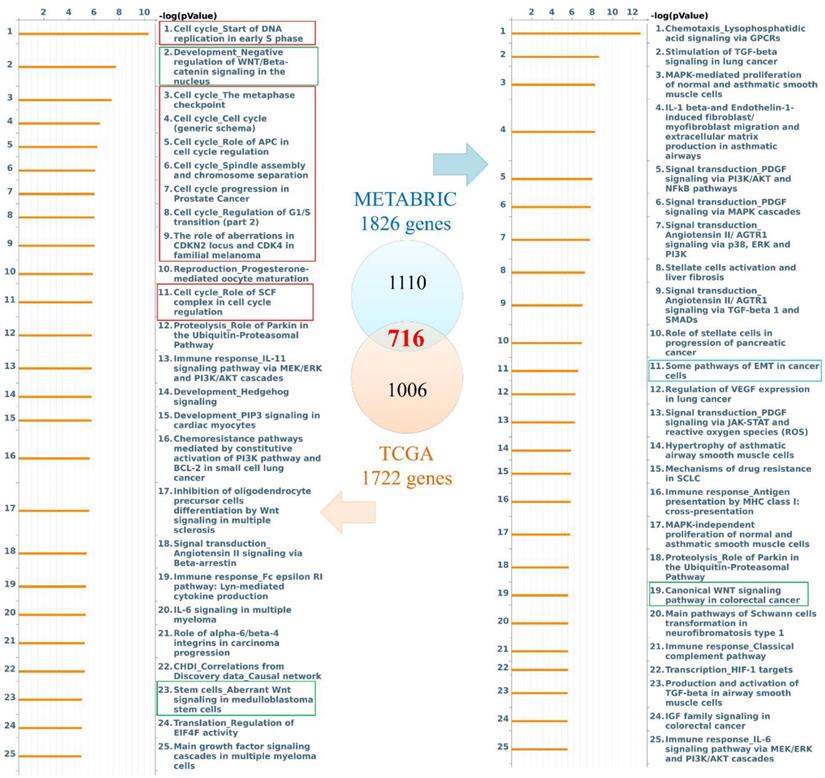
Genes coexpressed with PODXL2 were detected from TCGA and METABRIC datasets. We collected the top 10% interacting genes from TCGA and METABRIC datasets and analyzed them with the Metacore database (Figure 3). There were 716 coexpressed genes, and interacting networks were built. We found that genes coexpressed with PODXL2 mainly participated in the cell cycle, chemoresistance, cell growth, and the EMT (Figure 3). The MetaCore pathway analysis suggested that “signal transduction: angiotensin II/angiotensin II receptor type 1 (AGTR1) signaling via Ras homolog family member A (RhoA) and c-Jun N-terminal kinase (JNK)” had a high correlation with breast cancers exhibiting PODXL2 expression (Supplementary Figure 1).
High mRNA expression of PODXL2 in various cancer types, and both primary and metastatic breast cancer. (A) PODXL2 mRNA expression in 20 types of cancer relative to normal tissues from the Oncomine database. Red indicated increased expression of PODXL2 in cancer tissue comparison with normal tissue (left column), while blue indicated decreased expression of PODXL2 in cancer tissue with normal tissue (right column). The threshold was set at a p value of 10-4, a fold change of 1.5, and gene ranking of 10%. There was no analyses detected decreased expression of PODXL2 in cancer tissue and the right column were empty. (B) Heatmap representation from an analysis of a public high-throughput database. PODXL2 was found to have high expression in primary and metastatic breast cancers.
Expression of PODXL2 mRNA in cancers from the Oncomine database.
| Cancer | Cancer subtype | p value | Fold change | Gene rank (%) | Sample size | Ref. |
|---|---|---|---|---|---|---|
| Bladder | Superficial bladder cancer | 2.87E-15 | 2.551 | 1 | 194 | [58] |
| Bladder | Infiltrating bladder urothelial carcinoma | 7.73E-5 | 1.795 | 4 | 130 | [58] |
| Breast | Invasive ductal carcinoma | 1.07E-78 | 1.992 | 3 | 1700 | [36] |
| Breast | Medullary carcinoma | 9.36E-8 | 2.174 | 9 | 176 | [36] |
| Breast | Invasive carcinoma | 8.62E-5 | 1.633 | 9 | 165 | [36] |
| Breast | Invasive lobular carcinoma | 4.18E-20 | 1.678 | 10 | 292 | [36] |
| Breast | Invasive ductal carcinoma | 6.67E-21 | 2.114 | 8 | 450 | [37] |
| Breast | Invasive carcinoma | 1.45E-12 | 1.916 | 9 | 137 | [37] |
| Colorectal | Cecum adenocarcinoma | 3.48E-8 | 2.103 | 9 | 44 | [37] |
| Cervical | Cervical squamous cell carcinoma | 1.35E-8 | 5.005 | 2 | 56 | [59] |
| Esophageal | Barrett's esophagus | 3.90E-7 | 2.690 | 5 | 43 | [60] |
| Esophageal | Esophageal adenocarcinoma | 3.70E-9 | 1.919 | 7 | 103 | [60] |
| Lung | Lung adenocarcinoma | 6.78E-9 | 6.653 | 1 | 57 | [61] |
| Lung | Lung adenocarcinoma | 9.77E-18 | 4.084 | 2 | 116 | [62] |
| Lung | Lung adenocarcinoma | 7.88E-10 | 1.599 | 5 | 110 | [63] |
| Lung | Large cell lung carcinoma | 2.26E-6 | 2.182 | 5 | 84 | [63] |
| Melanoma | Cutaneous melanoma | 7.90E-7 | 7.322 | 4 | 52 | [64] |
| Prostate | Prostate adenocarcinoma | 2.42E-5 | 1.777 | 3 | 35 | [65] |
High expression of PODXL2 was associated with poor prognoses of patients with breast carcinoma. (A) mRNA levels of PODXL2 in the METABRIC database, and comparison between normal breast (n = 144, mean = 0.986) and invasive ductal carcinoma tissues (n = 1556, mean = 1.980). The mean difference and standard deviation (mean ± SD of difference) of PODXL2 between cancer and normal was 0.994 ± 3.010 with p-value 2.826E-40. (B) mRNA levels of PODXL2 in TCGA database, and comparison between normal breast (n = 61, mean = -1.314) and invasive ductal carcinoma tissues (n = 389, mean = -0.235). The mean ± SD of difference of PODXL2 between cancer and normal was 1.080 ± 2.524 with p-value 3.148E-18. (C) TCGA analysis of PODXL2 expression in different stages of breast cancer. PODXL2 levels increased as tumors progressed from stage 0 to stage IV cancer, the value from stage 0 (n = 253, mean = -0.600 ± 0.895), the value from stage I (n = 46, mean = -0.431 ± 0.883), the value from stage II (n = 205, mean = -0.251 ± 0.907), the value from stage III (n = 77, mean = -0.159 ± 0.882), the value from stage IV (n = 12, mean = -0.079 ± 0.853). Stage I vs. stage 0, p-value 0.2395; stage II vs. stage 0, p-value 0.00005; stage III vs. stage 0, p-value 0.0002; stage IV vs. stage 0, p-value 0.0496. (D) Disease-free survival curve from the Kaplan-Meier Plotter and evaluation of the impact from low (black line) and high (red line) expression of PODXL2. (E) Immuno-histochemical staining of PODXL2 protein in normal breast and breast cancer tissues from The Human Protein Atlas database. Two primary antibodies (CAB24934 and HPA042265) were tested in the different histological subtypes of breast cancer.
Expression of PODXL2 mRNA and Protein in Breast Cancer Cell Lines
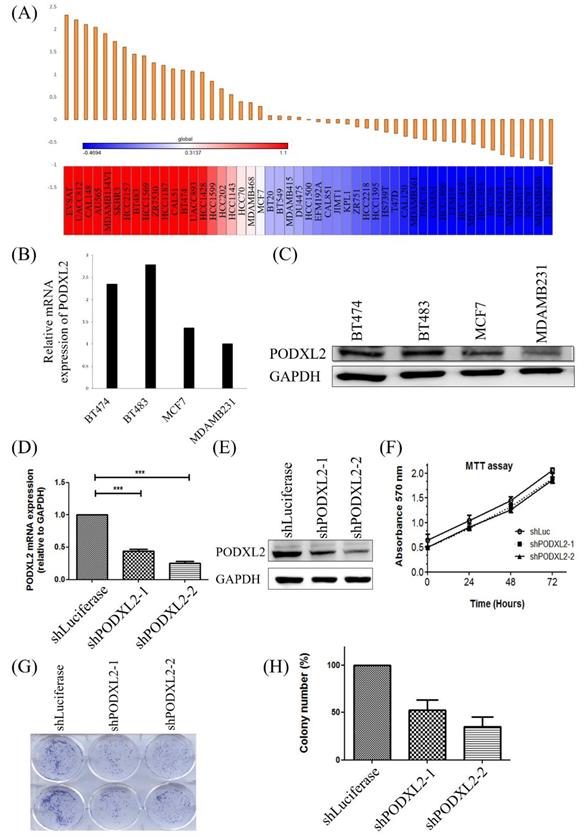
In addition to the coexpression study of PODXL2, we further assessed PODXL2 expression in the CCLE database. Results are shown as a heat map generated by GENE-E software (Figure 4A). According to the heat map, we chose four kinds of breast cancer cell lines with different expression levels of PODXL2, including BT474, BT483, MCF7, and MDA-MB-231 cells. The expression level of PODXL2 in the CCLE database from high to low was BT483, BT474, MCF4, and MDA-MB-231 cells. The mRNA level of PODXL2 was detected by CCLE dataset (Figure 4B), and protein levels were examined by Western blotting (Figure 4C). Results were consistent with the heat map results. The BT474 cell line was selected for a further in vitro investigation because of constitutional PODXL2 expression. Lentiviral transduction of PODXL2-shRNA was performed to inhibit PODXL2 expression in BT474 cells. The suppressive efficacy was confirmed by a qPCR (Figure 4D) and Western blotting (Figure 4E). We first measured the ability of cells to proliferate. The MTT assay detected no difference in short-term cell proliferation between PODXL2-silenced BT474 cells and control cells (Figure 4F). However, when we evaluated long-term cell proliferation by a colony-formation assay, results showed that the ability of cells to proliferate in PODXL2-silenced BT474 cells had decreased (Figure 4G, H). Therefore, it is likely that PODXL2 participates in long-term cell proliferation.
Diagram of expression pathways and networks of PODXL2 in breast cancer patients. The blue and orange circles represent the upper quartile of coexpressed genes in the METABRIC and TCGA databases, respectively. Total numbers of genes from each fraction of the two databases are also displayed. Lists of disease and biomarker networks that were significantly enriched according to the MetaCore database. Left: for TCGA; right: for METABRIC. The red rectangles meant the cell cycle-related pathways, green rectangles were stemness-regulated pathways, and the blue rectangles represented migration-associated pathway.
Regulation of Cell Migration by the PODXL2 Gene
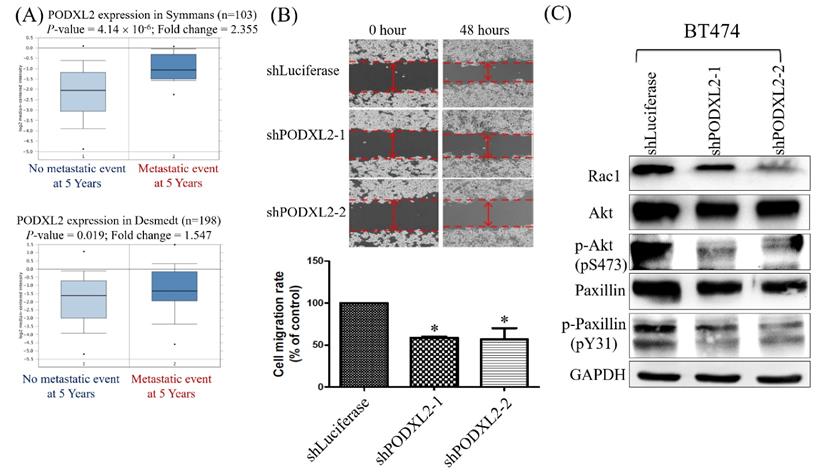
Two datasets were selected from the Oncomine database, and we found that patients with high PODXL2 expression had more metastatic events within the post-therapeutic 5 years (Figure 5A). The networks in Figure 3 imply that PODXL2 might be involved in the EMT. We evaluated the ability of cells to migrate by a wound-healing assay. The migratory ability of PODXL2-silenced BT474 cells was reduced compared to control cells (Figure 5B). A previous study showed that PODXL-promoted tumor metastasis was mediated by activating the Ras-related C3 botulinum toxin substrate 1 (Rac1) signaling cascade or PI3K/Akt signaling [43]. Downstream of Rac 1 is paxillin. Phosphorylation of paxillin at Tyr31, Tyr118, Ser188, and Ser190 was reported to promote migration [44]. Western blotting of migration-associated proteins was performed. Expression levels of Rac1, p-Akt (pS473), and p-paxillin (pY31) decreased after PODXL2 expression was downregulated (Figure 5C).
Expression levels of PODXL2 mRNA and protein in cancer cell lines and knockdown of the PODXL2 gene in BT474 breast cancer cells. (A) mRNA levels of PODXL2 from the CCLE database. Using R package GENE-E, we made a heat map of all cancer cell lines. Red indicates overexpression (left), while blue indicates downregulation (right) in cancer cell lines. (B) mRNA levels of PODXL2 were examined in four breast cancer cell lines by CCLE dataset. (C) Protein levels of PODXL2 were tested by Western blotting. Three independent experiments were performed, and GAPDH was used as an internal control. (D) Suppression of PODXL2 expression in BT474 cells by lentivirus-mediated PODXL2-shRNA. Detection of PODXL2 mRNA by a qPCR. (E) Examination of protein levels of PODXL2 by Western blotting. The suppressive efficacy in two stable clones was confirmed. (F) Cell proliferation was examined by an MTT assay at 0, 24, 48, and 72 h. Short-term cell proliferation was unchanged after knockdown of PODXL2, compared to control cells. (G) Anchorage-independent cell growth was tested by a colony-formation assay at 14 days. The colony was stained with a 2% methylene blue solution. (H) Quantification of the colony number in (G). Long-term cell proliferation was suppressed after knockdown of PODXL2, compared to control cells. Two independent experiments of an MTT assay and colony-formation assay were performed. shLuc, luciferase short-hairpin RNA, *p < 0.01; **p < 0.001; ***p < 0.0001.
Correlation between PODXL2 and CSC Characteristics in Breast Cancer
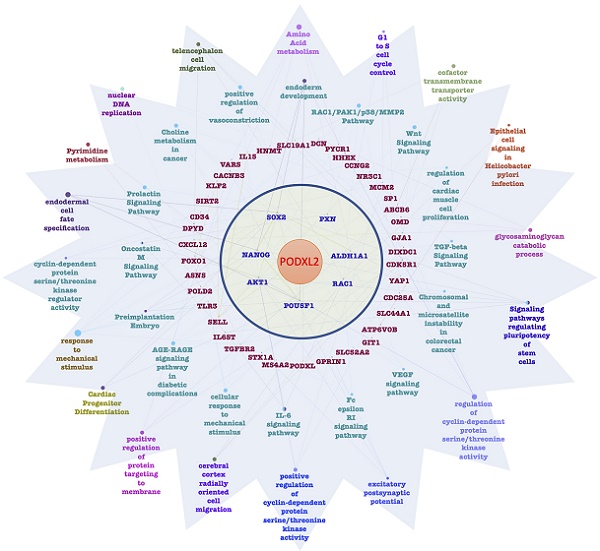
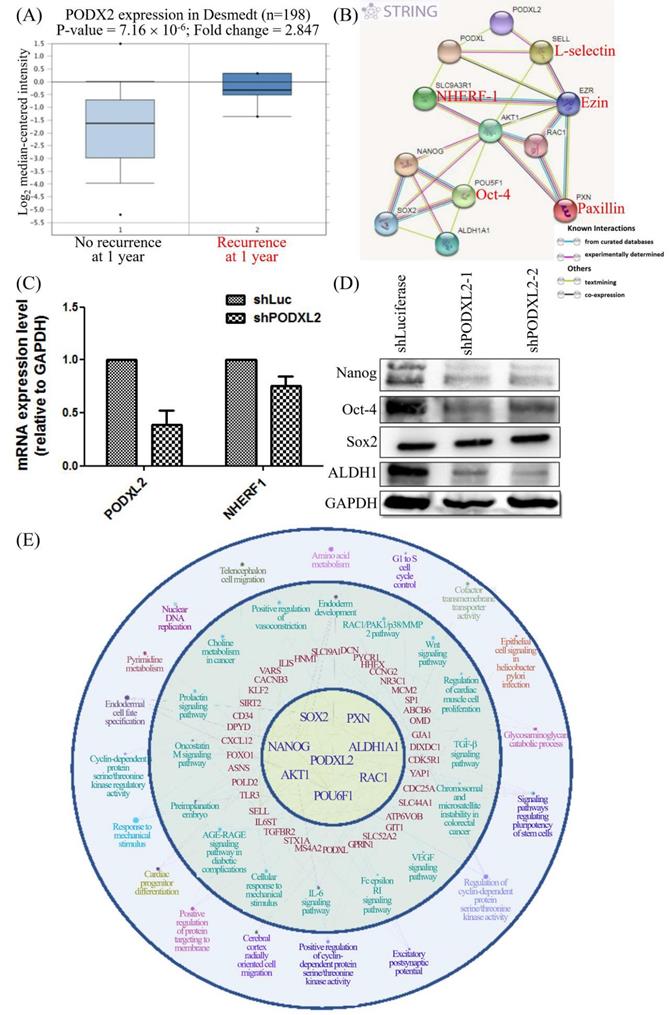
CD34 family proteins are markers of HSCs. Cancer metastasis and recurrence are also correlated with CSCs. High PODXL2 expression in the Desmedt dataset was associated with cancer recurrence at 1 year post-therapy (Figure 6A). We revealed PPIs in breast cancer by the STRING database. Potential interactions between PODXL, SLC9A3R1 (solute carrier family 9 subfamily A member 3 regulator 1, also named NHERF1), SELL (L-selectin), EZR (ezrin), AKT1, RAC1, PXN (paxillin), POU5F1 (POU class 5 homeobox 1, also named Oct-4), Nanog, SOX2, and ALDH1A1 (ALDH1) were found (Figure 6B), and some proteins presented with CSC characteristics. We investigated expression levels of the SC markers Nanog, Oct-4, and SOX2, and the breast CSC marker ALDH1 in PODXL2-silenced BT474 cells. Suppression of PODXL2 had no effect on the mRNA level of NHERF1 (Figure 6C), and thus, other signaling pathways should be considered. Downregulation of PODXL2 resulted in reduced expressions of Nanog, Oct-4, and ALDH1 according to Western blotting (Figure 6D). We suggest that increased PODXL2 expression could be used as a biomarker in patients with breast cancer. Our bioinformatics analysis and in vitro experiments revealed that PODXL2 plays an oncogenic role in breast cancer development via the Rac1 pathway. Moreover, downstream signaling of PODXL2 in breast cancer was simulated by the ClueGo and CluePedia databases in Figure 6E. A combination of the ClueGo, CluePedia, and GO pathways showed that PODXL2 interacted with NANOG, AKT1, and RAC1. PODXL2 also cooperated with CSC-related genes (SOX2, ALDH1A1, and POU5F1) and a migration-associated gene (PXN). The PODXL2 gene was located in the center of the networks, and the consequent phenotypes were drawn in the outer circle, which included regulation of the cell cycle, migration, and the polarity of SCs. The inferential network implied the function of PODXL2 in breast cancer.
Discussion
PODXL2 belongs to the CD34 family of proteins. PODXL2 is thought to play a crucial role in breast cancer, because PODXL2 is a transmembrane mucin with a PDZ domain that interacts with intracellular signal transduction. In the present study, PODXL2 expression was detected in breast cancer tissues. The bioinformatics analysis confirmed that high PODXL2 expression was correlated with poor prognoses of breast cancer patients. Knockdown of PODXL2 in breast cancer cell lines inhibited long-term cell proliferation in a colony-formation assay and suppressed migration in a wound-healing assay. PODXL2-knockdown also suppressed the downstream Rac1 level and reduced phosphorylated forms of Akt and paxillin. Downregulation of PODXL2 repressed expression of CSC-related proteins as occurs with other CD34 family proteins. Downstream signaling of PODXL2-interacting networks was speculated to be correlated with cell proliferation, migration, and CSC characteristics. The present study confirmed the function of PODXL2 in breast cancer.
Expression levels of PODXL2 mRNA associated with metastasis and migration. (A) mRNA levels of PODXL2 in the Symmans (upper row) and Desmedt (lower row) datasets, comparing primary breast cancer from patients without metastasis at postoperative 5 years (left column) and primary cancer from those with metastasis at 5 years (right column). (B) Cell migration was examined by a wound-healing assay at 0 and 48 h after seeding (upper row). Quantification of the wound distance is shown in the lower row. Cell migration was suppressed after knockdown of PODXL2, compared to control cells. Two independent experiments of a wound-healing assay were performed. (C) Rac1, Akt, phosphorylated (p)-Akt (s473), paxillin, and p-paxillin (Y31) protein expressions were detected by Western blotting in PODXL2-silenced BT474 cells and control cells, *P < 0.01; **P < 0.001; ***P < 0.0001.
PODXL2 is associated with recurrence and stemness in breast cancer. (A) mRNA levels of PODXL2 in the Desmedt datasets, comparing patients without recurrence at postoperative 1 year (left column) and those with recurrence at 1 year (right column). (B) Protein interaction network in the STRING database. Individual colored lines between proteins indicate different types of evidence illustrating the interaction. The evidence for these interactions was derived from curated databases (cyan-blue lines), experimental evidence (purple lines), text-mining evidence (green lines), and coexpression (dark lines). (C) mRNA levels of PODXL2 and downstream NHERF1 were examined by a qPCR in BT483 cells. Successful suppression of PODXL2 by shRNA was confirmed without obvious changes in NHERF1. (D) Expressions of stem cell proteins were detected by Western blotting in PODXL2-silenced BT474 cells and control cells, including Nanog, Oct-4, SOX2, and ALDH1. (E) Organized networks were drawn by ClueGO and the CluePedia database. In the center of the circle is the PODXL2 gene. The first ring includes the PXN, ALDH1A1, RAC1, POU6F1, AKT1, NANOG, and SOX2 genes. Interactions between the PODXL2 gene and first-ring genes deduced genes in the second circle (red-colored words). Correlated phenotypes are presented in the outer two circles. Connecting lines represent relationships between genes and/or pathways.
CD34 family proteins include CD34, PODXL, and PODXL2. All three of these proteins are transmembrane proteins and share the intracellular PDZ domain. CD34 and PODXL are overexpressed in several kinds of malignancies with characteristics of CSCs. Because of similar protein structures and genomic organization of PODXL2 with the two other CD34 family proteins, we hypothesized that functions of PODXL2 resemble those of PODXL and CD34. PODXL2 was overexpressed in six of 43 studies of breast cancer, primary and metastatic breast cancer, mRNA expression, and IHC staining of proteins. Through the bioinformatics analyzed from Symmans and Desmedt breast cancer datasets revealed that patients with metastatic events had higher PODXL2 expression levels. High PODXL2 expression was correlated with poor disease-free survival of breast cancer patients. Patients with cancer recurrence within postoperative one year had higher PODXL2 levels in the Desmedt breast cancer dataset. Our results confirmed that increased PODXL2 expression should play an important role in breast cancer.
PPIs are key points for studying protein functions. Previous studies indicated that PODXL2 shares an identical motif (PDZ domain-binding motif, DTHL) with PODXL and NHERF-1 as potential intracellular binding partners of PODXL2. The PODXL/NHERF-1 complex regulates PI3K/PTEN/AKT signaling and Rho-GTPase activity [45], and these small RAC1/GTPases of the Rho family have important roles in cell development through PI3K/Akt signaling [30, 31, 46]. We used the Oncomine, cBioportal, STRING, ClueGO, and CluePedia databases to study PODXL2-interacting proteins. The Oncomine and cBioPortal databases collect mRNA expression from several datasets of breast cancer, and genes coexpressed with PODXL2, these coexpressed genes were uploaded to MetaCore to analyze associated pathways. PODXL2-related pathways involved cell cycle progression, PIK3/AKT signaling, the EMT, cell proliferation, and chemoresistance. ATGR1/RhoA/JNK signaling was correlated with PODXL2 expression in breast cancer. STRING software selected the most closely associated genes with PODXL2 in cancer. CSC-related genes (POU5F1, NANOG, SOX2, and ALDH1A1), migration-associated genes (EZR and PXN), and RAC1/AKT1 signaling transduction genes were linked. ClueGo and CluePedia software further extended the networks to other genes and cellular phenotypes. All these bioinformatics tools are useful for predicting interactions and functions of PODXL2 in cancers. We achieved similar results after these analyses and confirmed that PODXL2 is essential for cell proliferation, migration, and maintenance of CSC characteristics.
In addition to bioinformatics predictions, we also performed an in vitro study with a breast cancer cell line to confirm the function of PODXL2. Stable clones of PODXL2-shRNA were established, and the suppressive efficacy was confirmed by a qPCR and Western blotting. Short-term cell proliferation did not change after silencing of PODXL2; however, long-term cell proliferation was suppressed in a colony-formation assay. The colony-formation assay indicates the ability of anchorage-independent growth, and cytoskeletal proteins of ezrin and paxillin were identified. Phosphorylation of paxillin is involved in cell migration.
Meanwhile, previous studies demonstrated that the distribution of distant metastases to certain organs is a non-random process known as "metastatic organotropism", which is regulated by multiple factors including subtypes of cancer, genetic signature of cancer cells, the microenvironment as well as the interactions network [47-51]. According to our bioinformatics and experimental data suggest that PODXL2 may also play a role in tropism. The present study demonstrated that PODXL2-silenced cells had a lower ability to migrate and decreased expressions of p-Akt and p-paxillin. CD34 family proteins are considered markers of SCs [52]. We examined expressions of CSC-related proteins in PODXL2-silenced cells, and we found that Nanog, Oct-4, and ALDH1 proteins were suppressed, however the NHERF1 might not be a hinge or focal point of signaling transduction, and Rac1/Akt is potential downstream signaling of PODXL2 via our experimental validation with Western blot. However, markers of breast CSCs not only include Nanog/Oct-4/ALDH1, but also CD133 and CD44 [53]. Previous studies used CD133+/CD44+/CD24- to sort breast CSCs for further investigation [22, 53-56]. Cell proliferation, migration, and CSC characteristics are associated with chemoresistance and cancer recurrence[57]. Our results proved the function of PODXL2 in breast cancer. Increased PODXL2 expression is a predictor of recurrence in patients with breast cancer, and PODXL2 may provide a potential target for therapy.
Conclusions
The present study identified that mRNA and protein levels of PODXL2 were highly expressed in breast carcinoma, which resulted in poor prognoses of breast cancer patients. In addition, coexpressed genes/proteins of PODXL2 mainly participated in cell proliferation and migration. Furthermore, PODXL2 is suggested to be involved in tumor metastasis and to play a role in breast CSCs. In summary, PODXL2 plays a significant role in breast cancer. Our results suggested that increased PODXL2 expression could serve as a biomarker and a therapeutic target in patients with breast cancer.
Abbreviation
AKT1: AKT serine/threonine kinase 1; ALDH1A1: aldehyde dehydrogenase 1 family member A1; EZR: ezrin; NANOG: Nanog homeobox; PODXL: podocalyxin-like protein 1; PODXL2: podocalyxin-like protein 2; POU5F1: POU class 5 homeobox 1 (Oct-4); PXN: paxillin; RAC1: Rac family small GTPase 1; SELL: L-selectin; SLC9A3R1: SLC9A3 regulator 1 (NHERF-1); SOX2: SRY-related HMG-box2.
Supplementary Material
Supplementary figures and tables.
Acknowledgements
Bioinformatics analyses and data mining were conducted at the Bioinformatics Core at National Cheng Kung University (Tainan, Taiwan). The authors are grateful for the support from the Human Biobank, Research Center of Clinical Medicine, National Cheng Kung University Hospital. The authors give special thanks for Dr. Dan Chamberlin of his professional English editing from the office of research and development in Taipei Medical University.
Author Contributions
Conceptualization, Y.-Y.L., C.-Y.W., H.-P.H., and M.-D.L.; methodology, Y.-Y.L., C.-Y.W., N.N.P., and C.-C.C.; validation, C.-Y.L. and Z.S.; formal analysis, Y.-Y.L. and C.-Y.W.; investigations, J.-H.H. and Y.-L.C.; resources, J.-H.H., Y.-L.C., M.-C. Y., and T.-Y.W.; data curation, Y.-Y.L., C.-Y.W., H.-P.H., and M.-D.L; writing—original draft preparation, Y.-Y.L., C.-Y.W., and N.N.P.; writing—review and editing, H.-P.H. and M.-D.L.; visualization, N.N.P., C.-C.C., and Z.S.; supervision, H.-P.H. and M.-D.L.; project administration, H.-P.H. and M.-D.L.; and funding acquisition, C.-Y.W., H.-P.H., and M.-D.L. All authors read and agreed to the published version of the manuscript.
Funding
The study was supported by the Ministry of Science and Technology (MOST) of Taiwan (grants MOST105-2325-B-006-003 to M-D.L., MOST 108-2314-B-006-082 to H-P.H., and MOST109-2320-B-038-009-MY2 to C-Y.W.), National Cheng Kung University Hospital (grant NCKUH-10601002 to M-D.L. and NCKUH10902031 to H-P.H.), and Taipei Medical University (grant TMU-108-AE1-B16 to C-Y.W.).
Competing Interests
The authors have declared that no competing interest exists.
References
1. Siegel RL, Miller KD, Jemal A. Cancer statistics, 2020. CA Cancer J Clin. 2020;70:7-30
2. Qian C-N, Mei Y, Zhang J. Cancer metastasis: issues and challenges. Chin J Cancer. 2017;36:38 -
3. Esmatabadi MJD, Bakhshinejad B, Motlagh FM, Babashah S, Sadeghizadeh M. Therapeutic resistance and cancer recurrence mechanisms: Unfolding the story of tumour coming back. Journal of Biosciences. 2016;41:497-506
4. O'Flaherty JD, Barr M, Fennell D, Richard D, Reynolds J, O'Leary J. et al. The Cancer Stem-Cell Hypothesis: Its Emerging Role in Lung Cancer Biology and Its Relevance for Future Therapy. Journal of Thoracic Oncology. 2012;7:1880-90
5. Hass R, von der Ohe J, Ungefroren H. Potential Role of MSC/Cancer Cell Fusion and EMT for Breast Cancer Stem Cell Formation. Cancers (Basel). 2019 11
6. Markopoulos GS, Roupakia E, Marcu KB, Kolettas E. Epigenetic Regulation of Inflammatory Cytokine-Induced Epithelial-To-Mesenchymal Cell Transition and Cancer Stem Cell Generation. Cells. 2019 8
7. Sonbol MB, Ahn DH, Bekaii-Saab T. Therapeutic Targeting Strategies of Cancer Stem Cells in Gastrointestinal Malignancies. Biomedicines. 2019 7
8. Lapidot T, Sirard C, Vormoor J, Murdoch B, Hoang T, Caceres-Cortes J. et al. A cell initiating human acute myeloid leukaemia after transplantation into SCID mice. Nature. 1994;367:645-8
9. Goodell MA, Rosenzweig M, Kim H, Marks DF, DeMaria M, Paradis G. et al. Dye efflux studies suggest that hematopoietic stem cells expressing low or undetectable levels of CD34 antigen exist in multiple species. Nat Med. 1997;3:1337-45
10. Schmidt JR, Rucker-Braun E, Heidrich K, von Bonin M, Stolzel F, Thiede C. et al. Pilot Study on Mass Spectrometry-Based Analysis of the Proteome of CD34(+)CD123(+) Progenitor Cells for the Identification of Potential Targets for Immunotherapy in Acute Myeloid Leukemia. Proteomes. 2018 6
11. Nielsen JS, McNagny KM. Novel functions of the CD34 family. Journal of Cell Science. 2008;121:3683-92
12. Doyonnas R, Kershaw DB, Duhme C, Merkens H, Chelliah S, Graf T. et al. Anuria, omphalocele, and perinatal lethality in mice lacking the CD34-related protein podocalyxin. J Exp Med. 2001;194:13-27
13. Somasiri A, Nielsen JS, Makretsov N, McCoy ML, Prentice L, Gilks CB. et al. Overexpression of the Anti-Adhesin Podocalyxin Is an Independent Predictor of Breast Cancer Progression. Cancer Research. 2004;64:5068-73
14. Cipollone JA, Graves ML, Köbel M, Kalloger SE, Poon T, Gilks CB. et al. The anti-adhesive mucin podocalyxin may help initiate the transperitoneal metastasis of high grade serous ovarian carcinoma. Clinical & Experimental Metastasis. 2012;29:239-52
15. Koch L-K, Zhou H, Ellinger J, Biermann K, Höller T, von Rücker A. et al. Stem cell marker expression in small cell lung carcinoma and developing lung tissue. Human Pathology. 2008;39:1597-605
16. Yu YP, Landsittel D, Jing L, Nelson J, Ren B, Liu L. et al. Gene Expression Alterations in Prostate Cancer Predicting Tumor Aggression and Preceding Development of Malignancy. Journal of Clinical Oncology. 2004;22:2790-9
17. Chen X, Higgins J, Cheung S-T, Li R, Mason V, Montgomery K. et al. Novel endothelial cell markers in hepatocellular carcinoma. Modern Pathology. 2004;17:1198-210
18. Thomas SN, Schnaar RL, Konstantopoulos K. Podocalyxin-like protein is an E-/L-selectin ligand on colon carcinoma cells: comparative biochemical properties of selectin ligands in host and tumor cells. Am J Physiol Cell Physiol. 2009;296:C505-C13
19. Taniuchi K, Furihata M, Naganuma S, Dabanaka K, Hanazaki K, Saibara T. Podocalyxin-like protein, linked to poor prognosis of pancreatic cancers, promotes cell invasion by binding to gelsolin. Cancer Sci. 2016;107:1430-42
20. Kusumoto H, Shintani Y, Kanzaki R, Kawamura T, Funaki S, Minami M. et al. Podocalyxin influences malignant potential by controlling epithelial-mesenchymal transition in lung adenocarcinoma. Cancer Sci. 2017;108:528-35
21. Amo L, Tamayo-Orbegozo E, Maruri N, Buqué A, Solaun M, Riñón M. et al. Podocalyxin-like protein 1 functions as an immunomodulatory molecule in breast cancer cells. Cancer Letters. 2015;368:26-35
22. Kim W-T, Ryu CJ. Cancer stem cell surface markers on normal stem cells. BMB Rep. 2017;50:285-98
23. Lee W-Y, Kuo C-C, Lin B-X, Cheng C-H, Chen K-C, Lin C-W. Podocalyxin-Like Protein 1 Regulates TAZ Signaling and Stemness Properties in Colon Cancer. Int J Mol Sci. 2017;18:2047
24. Sassetti C, Van Zante A, Rosen SD. Identification of Endoglycan, a Member of the CD34/Podocalyxin Family of Sialomucins. Journal of Biological Chemistry. 2000;275:9001-10
25. Fieger CB, Sassetti CM, Rosen SD. Endoglycan, a Member of the CD34 Family, Functions as an L-selectin Ligand through Modification with Tyrosine Sulfation and Sialyl Lewis x. Journal of Biological Chemistry. 2003;278:27390-8
26. Kerr SC, Fieger CB, Snapp KR, Rosen SD. Endoglycan, a member of the CD34 family of sialomucins, is a ligand for the vascular selectins. J Immunol. 2008;181:1480-90
27. Nicolini A, Ferrari P, Rossi G. Mucins and Cytokeratins as Serum Tumor Markers in Breast Cancer. Advances in Cancer Biomarkers: Springer Netherlands. 2015 p. 197-225
28. Perrais M, Pigny P, Buisine M-P, Porchet N, Aubert J-P, Van Seuningen-Lempire I. Aberrant Expression of Human Mucin GeneMUC5Bin Gastric Carcinoma and Cancer Cells. Journal of Biological Chemistry. 2001;276:15386-96
29. Song S, Byrd JC, Mazurek N, Liu K, Koo JS, Bresalier RS. Galectin-3 Modulates MUC2 Mucin Expression in Human Colon Cancer Cells at the Level of Transcription via AP-1 Activation. Gastroenterology. 2005;129:1581-91
30. Zhu G, Fan Z, Ding M, Zhang H, Mu L, Ding Y. et al. An EGFR/PI3K/AKT axis promotes accumulation of the Rac1-GEF Tiam1 that is critical in EGFR-driven tumorigenesis. Oncogene. 2015;34:5971-82
31. Murga C, Zohar M, Teramoto H, Gutkind JS. Rac1 and RhoG promote cell survival by the activation of PI3K and Akt, independently of their ability to stimulate JNK and NF-κB. Oncogene. 2002;21:207-16
32. Rhodes DR, Yu J, Shanker K, Deshpande N, Varambally R, Ghosh D. et al. ONCOMINE: a cancer microarray database and integrated data-mining platform. Neoplasia. 2004;6:1-6
33. Uhlen M, Zhang C, Lee S, Sjostedt E, Fagerberg L, Bidkhori G. et al. A pathology atlas of the human cancer transcriptome. Science. 2017 357
34. Li Q, Birkbak NJ, Gyorffy B, Szallasi Z, Eklund AC. Jetset: selecting the optimal microarray probe set to represent a gene. BMC bioinformatics. 2011;12:474
35. Gao J, Aksoy BA, Dogrusoz U, Dresdner G, Gross B, Sumer SO. et al. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci Signal. 2013;6:pl1-pl
36. Curtis C, Shah SP, Chin S-F, Turashvili G, Rueda OM, Dunning MJ. et al. The genomic and transcriptomic architecture of 2,000 breast tumours reveals novel subgroups. Nature. 2012;486:346
37. Cancer Genome Atlas N. Comprehensive molecular portraits of human breast tumours. Nature. 2012;490:61-70
38. Ciriello G, Gatza ML, Beck AH, Wilkerson MD, Rhie SK, Pastore A. et al. Comprehensive molecular portraits of invasive lobular breast cancer. Cell. 2015;163:506-19
39. Barretina J, Caponigro G, Stransky N, Venkatesan K, Margolin AA, Kim S. et al. The Cancer Cell Line Encyclopedia enables predictive modelling of anticancer drug sensitivity. Nature. 2012;483:603
40. Wang CY, Li CY, Hsu HP, Cho CY, Yen MC, Weng TY. et al. PSMB5 plays a dual role in cancer development and immunosuppression. Am J Cancer Res. 2017;7:2103-20
41. Wang CY, Chiao CC, Phan NN, Li CY, Sun ZD, Jiang JZ. et al. Gene signatures and potential therapeutic targets of amino acid metabolism in estrogen receptor-positive breast cancer. Am J Cancer Res. 2020;10:95-113
42. Wang CY, Chang YC, Kuo YL, Lee KT, Chen PS, Cheung CHA. et al. Mutation of the PTCH1 gene predicts recurrence of breast cancer. Sci Rep. 2019;9:16359
43. Lin CW, Sun MS, Liao MY, Chung CH, Chi YH, Chiou LT. et al. Podocalyxin-like 1 promotes invadopodia formation and metastasis through activation of Rac1/Cdc42/cortactin signaling in breast cancer cells. Carcinogenesis. 2014;35:2425-35
44. Webb DJ, Donais K, Whitmore LA, Thomas SM, Turner CE, Parsons JT. et al. FAK-Src signalling through paxillin, ERK and MLCK regulates adhesion disassembly. Nature Cell Biology. 2004;6:154-61
45. Schmieder S. Podocalyxin Activates RhoA and Induces Actin Reorganization through NHERF1 and Ezrin in MDCK Cells. Journal of the American Society of Nephrology. 2004;15:2289-98
46. Zins K, Lucas T, Reichl P, Abraham D, Aharinejad S. A Rac1/Cdc42 GTPase-specific small molecule inhibitor suppresses growth of primary human prostate cancer xenografts and prolongs survival in mice. PLoS One. 2013;8:e74924-e
47. Lu X, Kang Y. Organotropism of breast cancer metastasis. J Mammary Gland Biol Neoplasia. 2007;12:153-62
48. Kostic A, Lynch CD, Sheetz MP. Differential matrix rigidity response in breast cancer cell lines correlates with the tissue tropism. PLoS One. 2009;4:e6361
49. Zhao D, Najbauer J, Annala AJ, Garcia E, Metz MZ, Gutova M. et al. Human neural stem cell tropism to metastatic breast cancer. Stem Cells. 2012;30:314-25
50. Hakkarainen T, Sarkioja M, Lehenkari P, Miettinen S, Ylikomi T, Suuronen R. et al. Human mesenchymal stem cells lack tumor tropism but enhance the antitumor activity of oncolytic adenoviruses in orthotopic lung and breast tumors. Hum Gene Ther. 2007;18:627-41
51. Shinde A, Wilmanski T, Chen H, Teegarden D, Wendt MK. Pyruvate carboxylase supports the pulmonary tropism of metastatic breast cancer. Breast Cancer Res. 2018;20:76
52. Nielsen JS, McNagny KM. CD34 is a Key Regulator of Hematopoietic Stem Cell Trafficking to Bone Marrow and Mast Cell Progenitor Trafficking in the Periphery. Microcirculation. 2009;16:487-96
53. Martin T, Mansel R, Jiang W. Evaluation of the Distribution of Stem Cell Markers in Human Breast Cancer Reveals Correlation with Clinical Progression and Metastatic Disease in Ductal Carcinoma. Poster Session Abstracts: American Association for Cancer Research. 2009
54. Mine T, Matsueda S, Li Y, Tokumitsu H, Gao H, Danes C. et al. Breast cancer cells expressing stem cell markers CD44+ CD24 lo are eliminated by Numb-1 peptide-activated T cells. Cancer Immunol Immunother. 2009;58:1185-94
55. Kapucuoğlu N, Bozkurt KK, Başpınar Ş, Koçer M, Eroğlu HE, Akdeniz R. et al. The clinicopathological and prognostic significance of CD24, CD44, CD133, ALDH1 expressions in invasive ductal carcinoma of the breast. Pathology - Research and Practice. 2015;211:740-7
56. Laranjo M, Carvalho MJ, Costa T, Alves A, Oliveira RC, Casalta-Lopes J. et al. Mammospheres of hormonal receptor positive breast cancer diverge to triple-negative phenotype. The Breast. 2018;38:22-9
57. Phi LTH, Sari IN, Yang YG, Lee SH, Jun N, Kim KS. et al. Cancer Stem Cells (CSCs) in Drug Resistance and their Therapeutic Implications in Cancer Treatment. Stem Cells Int. 2018;2018:5416923
58. Lee JS, Leem SH, Lee SY, Kim SC, Park ES, Kim SB. et al. Expression Signature of E2F1 and Its Associated Genes Predict Superficial to Invasive Progression of Bladder Tumors. Journal of Clinical Oncology. 2010;28:2660-7
59. Scotto L, Narayan G, Nandula SV, Arias-Pulido H, Subramaniyam S, Schneider A. et al. Identification of copy number gain and overexpressed genes on chromosome arm 20q by an integrative genomic approach in cervical cancer: Potential role in progression. Genes, Chromosomes and Cancer. 2008;47:755-65
60. Kim SM, Park YY, Park ES, Cho JY, Izzo JG, Zhang D. et al. Prognostic biomarkers for esophageal adenocarcinoma identified by analysis of tumor transcriptome. PLoS One. 2010;5:e15074-e
61. Su LJ, Chang CW, Wu YC, Chen KC, Lin CJ, Liang SC. et al. Selection of DDX5 as a novel internal control for Q-RT-PCR from microarray data using a block bootstrap re-sampling scheme. BMC Genomics. 2007;8:140 -
62. Selamat SA, Chung BS, Girard L, Zhang W, Zhang Y, Campan M. et al. Genome-scale analysis of DNA methylation in lung adenocarcinoma and integration with mRNA expression. Genome research. 2012;22:1197-211
63. Hou J, Aerts J, Den Hamer B, Van Ijcken W, Den Bakker M, Riegman P. et al. Gene expression-based classification of non-small cell lung carcinomas and survival prediction. PLoS One. 2010;5:e10312
64. Talantov D. Novel Genes Associated with Malignant Melanoma but not Benign Melanocytic Lesions. Clinical Cancer Research. 2005;11:7234-42
65. Vanaja DK, Cheville JC, Iturria SJ, Young CY. Transcriptional silencing of zinc finger protein 185 identified by expression profiling is associated with prostate cancer progression. Cancer research. 2003;63:3877-82
Author contact
![]() Corresponding author: Hui-Ping Hsu (hphsuncku.edu.tw) and Ming-Derg Lai (a1211207ncku.edu.tw) Tel: +886-6-2353535 ext. 5272 (to Hui-Ping Hsu) and +886-6-2353535 ext. 5549 (to Ming-Derg Lai).
Corresponding author: Hui-Ping Hsu (hphsuncku.edu.tw) and Ming-Derg Lai (a1211207ncku.edu.tw) Tel: +886-6-2353535 ext. 5272 (to Hui-Ping Hsu) and +886-6-2353535 ext. 5549 (to Ming-Derg Lai).

 Global reach, higher impact
Global reach, higher impact