3.2
Impact Factor
ISSN: 1449-1907
Int J Med Sci 2020; 17(7):953-964. doi:10.7150/ijms.44377 This issue Cite
Research Paper
miR-19 regulates the expression of interferon-induced genes and MHC class I genes in human cancer cells
1. Guangdong Provincial Key Laboratory of Cancer Immunotherapy Research and Guangzhou Key Laboratory of Tumor Immunology Research, Cancer Research Institute, Southern Medical University, Guangzhou 510515, China
2. Zhongshan School of Medicine, Sun Yat-sen University, Guangzhou 510080, China
3. Institute of Comparative Medicine & Laboratory Animal Center, Southern Medical University, Guangzhou 510515, China
4. Department of Pharmacy, Nanfang Hospital, Southern Medical University, Guangzhou 510515, China
5. Radiotherapy Center, the First People's Hospital of Chenzhou, Chenzhou 423000, China
6. Department of Physiology, Faculty of Basic Medical Sciences, Guilin Medical University, Guilin 541004, China
7. Department of Pathology, the Second Affiliated Hospital, Guilin Medical University, Guilin 541199, China
*These authors contributed equally to this work.
Received 2020-1-30; Accepted 2020-3-19; Published 2020-4-6
Abstract
MicroRNA-19 (miR-19) is identified as the key oncogenic component of the miR-17-92 cluster. When we explored the functions of the dysregulated miR-19 in lung cancer, microarray-based data unexpectedly demonstrated that some immune and inflammatory response genes (i.e., IL32, IFI6 and IFIT1) were generally down-regulated by miR-19 overexpression in A549 cells, which prompted us to fully investigate whether the miR-19 family (i.e., miR-19a and miR-19b-1) was implicated in regulating the expression of immune and inflammatory response genes in cancer cells. In the present study, we observed that miR-19a or miR-19b-1 overexpression by miRNA mimics in the A549, HCC827 and CNE2 cells significantly downregulated the expression of interferon (IFN)-regulated genes (i.e., IRF7, IFI6, IFIT1, IFITM1, IFI27 and IFI44L). Furthermore, the ectopic miR-19a or miR-19b-1 expression in the A549, HCC827, CNE2 and HONE1 cells led to a general downward trend in the expression profile of major histocompatibility complex (MHC) class I genes (such as HLA-B, HLA-E, HLA-F or HLA-G); conversely, miR-19a or miR-19b-1 inhibition by the miRNA inhibitor upregulated the aforementioned MHC Class I gene expression, suggesting that miR-19a or miR-19b-1 negatively modulates MHC Class I gene expression. The miR-19a or miR-19b-1 mimics reduced the expression of interleukin (IL)-related genes (i.e., IL1B, IL11RA and IL6) in the A549, HCC827, CNE2 or HONE1 cells. The ectopic expression of miR-19a or miR-19b-1 downregulated IL32 expression in the A549 and HCC827 cells and upregulated IL32 expression in CNE2 and HONE1 cells. In addition, enforced miR-19a or miR-19b-1 expression suppressed IL-6 production by lung cancer and nasopharyngeal carcinoma (NPC) cells. Taken together, these findings demonstrate, for the first time, that miR-19 can modulate the expression of IFN-induced genes and MHC class I genes in human cancer cells, suggesting a novel role of miR-19 in linking inflammation and cancer, which remains to be fully characterized.
Keywords: miR-19a, miR-19b-1, lung cancer, nasopharyngeal carcinoma, MHC class I gene, interferon-inducible gene, interleukin-related gene
Introduction
MicroRNAs (miRNAs) are implicated in cancer initiation, progression, metastasis and angiogenesis [1]. The miR-17-92 gene cluster is frequently amplified in B-cell lymphomas and in a wide range of solid tumors, including lung and stomach cancers[1, 2]. The miR-19 family (including miR-19a and miR-19b-1) is considered a key component of the oncogenic miR-17-92 cluster[2].
Our previous study demonstrated that miR-19 triggered epithelial-mesenchymal transition (EMT), accompanied by the reduced proliferation of lung cancer cells[3]. When we explored the roles of dysregulated miR-19 in lung cancer, microarray-based gene expression data unexpectedly demonstrated a significant number of inflammatory and immune response genes up- or down-regulated by miR-19, including interferon(IFN)-stimulated genes such as GBP1, IFRD1, IFI35, IFI6 and PSMB9, and complement components, suggesting a strong relationship between miR-19 and inflammation in cancer. On the other hand, there are several lines of evidence that miR-19 family can act as regulators of inflammatory response in some diseases, including rheumatoid arthritis (RA)[4, 5], asthma[6], atherosclerosis[7, 8], nasal polyposis[9] and sepsis[10]. miR-17-92 was down-regulated in activated fibroblast-like synoviocytes (FLS) isolated from RA patients, and miR-19a and miR-19b-1 regulate Toll- like receptor 2 (TLR2) expression thereby reducing the inflammatory response induced by bacterial lipoprotein (BLP) in FLS and which is characterized by the secretion of IL-6 and Matrix metalloproteinases (MMP-3) [4, 11]. miR-19a upregulation in asthma is an indicator and a cause of increased TH2 cytokine production in the airways[6]. Moreover, the expression of interferon- regulated genes and MHC class I molecules are modulated by microRNAs, such as miR-9[12], miR-125a[13], miR-520b[14], let-7f-5p [15], let-7i-5p[15], miR-146b-5p[15] and miR-185-5p [15]. Our findings and published reports strongly indicate a novel role of miR-19 in linking inflammation and cancer, which remains to be fully characterized.
These aforementioned findings prompted us to fully explore whether the miR-19 family was involved in regulating the expression of IFN-related genes, major histocompatibility complex (MHC) class I genes and interleukin (IL)-related genes in cancer cells.
Materials and Methods
Cell line and cell culture
Human NPC cell lines (i.e., CNE2 and HONE1) were cultured in Roswell Park Memorial Institute 1640 medium (RPMI1640) (Corning) and lung cancer cell lines (i.e., A549 and HCC827) were cultured in Dulbecco's modified Eagle's medium (DMEM) (Corning), respectively, supplemented with 10% fetal bovine serum (FBS) (Biological Industries) in a humidified incubator with 5% CO2 at 37°C. All cells were approved by the Institutional Review Board of Southern Medical University.
miRNA transient transfection
Human miR-19 mimics (including miR-19a mimics and miR-19b-1 mimics), a nonspecific miRNA control (i.e., mimics control), human miR-19 inhibitors (including miR-19a inhibitor and miR-19b-1 inhibitor) and a nonspecific miRNA inhibitor control were purchased from Shanghai GenePharma Co., Ltd. (Shanghai, China). miRNA inhibitor which is a chemically modified single strand of RNA can sequester miRNAs avoiding their targeting to coding genes of endogenous target miRNA. miRNAs were transiently transfected into cells at a working concentration of 100 nmol/L using Lipofectamine 2000 reagent (Invitrogen) in accordance with the manufacturer's procedure. Cancer cells (i.e., A549, HCC827, CNE2 or HONE1 cells) were transfected with miR-19 mimics (100 nM) or miR-19 inhibitor (100 nM) for 48h, respectively, followed by evaluating the expression of the indicated IFN-inducible genes, MHC class I genes or IL-related genes via qRT-PCR.
Cytokine production assay
Lung cancer cells (A549 and HCC827) and NPC cells (CNE2 and HONE1) were transfected with 100 nM miR-19 mimics or its mimics control. 24 hours and 48 hours after culture, cell culture supernatants were collected and assayed for supernatant concentrations of IL6 by ELISA kit (MultiSciences, China) according to the manufacturer's instructions.
RNA isolation, reverse transcription and qRT-PCR
For miRNA and mRNA analyses, total RNA from cancer cells was extracted with Trizol Reagent (TaKaRa). Total RNA was reversely transcribed with the PrimeScript RT reagent Kit (TaKaRa). The expression levels of mature miRNA were determined by SYBR Green quantitative PCR amplifications performed on the Stratagene Mx3005P Real-Time PCR system (Agilent Technologies, Inc., USA). U6 was used for internal reference genes for miRNA[16]. Expression of mRNA analysis was performed using SYBR Green Master Mix (TaKaRa), and GAPDH was used for normalization. The primers used for the amplification of the indicated genes were listed in Table 1-4.
mRNA microarray analysis
As described in our paper[3], miR-19- and vector-expressing A549 cells were analyzed by Affymetrix arrays. Expression microarray analysis was carried out with commercially available Affymetrix Human Gene U133 Plus 2.0 array according to the Affymetrix standard protocol, which carries 47,000 transcripts representing 38,500 well-characterized human genes. Data analysis was performed using the Significance Analysis of Microarray software (SAM 3.0, Stanford University, USA; http://www-stat. stanford.edu). Heatmap is plotted using pheatmap R package.
Statistical analysis
Data were presented as mean±SD unless otherwise indicated of at least 3 independent experiments. Statistical analysis was performed using a SPSS 16.0 software (SPSS, North Chicago, IL) software package. Statistical significance was assessed by one-way analysis of variance (ANOVA) (*P<0.5; #P<0.01).
Results
miR-19 overexpression and downregulated endogenous miR-19 expression in cancer cells
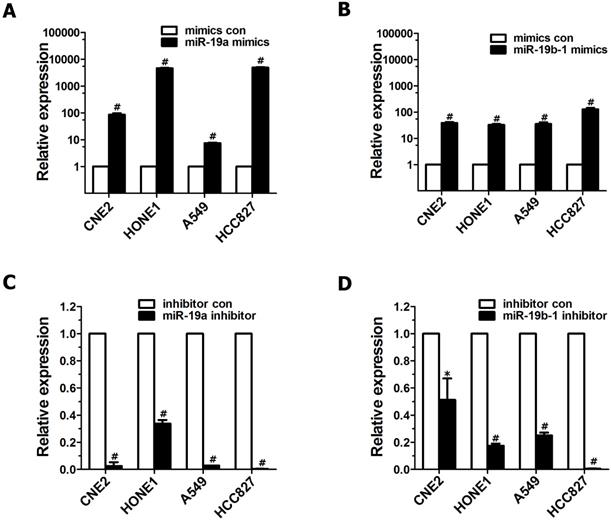
To explore the roles of miR-19 dysregulation in the pathogenesis of lung cancer and NPC, two methods were employed to overexpress miR-19 in cancer cells. First, A549 cells expressing both miR-19a and 19b-1 transgenes were simultaneously generated as described in our previous publication[3]. Next, miR-19a or miR-19b-1 mimics were transiently transfected into the indicated cancer cells. As shown in Figure 1, the levels of miR-19a (Figure 1A) or miR-19b-1 (Figure 1B) in indicated cells transfected with miR-19a or miR-19b-1 mimics were much higher than those in cancer cells transfected with control mimics. Moreover, the levels of miR-19a (Figure 1C) or miR-19b-1 (Figure 1D) in the indicated cancer cells transfected with the miR-19a or miR-19b-1 inhibitor were much lower than those in cells transfected with the control inhibitor.
Global analysis of gene expression changes in miR-19-expressing A549 cells
The global gene expression changes induced by miR-19 overexpression were determined by comparing the gene expression profiles between miR-19- and vector-expressing A549 cells based on microarray data. We found that a total of 352 and 501 genes were significantly downregulated and upregulated, respectively, by miR-19 overexpression [3]. Unexpectedly, among the 853 significantly changed genes, we found that some of the differentially expressed genes were involved in regulating immune and/or inflammatory responses, including IFN-inducible genes (Figure 2A, and Table 5) and IL-related genes (Figure 4A). Table 6 lists the GO terms representing biological processes related to immune and inflammatory responses. In summary, the microarray data suggest that miR-19 plays a significant role in regulating the expression of genes involved in immune and inflammatory responses.
Primers for qRT-PCR analysis of human miR-19
| Primer name | Primer sequence |
|---|---|
| U6 snRNA-RT | AACGCTTCACGAATTTGCGT |
| U6 snRNA-forward primer | CTCGCTTCGGCAGCACA |
| U6 snRNA-reverse primer | AACGCTTCACGAATTTGCGT |
| miR-19-RT-primer | GTCGTATCCAGTGCAGGGTCCGAGGTATTCGCACTGGATACGACTCAGT |
| miR-19a-forward primer | TGTGCAAATCTATGCAAA |
| miR-19a-reverse primer | GTGCAGGGTCCGAGGTATTC |
| miR-19b-1-forward primer | TGTGCAAATCCATGCAAA |
| miR-19b-1-reverse primer | GTGCAGGGTCCGAGGTATTC |
Primers for qRT-PCR analysis of interferon-regulated genes
| Gene | Forward primer (5'-3') | Reverse primer (5'-3') |
|---|---|---|
| IFI6 | GGTCTGCGATCCTGAATGGG | TCACTATCGAGATACTTGTGGGT |
| IFI16 | AGACTGAAGACTGAACCTGAAGA | GAACCCATTGCGGCAAACATA |
| IFI27 | TGCTCTCACCTCATCAGCAGT | CACAACTCCTCCAATCACAACT |
| IFI35 | GTGGACGTTCGGGAGCTAC | ACTGGCCGATTTGGCACAG |
| IFI44L | AGCCGTCAGGGATGTACTATAAC | AGGGAATCATTTGGCTCTGTAGA |
| IFIT1 | TTGATGACGATGAAATGCCTGA | CAGTCACCAGACTCCTCAC |
| IFIT2 | AAGCACCTCAAAGGGCAAAAC | TCGGCCCATGTGATAGTAGAC |
| IFITM1 | CCAAGGTCCACCGTGATTAAC | ACCAGTTCAAGAAGAGGGTGTT |
| IRF1 | ATGCCCATCACTCGGATGC | CCCTGCTTTGTATCGGCCTG |
| IRF7 | CCCACGCTATACCATCTACCT | GATGTCGTCATAGAGGCTGTTG |
| GAPDH | ACAACTTTGGTATCGTGGAAGG | GCCATCACGCCACAGTTTC |
Primers for qRT-PCR analysis of MHC class I genes
| Gene | Forward Primer (5'-3') | Reverse Primer (5'-3') |
|---|---|---|
| HLA-B | CAGTTCGTGAGGTTCGACAG | CAGCCGTACATGCTCTGGA |
| HLA-E | TTCCGAGTGAATCTGCGGAC | GTCGTAGGCGAACTGTTCATAC |
| HLA-F | TGGCCCTGACCGATACTTG | GCAGGAATTGCGTGTCGTC |
| HLA-G | GAGGAGACACGGAACACCAAG | GTCGCAGCCAATCATCCACT |
Primers for qRT-PCR analysis of interleukin-related genes
| Gene | Forward Primer (5'-3') | Reverse Primer (5'-3') |
|---|---|---|
| IL1B | CAGCTACGAATCTCCGACCAC | GGCAGGGAACCAGCATCTTC |
| IL6 | ACTCACCTCTTCAGAACGAATTG | CCATCTTTGGAAGGTTCAGGTTG |
| IL11RA | CAGCCAGATCAGCGGTTTAC | AGATGCTCTGCAAGCTCACAT |
| IL20RB | GGCCACTGTGCCATACAAC | TCTTTGGTGATCTCCATCCCA |
| IL32 | AGCTGGAGGACGACTTCAAA | AGAGCAGCAGAAACTCTGGA |
miR-19 overexpression and down-regulated endogenous miR-19 expression in human cancer cells. qRT-PCR-based assay was used to analyze the expression levels of miR-19 after human lung adenocarcinoma cell lines (A549 and HCC827) and NPC cell lines(CNE2 and HONE1) transfected with miR-19 mimics (100 nM) (A-B) or miR-19 inhibitor (100 nM) (C-D) for 48 hours (h). *P<0.5, #P<0.01 by one-way ANOVA.
Differentially expressed interferon-regulated genes
| Gene symbol | Annotation | Fold change |
|---|---|---|
| C3 | complement component 3 | 2.0557 |
| GBP1 | guanylate binding protein 1, interferon-inducible, 67kDa | 2.0482 |
| IFRD1 | interferon-related developmental regulator 1 | 1.9062 |
| IRF9 | interferon regulatory factor 9 | 1.5356 |
| IFNGR1 | interferon gamma receptor 1 | 1.4824 |
| IRF1 | interferon regulatory factor 1 | 1.1651 |
| IRF7 | interferon regulatory factor 7 | 1.0475 |
| IFI16 | interferon, gamma-inducible protein 16 | 0.8606 |
| IFITM2 | interferon induced transmembrane protein 2 (1-8D) | 0.8439 |
| PSMB10 | proteasome (prosome, macropain) subunit, beta type, 10 | 0.7610 |
| IRF9 /// RNF31 | interferon regulatory factor 9 /// ring finger protein 31 | 0.7605 |
| IFI35 | interferon-induced protein 35 | 0.6960 |
| PRKRA | protein kinase, interferon-inducible double stranded RNA dependent activator | 0.6525 |
| IFI6 | interferon, alpha-inducible protein 6 | 0.6417 |
| IFIT1 | interferon-induced protein with tetratricopeptide repeats 1 | 0.6346 |
| IFIT3 | interferon-induced protein with tetratricopeptide repeats 3 | 0.6016 |
| PSMB9 | proteasome (prosome, macropain) subunit, beta type, 9 (large multifunctional peptidase 2) | 0.5405 |
| PSMB8 | proteasome (prosome, macropain) subunit, beta type, 8 (large multifunctional peptidase 7) | 0.5074 |
| IRF2BP2 | interferon regulatory factor 2 binding protein 2 | 0.4720 |
| C1RL | complement component 1, r subcomponent-like | 0.4445 |
| C5 | complement component 5 | 0.2388 |
Gene ontology analysis of up- and down-regulated genes (related with immune and inflammation) from miR-19-expressing A549 cells to vector-expressing A549 cells
| GO ID | Biological process | Count | p-value | q-value |
|---|---|---|---|---|
| GO:0006954 | inflammatory response | 20 | 1.83E-25 | 5.78E-25 |
| GO:0006958 | complement activation, classical pathway | 4 | 1.60E-07 | 1.01E-07 |
| GO:0045087 | innate immune response | 4 | 1.00E-04 | 2.77E-05 |
| GO:0006955 | immune response | 8 | 1.21E-04 | 3.28E-05 |
| GO:0006957 | complement activation, alternative pathway | 2 | 2.74E-04 | 6.69E-05 |
| GO:0030101 | natural killer cell activation | 2 | 5.45E-04 | 1.19E-04 |
| GO:0002842 | positive regulation of T cell mediated immune response to tumor cell | 1 | 0.003256 | 4.26E-04 |
| GO:0002378 | immunoglobulin biosynthesis | 1 | 0.003256 | 4.26E-04 |
| GO:0002862 | negative regulation of inflammatory response to antigenic stimulus | 1 | 0.00488 | 5.70E-04 |
| GO:0002863 | positive regulation of inflammatory response to antigenic stimulus | 1 | 0.006502 | 6.77E-04 |
| GO:0050729 | positive regulation of inflammatory response | 1 | 0.033668 | 0.002274 |
| GO:0045089 | positive regulation of innate immune response | 1 | 0.036815 | 0.002446 |
| GO:0002768 | immune response-regulating cell surface receptor signaling pathway | 1 | 0.044637 | 0.002855 |
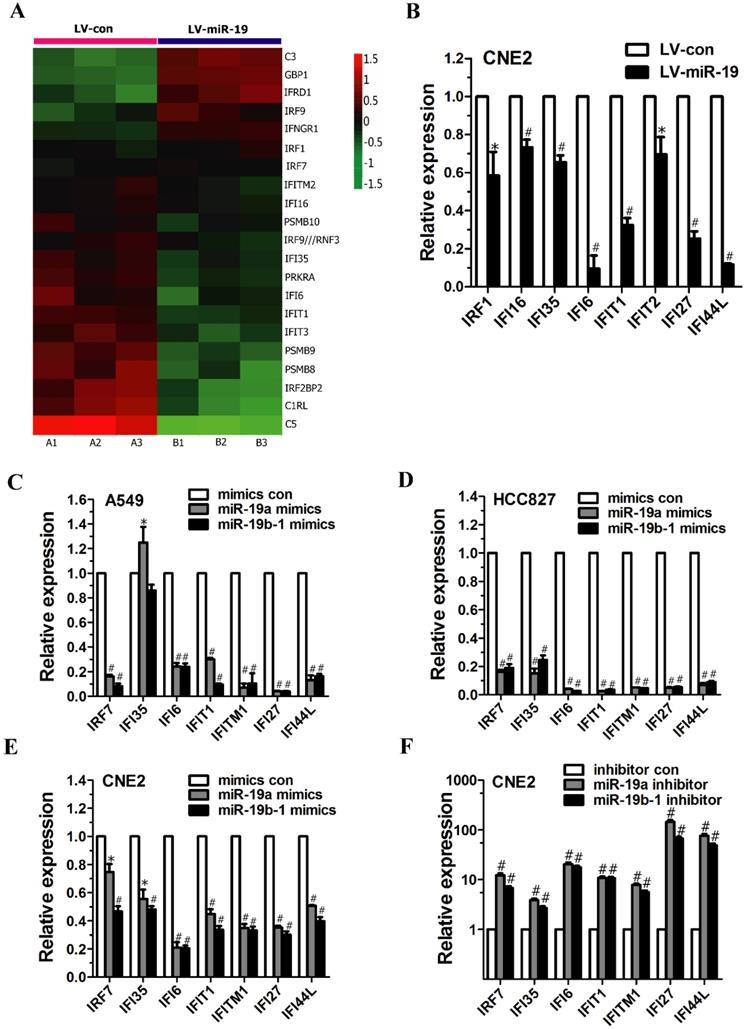
miR-19 regulated the expression of IFN-related genes in cancer cells. (A) Class comparison and hierarchical clustering of differentially expressed IFN-regulated genes between miR-19- and vector-expressing A549 cells. In the cluster heatmap of IFN-inducible gene, A1, A2 and A3 represented the total RNA isolated from different generations of vector-expressing A549 cells, and B1, B2 and B3 represented the total RNA isolated from different generations of miR-19-expressing A549 cells. Genes with increased and reduced expressions are shown in red and green, respectively. (B) qRT-PCR analyzed IFN-regulated gene expression in CNE2 cells expressing miR-19a and 19b-1 transgenes simultaneously. To generate stable cell line, the recombinant lentiviruses (i.e., LV-con and LV-miR-19) were used to infect CNE2 cells to generate vector- and miR-19-expressing CNE2 cells. (C-D) miR-19 modulated IFN-related gene expression in A549 (C) and HCC827 (D) cells. (E-F) miR-19 regulated IFN-related gene expression in CNE2 cells. *P<0.5, #P<0.01 by one-way ANOVA.
miR-19 altered IFN-regulated gene expression in cancer cells
As mentioned above, microarray analysis revealed the altered expression of IFN-regulated genes (e.g., GBP1, IRF9, IFI35, PRKRA, IFI6, IFIT1, IFIT3, PSMB9 and IRF2BP2) (Figure 2A). Moreover, qRT-PCR analysis demonstrated the significantly decreased expression of IFN-regulated genes (e.g., IRF1, IFI16, IFI35, IFI6, IFIT1, IFIT2, IFI27 and IFI44L) in CNE2 cells expressing both miR-19a and 19b-1 transgenes (Figure 2B).
To characterize the effects of the individual components of the miR-19 family on the IFN-regulated target genes, cancer cells were transfected with miR-19a (Figure 1A) and miR-19b-1 mimics (Figure 1B). As shown in Figure 2C and D, miR-19a and miR-19b-1 mimics significantly downregulated the expression of IFN-regulated genes (i.e., IRF7, IFI6, IFIT1, IFITM1, IFI27 and IFI44L) in both A549 and HCC827 cells. Next, we further explored whether miR-19 overexpression by miR-19a or miR-19b-1 mimics alters the expression of IFN-regulated genes in other cancer cells. qRT-PCR data revealed a general downward trend in the expression of IFN-regulated genes (i.e., IRF7, IFI35, IFI6, IFIT1, IFITM1, IFI27 and IFI44L) in CNE2 cells transfected with miR-19a or miR-19b-1 mimics (Figure 2E); conversely, endogenous miR-19 inhibition by the miR-19a or miR-19b-1 inhibitor in CNE2 cells correspondingly resulted in the upregulated expression of the aforementioned IFN-regulated genes (Figure 2F). Overall, miR-19a or miR-19b-1 overexpression leads to a general downward trend in the expression profile of the aforementioned genes involved in IFN induction in cancer cells.
Suppression of MHC class I gene expression by miR-19 in cancer cells
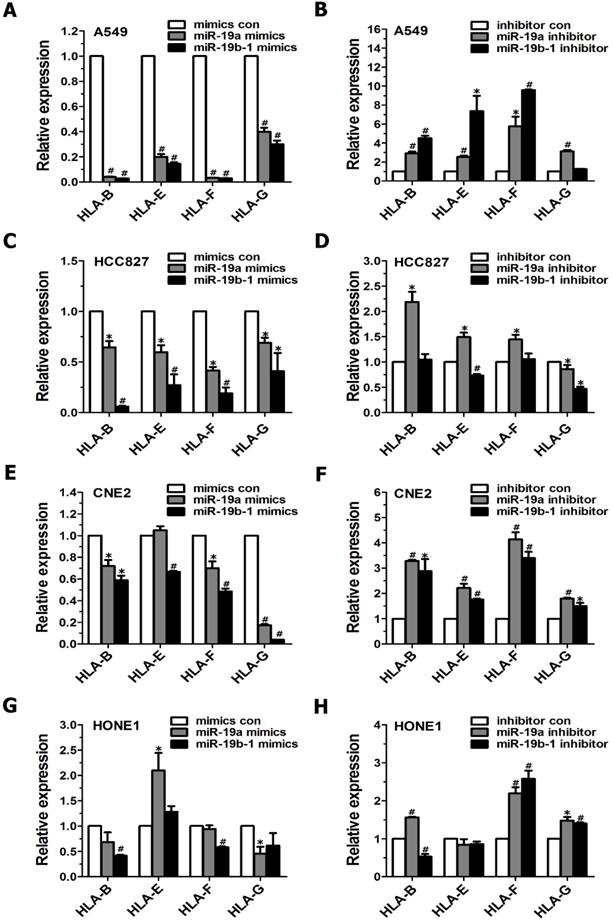
Although MHC class I molecules are constitutively expressed in essentially all nucleated cells, the cumulative evidence shows that tumor cells display downregulated MHC class I antigens, which enables them to evade immune surveillance[17]. Therefore, we further examined the influence of the miR-19 family on MHC class I gene expression in cancer cells. When cancer cells (i.e., A549, HCC827, CNE2 and HONE1 cells) were transiently transfected with miR-19a or miR-19b-1 mimics, qRT-PCR data revealed a general downward trend in the expression profile of the MHC Class I genes (such as HLA-B, HLA-F, HLA-G) (Figure 3A, C, E, G). Conversely, these tumor cells, when transfected with the miR-19a or miR-19b-1 inhibitor, showed a general upward trend in the indicated MHC Class I genes (Figure 3B, D, F, H). Taken together, the most significant change after the enforced miR-19a or miR-19b-1 expression in cancer cells was the generally decreased expression of the abovementioned MHC Class I genes.
miR-19 overexpression enhanced or reduced IL-related gene expression in cancer cells
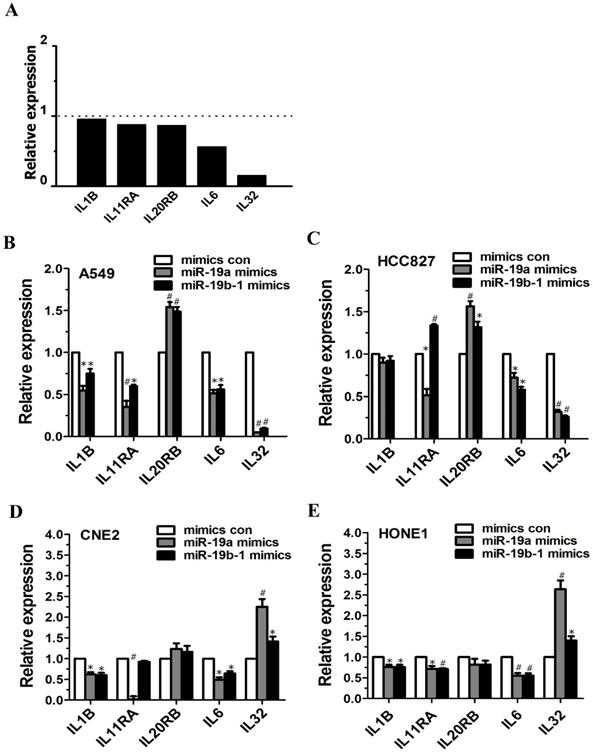
Microarray data demonstrated altered expression of IL6 (fold change: 0.5616) and IL32 (fold change: 0.1536) (Figure 4A). miR-19a or miR-19b-1 overexpression led to the downregulation of IL1B expression in A549, CNE2 and HONE1 cells (Figure 4B, D, E), and miR-19a overexpression reduced IL11RA expression in A549, HCC827, CNE2 and HONE1 cells (Figure 4B, C, D, E). The ectopic expression of miR-19a or miR-19b-1 upregulated IL20RB expression in A549 and HCC827 cells (Figure 4B, C) but not in CNE2 and HONE1 cells (Figure 4D, E). Moreover, miR-19a or miR-19b-1 overexpression reduced IL6 expression in the lung cancer and NPC cells examined (Figure 4B, C, D, E). miR-19a or miR-19b-1 mimics significantly downregulated IL32 expression in A549 and HCC827 cells (Figure 4B, C) and remarkably upregulated IL32 expression in CNE2 and HONE1 cells (Figure 4D, E). Together, ectopic miR-19a or miR-19b-1 expression alters IL-related gene expression.
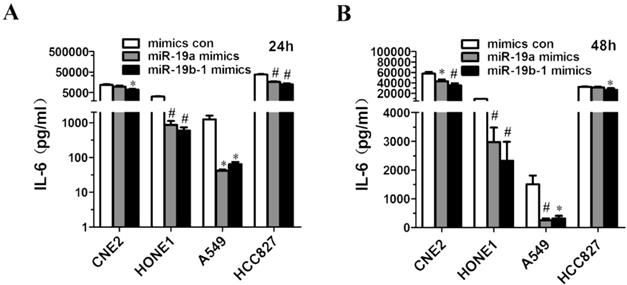
miR-19 overexpression resulted in decreased IL6 production by cancer cells
Given that miR-19a or miR-19b-1 mimics suppressed IL6 expression in lung cancer and NPC cells (Figure 4B, C, D, E), we further investigated the possibility that ectopic miR-19a or miR-19b-1 expression could affect IL6 production by these cancer cells. Strikingly, a marked decrease in the production of IL6 was observed in A549, HCC827, CNE2 and HONE1 cells 24 h (Figure 5A) and 48 h (Figure 5B) after mimic transfection. In summary, these findings demonstrate that miR-19 can modulate the IL6 production in cancer cells.
Discussion
Over the last few years, there has been increasing evidence that miR-19a or miR-19b-1 positively regulates cancer cell proliferation and tumorigenesis [18-20], stemness[21], and EMT, invasion and metastasis[18-20] by targeting different targets, suggesting that miR-19 plays oncogenic roles in cancer progression. Our microarray data unexpectedly revealed immune and inflammatory response genes down- or upregulated by miR-19 overexpression in lung cancer, which encouraged us to investigate the functions of onc-miR-19 in linking inflammation and cancer.
The miR-17-92 cluster regulates a broad spectrum of biological processes of T cell immunity, and is found to facilitate T cell proliferation, enhance antitumor activities and promote T cell-dependent antibody responses[22]. Moreover, some studies revealed the roles of miR-17-92 in the development of autoimmune diseases[22, 23]. miR-17-92 which is part of the CD28 costimulatory network regulated IL-10 production by Foxp3+ Tregs and control of experimental autoimmune encephalomyelitis (EAE)[23]. T cell-specific miR-17-92 deficiency reduced TH17 differentiation, which consequently ameliorated EAE symptoms[22]. Further studies demonstrated that miR-19b-1 is one of miRNAs in miR-17-92 cluster responsible for promoting TH17-mediated inflammation by targetedly repressing PTEN expression, thereby augmenting the PI3K-AKT-mTOR axis essential for proper TH17 differentiation[22].
miR-19 modulated the expression of MHC class I genes in cancer cells. Cancer cells were transfected with miR-19 mimics (100 nM)(A,C,E,G) or miR-19 inhibitor (100 nM)(B,D,F,H) for 48h, respectively, followed by evaluating MHC class I gene expression via qRT-PCR.
miR-19 overexpression enhanced or reduced the IL-related genes expression in cancer cells. (A) Graph illustrating the fold change in gene expression of representative differentially IL-related genes between LV-miR-19-infected A549 cells to LV-con-infected A549 cells. The horizontal dashed line marks a fold change of 1 (no change). (B-E) miR-19a or miR-19b-1 overexpression altered the IL-related gene expression in cancer cells. *P<0.5, #P<0.01 by one-way ANOVA.
miR-19a or miR-19b-1 overexpression resulted in the decreased IL6 production by cancer cells. The indicated cells were transfected with 100 nM miR-19 mimics or its mimics control, and incubated for 24h (A) and 48h (B). Supernatants were collected and assayed for IL6 levels. *P<0.5, #P<0.01 by one-way ANOVA.
Increasing evidence that miR-19 family (i.e., miR-19a and miR-19b-1) has been highly involved in immunity[4] and inflammatory diseases, including RA[4, 5, 11], asthma[6, 24, 25], atherosclerosis[7, 8], diabetic retinopathy[26], nasal polyposis[9], sepsis [10], encephalitis[27] and Crohn's disease[28-31]. miR-19b-1 positively regulated NF-κB signaling, which is one of the critical promoters of inflammation, while the positive regulation of NF-κB signaling by miR-19b-1 involves the coordinated suppression of negative regulators of NF-κB signaling, including A20/Tnfaip3, Rnf11, Fbxl11/Kdm2a and Zbtb16[4]. miR-17-92 was down-regulated in activated fibroblast-like synoviocytes (FLS) isolated from RA patients (RAFLS), miR-19b-1 enhanced basal IL6 and IL8 secretion by RAFLS to increase basal inflammation, and miR-19a and miR-19b-1 regulate IL6 and MMP-3 release by controlling TLR2 expression[4, 5, 11]. Elevated expression of miR-19a in human airway-infiltrating T cells of patients with asthma, and miR-19a promoted TH2 cytokine production in the airways and amplified inflammatory signaling by direct targeting of the inositol phosphatase PTEN, the signaling inhibitor SOCS1 and the deubiquitinase A20 [24], while miR-19b-1 reduced airway remodeling, airway inflammation and degree of oxidative stress by inhibiting Stat3 signaling through TSLP downregulation in a mouse asthma model[25]. Moreover, the endothelial hypoxia-inducible factor-1α promoted atherosclerosis and monocyte recruitment by upregulating miR-19a[7], while TNF-α or IFN-γ or IL-4 suppressed IL-10 in B cells (from patients with atherosclerosis) via upregulating miR-19a expression [8]. miR-19a negatively interfered with IL-10 expression in peripheral dendritic cells (DCs) of patients with nasal polyposis[19]. miR-19a directly regulated TNF-α expression in ulcerative colitis[28], and miR-19b-1 suppressed the inflammatory response by inhibiting SOCS3 to modulate chemokine production in intestinal epithelial cells (IECs) and thereby prevented the pathogenesis of Crohn's disease (CD)[29]. Japanese encephalitis virus-mediated inflammation via miR-19b-1, which plays important roles in immunity and inflammatory diseases.
Chronic inflammation is believed to have a crucial role in cancer development, and inflammation not only works as a tumor-promoting agent but also influences other steps of tumorigenesis by inducing DNA damage, angiogenesis, invasion and metastasis [32, 33]. There is increasing evidence that miRNAs drive tumor progression by regulating inflammation [34, 35]. The inhibition of miR-9, which was overexpressed in Hodgkin lymphoma, decreased the production of cytokines (i.e., TNF-α, CCL-5, IL-6 and IL-5) from L428 and L540 cells of Hodgkin lymphoma, followed by an impaired ability to attract normal inflammatory cells and then impairing tumor outgrowth in vivo[36]. Additionally, our previous study revealed that the most significant change following tumor suppressor miR-9 overexpression in NPC cells was the decreased expression of IL-related genes, including IL1B, IL11, IL1F8, IL1A, IL6 and IL7R [12]. miR-19a promoted colitis and colitis-associated colon cancer by downregulating TNFAIP3 in a targeted manner and constitutively activating NF-κB signaling[27]. miRNA-19a/b-1 exhibits oncogenic activity through negatively regulating SOCS3 via the JAK-STAT pathway together with the increased activation of SOCS3, IL6 and STAT3[37]. In this study, we revealed that ectopic expression of miR-19a or miR-19b-1 modulated the expression of IL-related genes (including IL1B, IL11RA, IL20RB, IL6 and IL32) and suppressed IL6 production in lung cancer and NPC cells. We found that miR-19a or miR-19b-1 overexpression resulted in decreased IL1B expression in lung cancer and NPC cells, while microglia-derived IL1B triggered p53-mediated cell cycle arrest and apoptosis in neural precursor cells[38], indicating that reduced IL1B expression induced by onco-miR-19 might contribute to cancer progression. In summary, these findings strongly indicate a novel role of miR-19 in linking inflammation and cancer, which remains to be characterized.
IL32 is a novel cytokine involved in inflammation and cancer development[39]. Various published data demonstrated that IL32 promotes or decreases tumor development[39]. For example, IL32 contributes to gastric cancer progression by increasing the metastatic potential resulting from AKT, β-catenin and HIF-1α activation[40]. IL-32 inhibits tumor growth via inhibition of NF-κB and STAT3 signals in colon and prostate cancer[39]. In this study, we found that miR-19a or miR-19b-1 overexpression remarkably downregulated IL32 expression in lung cancer cells and led to an increase in IL32 expression in NPC cells. These findings from this study and other published reports strongly suggest that modulating IL32 expression by miR-19 might contribute to the pathogenesis of lung cancer and NPC, which remains to be examined.
IL6, a multifunctional cytokine, is involved in the host immune defense mechanism as well as the modulation of growth and differentiation in various malignancies[41]. The deregulated overexpression of IL6 is associated with tumor progression through inhibition of cancer cell apoptosis, stimulation of angiogenesis and drug resistance[41]. Clinical studies revealed that increased serum IL6 concentrations in patients are associated with advanced tumor stages of various cancers (e.g., lung and colorectal cancers) and short survival in patients[41]. Thus, blocking IL6 signaling is a potential therapeutic strategy for cancers (i.e., anti-IL6 therapy) characterized by pathological IL6 overproduction. miR-19a and miR-19b-1 repressed the release of the cytokines IL6 and MMP-3 in BLP-activated RAFLS by controlling TLR2 expression, suggesting that miR-19a/b-1 can act as negative regulators of inflammation in humans[11]. Additionally, our present study revealed that miR-19a or miR-19b-1 overexpression reduced IL6 expression and production in cancer cells, which contradicts the pathological IL6 overproduction observed in various cancers described above. In summary, the aforementioned findings encourage us to further explore how the decreased IL6 expression and production resulting from the miR-19 overexpression in lung cancer and NPC cells contributes to cancer progression.
Increasing evidence supports that the IFN-inducible genes that are expressed in tumor cells regulate cancer cell proliferation and tumorigenesis[42] and metastasis[43]. IFITM1 is a negative regulator of cell proliferation and tumorigenesis in hepatocellular carcinoma (HCC), while the anti-proliferative action of interferon-γ is mediated by IFITM1[42]. The silencing of IRF7 in breast cancer cells promoted bone metastasis through tumor immune escape, while the IRF7-driven suppression of metastasis was reliant on IFN signaling to host immune cells [43]. In this study, we revealed for the first time that the ectopic expression of miR-19a or miR-19b-1 in cancer cells significantly downregulated the expression of PSMB8, PSMB9 and IFN-regulated genes (i.e., IRF7, IFI6, IFIT1, IFITM1, IFI27 and IFI44L), which have never been reported in other physiological and pathological processes. Additionally, the expression of IFN-induced protein family members with tetratricopeptide repeats (IFIT), including IFIT1, IFIT2, IFIT3 and IFIT5, was decreased in HCC tissues[44]. Higher IFIT3 expression in HCC tissues predicts a better response to IFN-a therapy in HCC patients, and IFIT3 promotes IFN-a effector responses and therapeutic effects by strengthening IFN-a effector signaling in HCC[44]. In summary, these findings from this study and other published reports strongly suggest that modulating the expression of IFN-inducible genes in cancer cells by miR-19 might contribute to the pathogenesis of lung cancer and NPC, which remains to be thoroughly investigated.
MHC class I molecules, including the classical MHC class I molecules [i.e., HLA(human leukocyte antigen)-A, HLA-B and HLA-C] and non-classical MHC class I molecules (i.e., HLA-E, HLA-F, HLA-G, HLA-H and HLA-J), are constitutively expressed in essentially all nucleated cells[17]. Many cancer cells display downregulated MHC class I antigen (MHC-I), which enables them to evade immune surveillance, while the downregulation of HLA class I expression contributes to a poor prognosis in cancer patients [17]. The MHC class I promoter is known to be activated by several transcription factors, including CIITA[45-48] and USF1[49, 50]. CXCR4-mediated cell surface MHC-I downregulation in cancer progression facilitated tumor evasion of immune surveillance[51]. Kinases, including MAP2K1 (MEK1), EGFR and RET, were validated as negative regulators of human HLA expression in multiple cancer types, and activated MAPK signaling in mouse tumors in vivo suppressed components of MHC-I, while the pharmacologic inhibition of MAPK signaling led to improved peptide/MHC target recognition and killing by T cells and TCR-mimic antibodies[52]. EGFR tyrosine kinase inhibitors augmented the expression of MHC-I and MHC-II molecules in primary and malignant human keratinocytes[53]. The inhibition of the MAPK pathway induced the upregulation of HLA-A expression and enhanced the sensitivity of targeted tumors to Ag-specific CTL lysis in esophageal and gastric cancer[54]. Additionally, miR-9, which functions as a tumor suppressor in NPC[55], positively regulated the expression of MHC class I genes (such as HLA-B and HLA-F) in NPC[12]. The previous studies revealed that the non-classical HLA-F and HLA-G act as the important mediators of immune escape[56, 57]. This study demonstrated that miR-19a or miR-19b-1 overexpression in cancer cells led to a general downward trend in the expression profile of MHC Class I molecules (such as HLA-B, HLA-E, HLA-F, HLA-G or HLA-J), which has never been reported in other physiological and pathological processes. As described in the discussion section, miR-19a and miR-19b-1 function as oncomiR in various cancer progressions. Taken together, these findings shed new light on the role of the miR-19 family in tumor evasion of immune surveillance by inducing the downregulation of cell surface HLA class I expression, which remains to be fully characterized.
Conclusion
We have demonstrated that miR-19 can regulate IL-related gene expression in lung cancer cells and NPC cells, and we have found for the first time that miR-19 overexpression and/or inhibition alter the expression of IFN-induced genes and MHC class I genes in cancer cells. However, the pathological consequences of the altered expression of immune- or inflammatory-related genes by miR-19 in cancer cells remain to be examined. Future studies are required to clarify the potential contribution of miR-19 to the proliferation, EMT, invasion, metastasis or tumor angiogenesis of cancer cells by playing critical roles in modulating the aforementioned genes involved in immune and inflammation responses. Collectively, we suspect that miR-19 might function as an oncomiR to promote tumor progression by regulating the expression of the above-mentioned genes involved in immunity and inflammation.
Abbreviations
qRT-PCR: quantitative real-time polymerase chain reaction; IFN: interferon; MHC: major histocompatibility complex; IL: interleukin; NPC: nasopharyngeal carcinoma.
Acknowledgements
We thank Prof. Andrea Ventura (Memorial Sloan Kettering Cancer Center) and Prof. Jeng-Shin Lee (Harvard Gene Therapy Initiative, Harvard Medical School) for generously providing plasmids. This work was supported by the National Natural Science Foundation of China (Grant No. 81872209, 81672689, 81372896 and 81172587, to D. Xiao; Grant No. 81600086 and 81770100, to Y. Sun; Grant No. 81600488 and 81870602, to X.-L. Lin; Grant No. 81702778, to J.S. Jia; Grant No. 81560441 and 81760491, to S.J. Xiao; Grant No. 81660485, to X.L. Zhang), the Natural Science Foundation of Guangdong Province of China (Grant No. 2014A030313294 to D. Xiao), the Science and Technology Planning Project of Guangdong Province of China (Grant No. 2009B060300008, 2013B060300013 and 2017A010105017, to D. Xiao; Grant No. 2017A030303018, to J.S. Jia; Grant No. 2015A030302024, to X.-L. Lin), the China Postdoctoral Science Foundation (Grant No. 2015M572338, 2016T90792, 2017M622740 and 2018T110884, to X.-L. Lin) and the Medical Scientific Research Foundation of Guangdong Province of China (Grant No. A2017420, to J.S. Jia).
Author contributions
D Xiao and Y Sun conceived and designed the study; J Li, TY Lin, L Chen, Y Liu, MJ Dian, WC Hao, XL Lin, XY Li, YL Li, M Lian, HW Chen, JS Jia, SJ Xiao, XL Zhang performed the experiments; D Xiao, Y Sun and J Li analyzed the data; XL Lin and SJ Xiao contributed essential reagents or tools; D Xiao, Y Sun, J Li, SJ Xiao and TY Lin wrote the paper. All authors contributed to data analysis, drafting and revising the article, gave final approval of the version to be published, and agree to be accountable for all aspects of the work.
Competing Interests
The authors have declared that no competing interest exists.
References
1. Rupaimoole R, Slack FJ. MicroRNA therapeutics: towards a new era for the management of cancer and other diseases. Nature reviews Drug discovery. 2017;16:203-22
2. van Haaften G, Agami R. Tumorigenicity of the miR-17-92 cluster distilled. Genes & Development. 2010;24:1-4
3. Li J, Yang S, Yan W, Yang J, Qin YJ, Lin XL. et al. MicroRNA-19 triggers epithelial-mesenchymal transition of lung cancer cells accompanied by growth inhibition. Laboratory investigation; a journal of technical methods and pathology. 2015;95:1056-70
4. Gantier MP, Stunden HJ, McCoy CE, Behlke MA, Wang D, Kaparakis-Liaskos M. et al. A miR-19 regulon that controls NF-kappaB signaling. Nucleic acids research. 2012;40:8048-58
5. Philippe L, Alsaleh G, Bahram S, Pfeffer S, Georgel P. The miR-17 ∼ 92 Cluster: A Key Player in the Control of Inflammation during Rheumatoid Arthritis. Frontiers in immunology. 2013 4
6. Simpson LJ, Patel S, Bhakta NR, Choy DF, Brightbill HD, Ren X. et al. A microRNA upregulated in asthma airway T cells promotes TH2 cytokine production. 2014; 15: 1162-70.
7. Akhtar S, Hartmann P, Karshovska E, Rinderknecht FA, Subramanian P, Gremse F. et al. Endothelial Hypoxia-Inducible Factor-1alpha Promotes Atherosclerosis and Monocyte Recruitment by Upregulating MicroRNA-19a. Hypertension (Dallas, Tex: 1979). 2015;66:1220-6
8. Ren ZQ, Liu N, Zhao K. Micro RNA-19a suppresses IL-10 in peripheral B cells from patients with atherosclerosis. Cytokine. 2016;86:86-91
9. Luo XQ, Shao JB, Xie RD, Zeng L, Li XX, Qiu SQ. et al. Micro RNA-19a interferes with IL-10 expression in peripheral dendritic cells of patients with nasal polyposis. Oncotarget. 2017;8:48915-21
10. Jiang Y, Zhou H, Ma D, Chen ZK, Cai X. MicroRNA-19a and CD22 Comprise a Feedback Loop for B Cell Response in Sepsis. Medical science monitor: international medical journal of experimental and clinical research. 2015;21:1548-55
11. Philippe L, Alsaleh G, Suffert G, Meyer A, Georgel P, Sibilia J. et al. TLR2 expression is regulated by microRNA miR-19 in rheumatoid fibroblast-like synoviocytes. Journal of immunology. 2012;188:454-61
12. Gao F, Zhao ZL, Zhao WT, Fan QR, Wang SC, Li J. et al. miR-9 modulates the expression of interferon-regulated genes and MHC class I molecules in human nasopharyngeal carcinoma cells. Biochemical and biophysical research communications. 2013;431:610-6
13. Mari L, Hoefnagel SJM, Zito D, van de Meent M, van Endert P, Calpe S. et al. microRNA 125a Regulates MHC-I Expression on Esophageal Adenocarcinoma Cells, Associated With Suppression of Antitumor Immune Response and Poor Outcomes of Patients. Gastroenterology. 2018;155:784-98
14. Yadav D, Ngolab J, Lim RS, Krishnamurthy S, Bui JD. Cutting edge: down-regulation of MHC class I-related chain A on tumor cells by IFN-gamma-induced microRNA. Journal of immunology. 2009;182:39-43
15. Codolo G, Toffoletto M, Chemello F, Coletta S, Soler Teixidor G, Battaggia G. et al. Helicobacter pylori Dampens HLA-II Expression on Macrophages via the Up-Regulation of miRNAs Targeting CIITA. Frontiers in immunology. 2019;10:2923
16. Duan ZY, Cai GY, Li JJ, Bu R, Wang N, Yin P. et al. U6 can be used as a housekeeping gene for urinary sediment miRNA studies of IgA nephropathy. Scientific reports. 2018;8:10875
17. Jasinski-Bergner S, Mandelboim O, Seliger B. The role of microRNAs in the control of innate immune response in cancer. Journal of the National Cancer Institute. 2014 106
18. Fan Y, Yin S, Hao Y, Yang J, Zhang H, Sun C. et al. miR-19b promotes tumor growth and metastasis via targeting TP53. Rna. 2014;20:765-72
19. Liu Y, Liu R, Yang F, Cheng R, Chen X, Cui S. et al. miR-19a promotes colorectal cancer proliferation and migration by targeting TIA1. Molecular cancer. 2017;16:53
20. Zhang J, Xiao Z, Lai D, Sun J, He C, Chu Z. et al. miR-21, miR-17 and miR-19a induced by phosphatase of regenerating liver-3 promote the proliferation and metastasis of colon cancer. British journal of cancer. 2012;107:352-9
21. Zhu J, Wang S, Chen Y, Li X, Jiang Y, Yang X. et al. miR-19 targeting of GSK3beta mediates sulforaphane suppression of lung cancer stem cells. The Journal of nutritional biochemistry. 2017;44:80-91
22. Liu SQ, Jiang S, Li C, Zhang B, Li QJ. miR-17-92 cluster targets phosphatase and tensin homology and Ikaros Family Zinc Finger 4 to promote TH17-mediated inflammation. The Journal of biological chemistry. 2014;289:12446-56
23. de Kouchkovsky D, Esensten JH, Rosenthal WL, Morar MM, Bluestone JA, Jeker LT. microRNA-17-92 regulates IL-10 production by regulatory T cells and control of experimental autoimmune encephalomyelitis. Journal of immunology. 2013;191:1594-605
24. Haj-Salem I, Fakhfakh R, Berube JC, Jacques E, Plante S, Simard MJ. et al. MicroRNA-19a enhances proliferation of bronchial epithelial cells by targeting TGFbetaR2 gene in severe asthma. Allergy. 2015;70:212-9
25. Ye L, Mou Y, Wang J, Jin ML. Effects of microRNA-19b on airway remodeling, airway inflammation and degree of oxidative stress by targeting TSLP through the Stat3 signaling pathway in a mouse model of asthma. Oncotarget. 2017;8:47533-46
26. Wang C, You Q, Cao X, Guo H, Gao X, Peng X. Micro RNA-19a suppresses interleukin-10 in peripheral B cells of patients with diabetic retinopathy. American journal of translational research. 2017;9:1410-7
27. Wang T, Xu X, Xu Q, Ren J, Shen S, Fan C. et al. miR-19a promotes colitis-associated colorectal cancer by regulating tumor necrosis factor alpha-induced protein 3-NF-kappaB feedback loops. Oncogene. 2017;36:3240-51
28. Chen B, She S, Li D, Liu Z, Yang X, Zeng Z. et al. Role of miR-19a targeting TNF-alpha in mediating ulcerative colitis. Scandinavian journal of gastroenterology. 2013;48:815-24
29. Cheng X, Zhang X, Su J, Zhang Y, Zhou W, Zhou J. et al. miR-19b downregulates intestinal SOCS3 to reduce intestinal inflammation in Crohn's disease. Scientific reports. 2015;5:10397
30. Deng J, Yang S, Yuan Q, Chen Y, Li D, Sun H. et al. Acupuncture Ameliorates Postoperative Ileus via IL-6-miR-19a-KIT Axis to Protect Interstitial Cells of Cajal. The American journal of Chinese medicine. 2017;45:737-55
31. Schaefer JS, Montufar-Solis D, Vigneswaran N, Klein JR. Selective upregulation of microRNA expression in peripheral blood leukocytes in IL-10-/- mice precedes expression in the colon. Journal of immunology. 2011;187:5834-41
32. Marelli G, Sica A, Vannucci L, Allavena P. Inflammation as target in cancer therapy. Current opinion in pharmacology. 2017;35:57-65
33. Zitvogel L, Pietrocola F, Kroemer G. Nutrition, inflammation and cancer. Nature immunology. 2017;18:843-50
34. Davidson-Moncada J, Papavasiliou FN, Tam W. MicroRNAs of the immune system: roles in inflammation and cancer. Annals of the New York Academy of Sciences. 2010;1183:183-94
35. Tili E, Michaille JJ, Croce CM. MicroRNAs play a central role in molecular dysfunctions linking inflammation with cancer. Immunological reviews. 2013;253:167-84
36. Leucci E, Zriwil A, Gregersen LH, Jensen KT, Obad S, Bellan C. et al. Inhibition of miR-9 de-represses HuR and DICER1 and impairs Hodgkin lymphoma tumour outgrowth in vivo. Oncogene. 2012;31:5081-9
37. Christopher AF, Gupta M, Bansal P. Micronome revealed miR-19a/b as key regulator of SOCS3 during cancer related inflammation of oral squamous cell carcinoma. Gene. 2016;594:30-40
38. Guadagno J, Swan P, Shaikh R, Cregan SP. Microglia-derived IL-1beta triggers p53-mediated cell cycle arrest and apoptosis in neural precursor cells. Cell death & disease. 2015;6:e1779
39. Hong JT, Son DJ, Lee CK, Yoon DY, Lee DH, Park MH. Interleukin 32, inflammation and cancer. Pharmacology & therapeutics. 2017;174:127-37
40. Tsai CY, Wang CS, Tsai MM, Chi HC, Cheng WL, Tseng YH. et al. Interleukin-32 increases human gastric cancer cell invasion associated with tumor progression and metastasis. Clinical cancer research: an official journal of the American Association for Cancer Research. 2014;20:2276-88
41. Rossi JF, Lu ZY, Jourdan M, Klein B. Interleukin-6 as a therapeutic target. Clinical cancer research: an official journal of the American Association for Cancer Research. 2015;21:1248-57
42. Yang G, Xu Y, Chen X, Hu G. IFITM1 plays an essential role in the antiproliferative action of interferon-gamma. Oncogene. 2007;26:594-603
43. Bidwell BN, Slaney CY, Withana NP, Forster S, Cao Y, Loi S. et al. Silencing of Irf7 pathways in breast cancer cells promotes bone metastasis through immune escape. Nature medicine. 2012;18:1224-31
44. Yang Y, Zhou Y, Hou J, Bai C, Li Z, Fan J. et al. Hepatic IFIT3 predicts interferon-alpha therapeutic response in patients of hepatocellular carcinoma. Hepatology (Baltimore, Md). 2017;66:152-66
45. Gobin SJ, Peijnenburg A, Keijsers V, van den Elsen PJ. Site alpha is crucial for two routes of IFN gamma-induced MHC class I transactivation: the ISRE-mediated route and a novel pathway involving CIITA. Immunity. 1997;6:601-11
46. Martin BK, Chin KC, Olsen JC, Skinner CA, Dey A, Ozato K. et al. Induction of MHC class I expression by the MHC class II transactivator CIITA. Immunity. 1997;6:591-600
47. Raval A, Howcroft TK, Weissman JD, Kirshner S, Zhu XS, Yokoyama K. et al. Transcriptional coactivator, CIITA, is an acetyltransferase that bypasses a promoter requirement for TAF(II)250. Molecular cell. 2001;7:105-15
48. Howcroft TK, Raval A, Weissman JD, Gegonne A, Singer DS. Distinct transcriptional pathways regulate basal and activated major histocompatibility complex class I expression. Molecular and cellular biology. 2003;23:3377-91
49. Boodhoo A, Wong AM, Williamson D, Voon D, Lee S, Allcock RJ. et al. A promoter polymorphism in the central MHC gene, IKBL, influences the binding of transcription factors USF1 and E47 on disease-associated haplotypes. Gene expression. 2004;12:1-11
50. Lu XB, Wang ZX, Liu SB, Zhang XY, Lu LF, Li S. et al. Interferon Regulatory Factors 1 and 2 Play Different Roles in MHC II Expression Mediated by CIITA in Grass Carp, Ctenopharyngodon idella. Frontiers in immunology. 2019;10:1106
51. Wang Z, Zhang L, Qiao A, Watson K, Zhang J, Fan GH. Activation of CXCR4 triggers ubiquitination and down-regulation of major histocompatibility complex class I (MHC-I) on epithelioid carcinoma HeLa cells. The Journal of biological chemistry. 2008;283:3951-9
52. Brea EJ, Oh CY, Manchado E, Budhu S, Gejman RS, Mo G. et al. Kinase Regulation of Human MHC Class I Molecule Expression on Cancer Cells. Cancer immunology research. 2016;4:936-47
53. Pollack BP, Sapkota B, Cartee TV. Epidermal growth factor receptor inhibition augments the expression of MHC class I and II genes. Clinical cancer research: an official journal of the American Association for Cancer Research. 2011;17:4400-13
54. Mimura K, Shiraishi K, Mueller A, Izawa S, Kua LF, So J. et al. The MAPK pathway is a predominant regulator of HLA-A expression in esophageal and gastric cancer. Journal of immunology. 2013;191:6261-72
55. Lu J, Luo H, Liu X, Peng Y, Zhang B, Wang L. et al. miR-9 targets CXCR4 and functions as a potential tumor suppressor in nasopharyngeal carcinoma. Carcinogenesis. 2014;35:554-63
56. Friedrich M, Jasinski-Bergner S, Lazaridou MF, Subbarayan K, Massa C, Tretbar S. et al. Tumor-induced escape mechanisms and their association with resistance to checkpoint inhibitor therapy. 2019; 68: 1689-700.
57. Hayakawa S. No cancer in cancers: evolutionary trade-off between successful viviparity and tumor escape from the adaptive immune system. Medical hypotheses. 2006;66:888-97
Author contact
![]() Corresponding authors: Yan Sun, suny69sysu.edu.cn; Dong Xiao, xiaodongedu.cn, Xiao_dcom; Sheng-Jun Xiao, xiaoshengjunedu.cn.
Corresponding authors: Yan Sun, suny69sysu.edu.cn; Dong Xiao, xiaodongedu.cn, Xiao_dcom; Sheng-Jun Xiao, xiaoshengjunedu.cn.

 Global reach, higher impact
Global reach, higher impact