Impact Factor
ISSN: 1449-1907
Int J Med Sci 2020; 17(3):292-301. doi:10.7150/ijms.37804 This issue Cite
Research Paper
Altered Long Non-coding RNAs Involved in Immunological Regulation and Associated with Choroidal Neovascularization in Mice
1. Department of Ophthalmology, The Second Xiangya Hospital, Central South University, Changsha, Hunan, China
2. Hunan Clinical Research Center of Ophthalmic Disease, Changsha, Hunan, China
3. Centre for Eye Research Australia, Royal Victorian Eye and Ear Hospital, East Melbourne, Victoria, Australia
4. Ophthalmology, Department of Surgery, University of Melbourne, East Melbourne, Victoria, Australia
5. Department of Ophthalmology, Kurume University School of Medicine, Kurume, Fukuoka, Japan
Received 2019-6-21; Accepted 2019-11-14; Published 2020-1-16
Abstract
Choroidal neovascularization (CNV) is a severe complication of the wet form of age-related macular degeneration (AMD). Long non-coding RNAs (lncRNAs) have been implicated in the pathogenesis of different ocular neovascular diseases. To identify the function and therapeutic potential of lncRNAs in CNV, we assessed lncRNAs and mRNA expression profile in a mouse model of laser-induced CNV by microarray analysis. The results of altered lncRNAs were validated by qRT-PCR. Bioinformatics analyses, including Gene Ontology (GO) analysis and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway analysis, were performed to clarify the potential biological functions and signaling pathways with which altered genes are most closely related. Moreover, to identify the interaction of lncRNAs and mRNAs, we constructed a coding-non-coding gene co-expression (CNC) network. By microarray analysis, we identified 716 altered lncRNAs and 821 altered mRNAs in CNV mice compared to controls. A CNC network profile based on 7 validated altered lncRNAs (uc009ewo.1, AK148935, uc029sdr.1, ENSMUST00000132340, AK030988, uc007mds.1, ENSMUST00000180519) as well as 282 interacted and altered mRNAs, and were connected by 713 edges. GO and KEGG analyses suggested that altered mRNAs, as well as those lncRNA-interacted mRNAs were enriched in immune system process and chemokine signaling pathway. Thus, lncRNAs are significantly altered in this mouse model of CNV and are involved in immunological regulation, suggesting that lncRNAs may play a critical role in the pathogenesis of CNV. Thus, dysregulated lncRNAs and their target genes might be promising therapeutic targets to suppress CNV in AMD.
Keywords: long non-coding RNA, choroidal neovascularization, angiogenesis, age-related macular degeneration, immunological regulation
Introduction
Age-related macular degeneration (AMD) is one of the major causes of visual impairments and blindness in developed regions [1]. Choroidal neovascularization (CNV) is a severe complication of the neovascular or wet AMD [2]. It is well known that vascular endothelial growth factor (VEGF) plays key roles in the pathogenesis of neovascular AMD [3, 4]. Numerous studies have demonstrated that anti-VEGF therapy is effective and safe for patients with CNV due to AMD for which it has been widely used in clinics [5, 6]. However, recent studies suggest that some VEGF neutralizing proteins are not effective in some patients [7, 8]. There is increasing evidence that a number of factors may be responsible for non-responses including tolerance or tachyphylaxis (a term refers to the sudden decrease in response to a drug after its administration) of anti-VEGF therapies [9, 10]. Therefore, there is an urgent need for the mechanistic understanding of CNV, to explore novel therapeutic targets for early intervention of CNV, which would provide potential alternatives to anti-VEGF therapies.
Long non-coding RNAs (lncRNAs) is a subtype of non-coding RNAs with the transcripts of more than 200 nucleotides [11]. Although lncRNAs have no protein-coding capacity, they may regulate physiological functions as well as pathological processes in many diseases [12-15]. In particular, the activation of lncRNAs might regulate the expression of protein-coding genes through sophisticated mechanisms in ocular disorders [16-19]. It has been reported that lncRNAs have differential expression profiles in a mouse model of ischemia-induced retinal neovascularization[20]. The role of lncRNAs in AMD has also been investigated in a number of studies. A study showed that lncRNAs are differentially expressed in RPE/choroid samples in patients with early AMD compared to controls and might be involved in important regulative functions [21]. Among altered lncRNAs in patients with early AMD, RP11-234O6.2 has been demonstrated as having protective effects in the aging RPE model [21]. A few in vitro studies investigated the roles of several certain lncRNAs (such as ZNF503-AS1 and BANCR) in retinal pigment epithelium (RPE) cells [22, 23]. Nevertheless, it still remains unclear that the expression profiles, targets, and effects of lncRNAs and their contribution to the pathogenesis of CNV as well as neovascular AMD.
Laser-induced CNV is a well-established mouse model to investigate the pathogenesis of CNV and neovascular AMD [24]. Previous studies using the CNV mouse model to examine the role of VEGF and many other molecules [25-27]. We recently identified altered microRNAs and tRNA-derived small RNAs in the laser-induced CNV model [28].
In the present study, microarray analyses were applied to clarify the expression profiles of lncRNAs and mRNAs in the CNV mouse model. In addition, bioinformatics analyses, including gene ontology (GO) analysis and Kyoto Encyclopedia of Genes and Genomes (KEGG) analysis, were conducted to explore the related biological functions and potential signaling pathways of altered genes. Moreover, a coding-non-coding gene co-expression (CNC) network was established to investigate the correlation of differentially expressed lncRNAs and mRNAs, and to further predict the possible roles of differentially expressed lncRNAs in CNV.
Materials and Methods
Animals
C57BL/6J mice (male, aged 7-8 weeks old) (SJA Laboratory Animal Co., Ltd., Hunan, China) were used for experiments in accordance to the ARVO Statement for the Use of Animals in Ophthalmic and Vision Research. The procedures were proved by the Institutional Animal Care and Use Committee of Central South University.
Laser-induced CNV mouse model
As previously described, the CNV mouse model was induced by laser photocoagulation [27-29] with some modification. In brief, the photocoagulation was performed with 25 spots in each eye using a 532-nm diode laser (100 mW, 0.1 s duration, 50 μm). On day 7 after laser photocoagulation treatment, the eyes were enucleated. Age-matched C57BL/6J mice without the treatment were used as controls.
LncRNA and mRNA microarray analysis
Tissues of RPE-choroid-sclera complexes were collected from 4 eyes to be pooled as one sample, and a total of 6 samples (3 CNV samples and 3 control samples) were analyzed. Total RNA was isolated by using Trizol RNA extraction kit (Invitrogen, Carlsbad, CA, USA). The microarray analysis was performed by mouse lncRNA Microarray V3.0 (Arraystar, Rockville, MD, USA) as described [20]. The raw data of microarray has been uploaded to the Gene Expression Omnibus database (http://www.ncbi.nlm.nih.gov/geo/) for public access with the accession number GSE129743. Altered lncRNAs and mRNAs expression were identified by a Volcano Plot filtering [fold change (FC) ≥1.5] and P<0.05.
Quantitative real-time reverse transcription-polymerase chain reaction (qRT-PCR)
The validation of lncRNA and mRNA microarray results was further processed by qRT-PCR [30]. RNA was transcribed into cDNA utilizing SuperScript Ⅲ Reverse Transcriptase (Invitrogen) following instructions of the manufacturer by Gene Amp PCR System 9700 (Applied Biosystems, Foster City, CA, USA). The qRT-PCR was performed by the ViiA 7 RT PCR System (Applied Biosystems) with the 2 × PCR Master Mix (Arraystar). LncRNAs' relative expression levels were normalized with GAPDH [31, 32]. Primer used for qRT-PCR are shown in Table 1. P<0.05 was considered statistically significant differences.
GO and KEGG pathway analyses
GO analysis (http://www.geneontology.org/) and KEGG pathway analysis (http://www.genome.jp/kegg/) were used to investigate the potential biological functions and significant pathways of the altered mRNAs or CNC-associated mRNAs.
Construction of lncRNA-mRNA co-expression network
According to validated altered lncRNAs and their related mRNAs, we established a CNC network profile to explore the relationship between lncRNAs and mRNAs. We selected Pearson correlation coefficients (PCCs)≥ 0.97 to construct the network utilizing Cytoscape version 2.8.1 software (The Cytoscape Consortium, San Diego, CA, USA) based on the PCCs of correlation analysis of lncRNA and mRNA.
Results
Expression profiles of lncRNA and mRNA in CNV mice
To assess the expressions of lncRNAs in CNV compared to control mice, microarray analysis was conducted in the collected RPE-choroid-sclera complexes. Our results showed that 716 lncRNAs were significantly altered in CNV mice compared to control mice: 442 were upregulated and 274 were downregulated (FC≥1.5, P<0.05). The top 20 up- and downregulated lncRNAs are listed in Table 2-3. Among the differentially expressed lncRNA transcripts, AK036888 tops among upregulated lncRNAs with an FC of 13.38, whereas ENSMUST00000135495 tops among downregulated lncRNAs with an FC of 7.47. The clustering analysis demonstrated the relevance of lncRNA expression patterns in CNV and control samples by showing the top 20 up- and downregulated lncRNAs (Fig. 1A). The variation of lncRNA expression between CNV and control mice is shown in a volcano plot (Fig. 1C) and a scatter plot (Fig. 1E).
We also identified that 821 mRNAs were significantly altered in CNV mice compared to control mice: 588 were upregulated and 233 were downregulated (FC≥1.5 , P<0.05). The top 20 significantly altered mRNA are shown in Table 4-5. Among those upregulated mRNAs, Aplnr (NM_011784) tops with an FC of 11.11, while expression of Prpmp5 (NM_001024705) tops among downregulated genes with FC of 3.61. The heatmap plot revealed the clustering analysis among mRNA expression patterns by presenting the top 20 up- and downregulated mRNAs in the collected samples (Fig. 1B). A volcano plot (Fig. 1D) and a scatter plot (Fig. 1F) showed the variation of mRNA expression between CNV and control mice.
Sequence of the primers for lncRNAs.
| Forward and reverse primer | Tm (℃) | Product length (bp) | |
|---|---|---|---|
| GAPDH (MOUSE) | F:5' CACTGAGCAAGAGAGGCCCTAT3' R:5' GCAGCGAACTTTATTGATGGTATT3' | 60 | 144 |
| uc009ewo.1 | F:5' GGTAAGTCCCCACTATCATTCTC 3' R:5' AAACACCTTTGCCCTCCTC 3' | 60 | 184 |
| AK148935 | F:5' TTTGTTGGCTGCCTTCTTTC 3' R:5' TGACTAACCTGTGAGTGTCCCTA 3' | 60 | 72 |
| uc029sdr.1 | F:5' CTCTTGATGTATCCCAGGGTG 3' R:5' CAGGAAACACAATGCTACTCTC 3' | 60 | 210 |
| ENSMUST00000132340 | F:5' CGGAATGTTACTGCCCATAG 3' R:5' TGGTCATTAGGATAAGTTCTGG 3' | 60 | 254 |
| AK030988 | F:5' TGGGACCTAAGGATGGAAGA 3' R:5' AGACCAGAGCCAATGTGAGC 3' | 60 | 167 |
| uc007mds.1 | F:5' TTCAGCCCCGACGAGCAC 3' R:5' GAAAGGGTTTGTTCGGCTCAC 3' | 60 | 187 |
| ENSMUST00000180519 | F:5' TCTGGTTCTCGGCTATGTGC 3' R:5' AACAACAGCACAAACAGGGT 3' | 60 | 129 |
Tm: temperature. bp: base pair.
Top 20 significantly up-regulated lncRNAs.
| Sequence Name | P-value | FDR | Fold Change | Regulation | Strand | Relationship | CNV 1 | CNV 2 | CNV 3 | Control 1 | Control 2 | Control 3 |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| AK036888 | 0.000171 | 0.150561 | 13.378940 | up | - | intergenic | 6.585418 | 6.480785 | 6.071276 | 3.077727 | 2.512265 | 2.321811 |
| uc007pgi.1 | 0.014847 | 0.390976 | 9.578848 | up | - | intergenic | 5.211190 | 7.107776 | 4.430212 | 2.325983 | 2.321827 | 2.321811 |
| AK078702 | 0.000536 | 0.150561 | 7.117427 | up | - | intergenic | 6.359641 | 6.132188 | 6.161450 | 3.862077 | 3.371296 | 2.925838 |
| AK035526 | 0.005083 | 0.298875 | 6.657825 | up | + | natural antisense | 5.724644 | 7.089529 | 7.076339 | 4.231499 | 3.878298 | 3.575563 |
| uc009ewo.1 | 0.000984 | 0.182037 | 5.719433 | up | - | intron sense-overlapping | 8.163315 | 7.829017 | 7.371524 | 5.300458 | 5.566779 | 4.949003 |
| AK036625 | 0.005126 | 0.298875 | 5.353162 | up | - | intergenic | 5.726749 | 5.680111 | 5.143081 | 2.325983 | 3.345529 | 3.617254 |
| ENSMUST00000148005 | 0.001561 | 0.218932 | 5.264145 | up | + | exon sense-overlapping | 6.509248 | 6.302205 | 5.601678 | 3.998450 | 3.479648 | 3.746437 |
| ENSMUST00000153541 | 0.000164 | 0.150561 | 4.957192 | up | + | exon sense-overlapping | 11.456490 | 11.349358 | 10.999685 | 9.005207 | 9.098501 | 8.773256 |
| AK008836 | 0.004393 | 0.293747 | 4.888444 | up | - | intronic antisense | 7.371524 | 6.772026 | 6.602773 | 4.573347 | 5.202004 | 4.102845 |
| ENSMUST00000146254 | 0.000028 | 0.106609 | 4.640225 | up | - | exon sense-overlapping | 7.526886 | 7.218386 | 7.363228 | 5.129217 | 5.254380 | 5.082319 |
| NR_026843 | 0.001265 | 0.203615 | 4.591574 | up | - | natural antisense | 6.264922 | 6.398417 | 5.991045 | 4.478630 | 3.930072 | 3.648715 |
| ENSMUST00000122340 | 0.001105 | 0.191468 | 4.442445 | up | - | intronic antisense | 8.984519 | 8.526921 | 8.527271 | 6.376287 | 6.936308 | 6.272055 |
| AK082144 | 0.000016 | 0.106609 | 4.424734 | up | - | intergenic | 6.554079 | 6.639836 | 6.746826 | 4.380675 | 4.513834 | 4.609461 |
| AK048117 | 0.001609 | 0.221747 | 4.382340 | up | - | intergenic | 9.343423 | 9.431755 | 9.431267 | 7.627238 | 6.720520 | 7.463583 |
| ENSMUST00000143423 | 0.000941 | 0.181475 | 4.307678 | up | + | exon sense-overlapping | 4.561347 | 5.009892 | 4.564078 | 2.963872 | 2.321827 | 2.528887 |
| AK148935 | 0.000638 | 0.156222 | 4.263167 | up | - | intergenic | 8.259856 | 8.525405 | 8.333067 | 6.682964 | 6.086507 | 6.073080 |
| AK047865 | 0.001065 | 0.191468 | 4.256438 | up | - | intergenic | 9.039629 | 8.963352 | 9.098866 | 7.275191 | 6.469285 | 7.088430 |
| ENSMUST00000141975 | 0.000283 | 0.150561 | 4.232972 | up | - | exon sense-overlapping | 6.736901 | 6.384502 | 6.153523 | 4.416513 | 4.338800 | 4.274602 |
| uc029sdr.1 | 0.000052 | 0.120829 | 4.185189 | up | + | intergenic | 10.768113 | 10.641598 | 10.589025 | 8.507404 | 8.798939 | 8.496515 |
| NR_045384 | 0.010196 | 0.362008 | 4.172997 | up | + | intergenic | 5.724329 | 5.884554 | 5.893274 | 4.532355 | 3.801238 | 2.985313 |
P-values were calculated using unpaired t-test. FDR: false discovery rate. Fold change: the absolute ratio (no log scale) of average normalized intensities between CNV group and control group. CNV 1 to 3 and Control 1 to 3: each sample's normalized intensity (log2 scale). Hereinafter the same.
Top 20 significantly down-regulated lncRNAs.
| Sequence Name | P-value | FDR | Fold Change | Regulation | Strand | Relationship | CNV1 | CNV2 | CNV3 | Control1 | Control2 | Control3 |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ENSMUST00000135495 | 0.044869 | 0.516189 | 7.465422 | down | - | exon sense-overlapping | 4.264994 | 6.259270 | 3.847529 | 8.927113 | 6.595256 | 7.550095 |
| ENSMUST00000151832 | 0.042431 | 0.506906 | 3.682991 | down | + | exon sense-overlapping | 3.973213 | 5.239059 | 4.472576 | 5.519285 | 6.476424 | 7.331772 |
| AK079094 | 0.000472 | 0.150561 | 2.875171 | down | - | natural antisense | 4.097720 | 3.991172 | 3.967711 | 5.716774 | 5.645347 | 5.265424 |
| ENSMUST00000171338 | 0.003991 | 0.284707 | 2.831007 | down | - | exon sense-overlapping | 4.347292 | 4.591844 | 4.052868 | 6.017756 | 5.435850 | 6.042344 |
| ENSMUST00000147716 | 0.002243 | 0.244886 | 2.805529 | down | - | intergenic | 7.029440 | 6.772935 | 7.329016 | 8.414517 | 8.813207 | 8.368486 |
| AK030988 | 0.004724 | 0.293747 | 2.720887 | down | - | intergenic | 7.828629 | 8.353325 | 8.177764 | 9.318910 | 9.964228 | 9.408810 |
| ENSMUST00000148202 | 0.005363 | 0.303718 | 2.572706 | down | - | intergenic | 3.028002 | 2.478576 | 3.041757 | 3.901149 | 4.463217 | 4.273829 |
| AK005624 | 0.012396 | 0.380110 | 2.559854 | down | + | bidirectional | 4.166821 | 3.256077 | 3.443728 | 5.159213 | 5.085467 | 4.690131 |
| AK009740 | 0.014086 | 0.383884 | 2.485166 | down | - | bidirectional | 2.530040 | 3.314212 | 2.545198 | 4.152919 | 4.397770 | 3.778787 |
| ENSMUST00000155217 | 0.000964 | 0.182037 | 2.460863 | down | - | intronic antisense | 7.949919 | 8.236430 | 8.067902 | 9.477743 | 9.536148 | 9.137853 |
| NR_037995 | 0.007062 | 0.335485 | 2.453643 | down | - | bidirectional | 7.028992 | 6.600705 | 6.301580 | 8.136388 | 8.017734 | 7.661932 |
| ENSMUST00000155625 | 0.004510 | 0.293747 | 2.406930 | down | - | exon sense-overlapping | 2.323709 | 2.787010 | 2.712476 | 4.039022 | 4.044215 | 3.541541 |
| uc007lit.1 | 0.039689 | 0.498925 | 2.389210 | down | + | natural antisense | 3.383898 | 2.440709 | 2.323922 | 3.760049 | 4.469326 | 3.688754 |
| ENSMUST00000141135 | 0.027081 | 0.445563 | 2.334129 | down | + | exon sense-overlapping | 3.241041 | 2.759476 | 2.558381 | 3.748812 | 3.811546 | 4.667192 |
| NR_033584 | 0.001977 | 0.240414 | 2.331771 | down | + | intergenic | 4.933888 | 5.203815 | 5.150325 | 6.451165 | 6.479938 | 6.021205 |
| TCONS_00016118 | 0.007575 | 0.340233 | 2.312752 | down | - | intronic antisense | 5.561865 | 5.562532 | 4.940521 | 6.790447 | 6.549662 | 6.353641 |
| AK029573 | 0.013265 | 0.380110 | 2.305416 | down | + | natural antisense | 4.500548 | 3.852448 | 3.562495 | 5.301173 | 5.106782 | 5.122617 |
| ENSMUST00000144044 | 0.040861 | 0.503207 | 2.225878 | down | + | exon sense-overlapping | 4.548734 | 5.325275 | 4.294675 | 6.335644 | 5.575793 | 5.720369 |
| NR_033469 | 0.005124 | 0.298875 | 2.225231 | down | + | intergenic | 6.507724 | 6.533446 | 6.704603 | 7.941327 | 7.926619 | 7.339692 |
| AK013439 | 0.011666 | 0.378564 | 2.208164 | down | + | intergenic | 6.454914 | 6.874332 | 6.432539 | 7.343084 | 8.090536 | 7.756705 |
Top 20 significantly up-regulated mRNAs.
| Sequence Name | Gene Symbol | P-value | Fold Change | Regulation | Chrom | CNV 1 | CNV 2 | CNV 3 | Control 1 | Control 2 | Control 3 |
|---|---|---|---|---|---|---|---|---|---|---|---|
| NM_011784 | Aplnr | 0.000012 | 11.110603 | up | chr2 | 7.033988 | 6.843315 | 7.167412 | 3.528313 | 3.702581 | 3.392226 |
| NM_011646 | Try4 | 0.001518 | 10.794919 | up | chr6 | 5.868813 | 6.207469 | 6.427937 | 3.563739 | 2.321827 | 2.321811 |
| NM_008372 | Il7r | 0.000099 | 9.886701 | up | chr15 | 6.023879 | 5.297020 | 5.565189 | 2.325983 | 2.321827 | 2.321811 |
| NM_177843 | Gm14461 | 0.001185 | 8.779374 | up | chr2 | 7.331272 | 7.067940 | 6.677774 | 3.347988 | 4.486817 | 3.839826 |
| NM_001003405 | Try5 | 0.000012 | 8.310638 | up | chr6 | 5.221975 | 5.431142 | 5.596919 | 2.325983 | 2.437364 | 2.321811 |
| NM_206535 | Cd200r2 | 0.000381 | 7.767570 | up | chr16 | 6.312686 | 6.182743 | 5.670891 | 2.734021 | 3.294071 | 3.265839 |
| NM_175406 | Atp6v0d2 | 0.001067 | 7.672187 | up | chr4 | 10.914325 | 10.474780 | 10.348221 | 7.376840 | 8.241582 | 7.299991 |
| NM_011089 | Pira2 | 0.000091 | 7.671351 | up | chr7 | 6.094800 | 5.933179 | 5.760588 | 2.792479 | 3.300848 | 2.876798 |
| NM_207244 | Cd200r4 | 0.006075 | 7.548943 | up | chr16 | 6.464434 | 6.282340 | 5.594127 | 2.325983 | 3.989506 | 3.276587 |
| NM_008311 | Htr2b | 0.000706 | 7.192641 | up | chr1 | 10.557041 | 10.345577 | 9.884406 | 7.819662 | 7.395292 | 7.032505 |
| NM_011088 | Pira11 | 0.000196 | 6.976422 | up | chr7 | 10.466558 | 10.217773 | 10.155017 | 7.762837 | 7.555972 | 7.113078 |
| NM_011094 | Pira7 | 0.000161 | 6.963833 | up | chr7 | 10.961688 | 10.969189 | 10.845609 | 8.455120 | 8.155355 | 7.766367 |
| NM_028973 | Lrrc15 | 0.012774 | 6.827810 | up | chr16 | 8.322155 | 8.931940 | 10.139378 | 6.901063 | 6.511550 | 5.666592 |
| NM_178241 | Cxcr1 | 0.003685 | 6.805119 | up | chr1 | 6.005635 | 5.869263 | 5.788451 | 3.879585 | 2.321827 | 3.162076 |
| NM_001166672 | Gm14548 | 0.000097 | 6.673624 | up | chr7 | 11.580840 | 11.296632 | 11.179657 | 8.847641 | 8.587764 | 8.406313 |
| NM_019948 | Clec4e | 0.000563 | 6.596644 | up | chr6 | 5.670682 | 5.027602 | 5.075513 | 2.887439 | 2.399350 | 2.321811 |
| NM_011087 | Pira1 | 0.003576 | 6.187779 | up | chr7 | 5.488713 | 5.426346 | 5.204069 | 2.325983 | 3.583068 | 2.321811 |
| NM_001267695 | Ctss | 0.000957 | 5.934620 | up | chr3 | 9.404564 | 8.806512 | 8.751737 | 6.192754 | 6.833984 | 6.228608 |
| NM_008848 | Pira6 | 0.000255 | 4.851621 | up | chr7 | 4.801415 | 4.635361 | 4.962917 | 2.325983 | 2.838959 | 2.399350 |
| NM_008404 | Itgb2 | 0.001909 | 4.841209 | up | chr10 | 9.670052 | 8.907650 | 8.869934 | 6.768681 | 7.213496 | 6.639358 |
Top 20 significantly down-regulated mRNAs.
| Sequence Name | Gene Symbol | P-value | Fold Change | Regulation | Chrom | CNV 1 | CNV 2 | CNV 3 | Control 1 | Control 2 | Control 3 |
|---|---|---|---|---|---|---|---|---|---|---|---|
| NM_001024705 | Prpmp5 | 0.014868 | 3.605863 | down | chr6 | 9.067530 | 8.986222 | 9.954781 | 11.192267 | 11.751723 | 10.615577 |
| NM_019840 | Pde4b | 0.001704 | 2.927001 | down | chr4 | 3.256077 | 3.624609 | 3.076331 | 5.079052 | 4.632161 | 4.894074 |
| NM_175296 | Mael | 0.013959 | 2.785800 | down | chr1 | 5.162247 | 5.565056 | 5.125866 | 7.062294 | 7.111513 | 6.113637 |
| NM_008623 | Mpz | 0.008683 | 2.769509 | down | chr1 | 12.174806 | 13.137897 | 12.638795 | 13.881432 | 14.154913 | 14.324043 |
| NM_183160 | Tmem252 | 0.006698 | 2.677545 | down | chr19 | 3.883402 | 4.178289 | 4.299822 | 5.111757 | 5.964037 | 5.548451 |
| NM_207280 | Ccdc121 | 0.014313 | 2.450424 | down | chr1 | 2.805933 | 2.324675 | 2.323922 | 3.639443 | 3.399682 | 4.294497 |
| NM_001025255 | Mbp | 0.000151 | 2.427744 | down | chr18 | 2.949733 | 3.182399 | 3.054139 | 4.463824 | 4.263468 | 4.297829 |
| NM_026925 | Pnlip | 0.028181 | 2.426672 | down | chr19 | 3.116513 | 2.324675 | 2.323922 | 3.895614 | 3.380287 | 4.326145 |
| NM_001113373 | Shank2 | 0.023401 | 2.379186 | down | chr7 | 3.648482 | 3.438832 | 2.672217 | 4.131874 | 4.666463 | 4.712598 |
| NM_001081153 | Unc13c | 0.034934 | 2.372052 | down | chr9 | 11.000760 | 10.198641 | 11.528724 | 12.222275 | 12.268342 | 11.975915 |
| NM_001122594 | Phlpp2 | 0.001196 | 2.356591 | down | chr8 | 3.131635 | 2.660591 | 2.931842 | 4.163803 | 4.026565 | 4.243805 |
| NM_001161722 | Tfeb | 0.003061 | 2.335554 | down | chr17 | 2.690098 | 3.230295 | 2.904800 | 4.046248 | 4.066880 | 4.383359 |
| NM_028807 | Exoc3l4 | 0.026008 | 2.331869 | down | chr12 | 4.272006 | 3.720309 | 3.146549 | 5.178742 | 4.693262 | 4.931320 |
| NM_001127685 | BC048943 | 0.013403 | 2.324064 | down | chr12 | 9.447387 | 8.766418 | 8.507593 | 10.125555 | 10.236406 | 10.009386 |
| NM_001204253 | Clec1b | 0.008321 | 2.321635 | down | chr6 | 4.644223 | 5.290836 | 4.979474 | 5.859919 | 6.408708 | 6.291329 |
| NM_007545 | Hrk | 0.003712 | 2.290870 | down | chr5 | 7.678339 | 8.078760 | 7.425854 | 8.965889 | 8.983374 | 8.821377 |
| NM_010220 | Fkbp5 | 0.029949 | 2.281321 | down | chr17 | 6.708265 | 6.429901 | 7.553089 | 8.247108 | 8.176554 | 7.837201 |
| NM_020599 | Rlbp1 | 0.007127 | 2.235286 | down | chr7 | 8.194874 | 8.097004 | 7.941646 | 9.260371 | 9.602041 | 8.852490 |
| NM_147110 | Olfr570 | 0.003044 | 2.223418 | down | chr7 | 5.491363 | 5.307785 | 5.178571 | 6.613100 | 6.653939 | 6.169017 |
| NM_027790 | Dhrs2 | 0.003418 | 2.203722 | down | chr14 | 5.776111 | 6.199949 | 5.806094 | 6.968227 | 7.311111 | 6.922644 |
Altered expressions of lncRNAs and mRNAs between CNV group and control group by microarray. Heat map and hierarchical clustering analysis of the top 20 up- and downregulated lncRNAs (A) and mRNAs (B) between CNV and control samples. The top column represents lncRNA and mRNA relative expression varies according to the color scale. The volcano plot presents all identified lncRNA (C) and mRNA (D) expression variation between the CNV and control samples. The horizontal green line shows the default 1.5-fold change and the vertical green line represents a P-value of 0.05. The red and green plots represent up- and downregulated RNAs with FC≥1.5 and P<0.05. The scatter plot presents all identified lncRNA (E) and mRNA (F) expression variation between the CNV and control samples. The x-axis and y-axis values are each sample's normalized values (log2 transformed). The gray line shows the default 1.5-fold changes. The red and green plots represent altered RNAs with FC≥1.5. The qRT-PCR validation of seven randomly selected lncRNAs: AK148935, ENSMUST00000132340, uc009ewo.1, uc029sdr.1, AK030988, ENSMUST00000180519, uc007mds.1 (G).
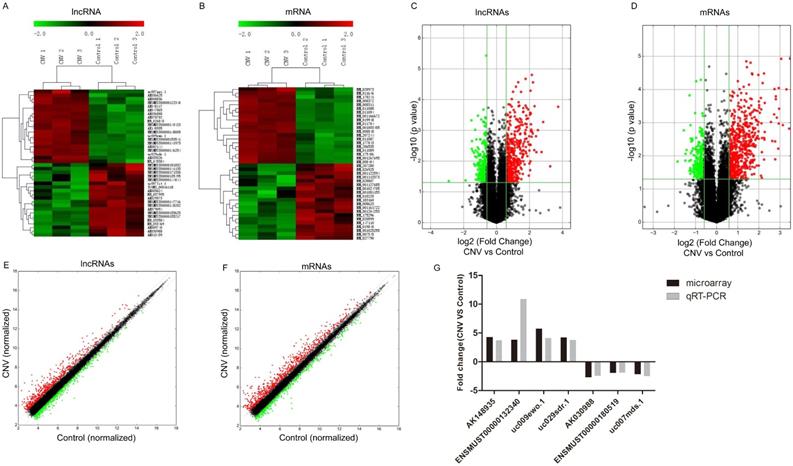
Validation of the microarray data of lncRNAs by qRT-PCR
To validate the accuracy and reliability of the microarray profiling data, seven lncRNAs (uc009ewo.1, AK148935, uc029sdr.1, ENSMUST00000132340, AK030988, uc007mds.1, ENSMUST00000180519) were randomly selected for qRT-PCR. The qRT-PCR results are consistent with the microarray analyses where expression of lncRNAs AK148935, ENSMUST00000132340, uc009ewo.1 and uc029sdr.1 were significantly upregulated and expression of lncRNAs AK030988, ENSMUST00000180519 and uc007mds.1 were significantly downregulated in CNV mice compared to control mice (Fig. 1G).
GO and KEGG pathway analyses of altered genes
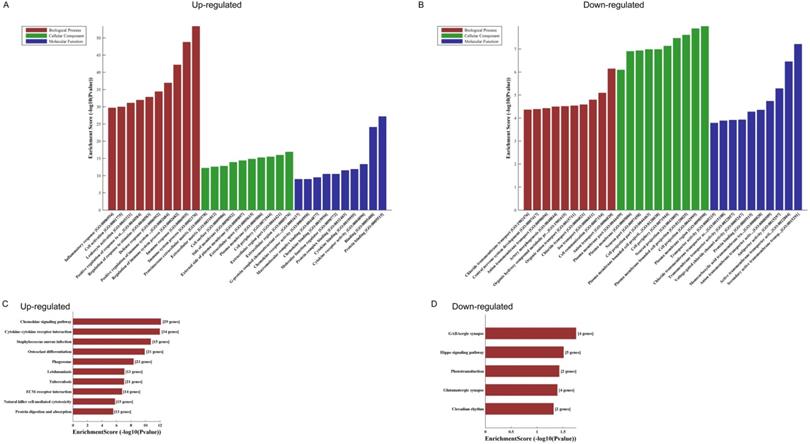
GO analyses of the 821 altered mRNAs showed that upregulated genes were enriched in immune system process (ontology: biological process, GO: 0002376), extracellular region (ontology: cellular component, GO: 0005576) and protein binding (ontology: molecular function, GO: 0005515) (Fig. 2A); while downregulated genes were enriched in anion transport (ontology: biological process, GO:0006820), plasma membrane region (ontology: cellular component, GO: 0098590) and secondary active transmembrane transporter activity (ontology: molecular function, GO: 0015291) (Fig. 2B).
KEGG pathway analysis of those differentially expressed mRNAs found that the upregulated genes were enriched in chemokine signaling pathway, cytokine-cytokine receptor interaction and staphylococcus aureus infection (Fig. 2C), while the downregulated genes were enriched in GABAergic synapse, Hippo signaling pathway and phototransduction (Fig. 2D).
CNC networks with GO and KEGG analyses
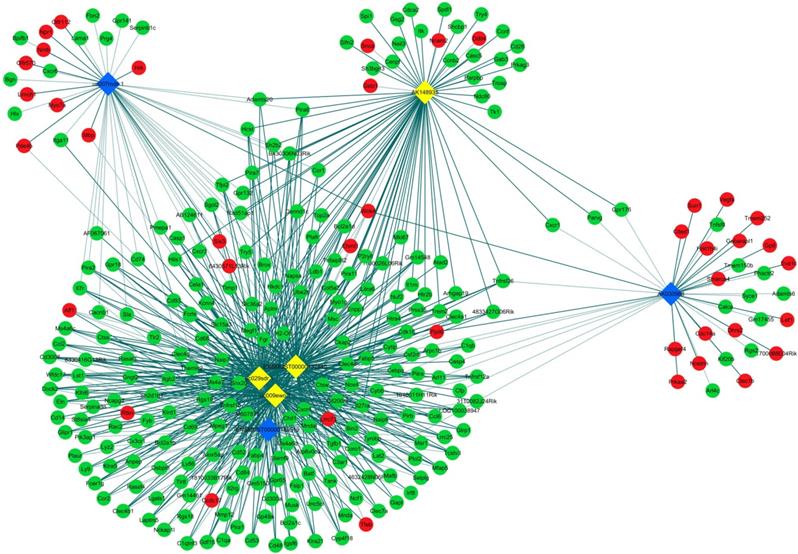
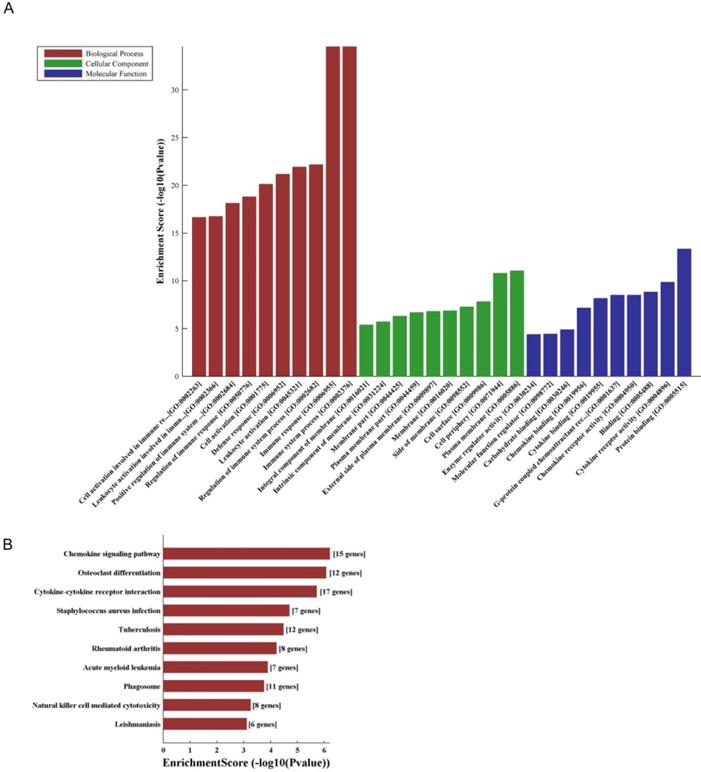
LncRNA and mRNA co-expression network was established based on 7 validated altered lncRNAs as well as 282 interacted mRNAs which were differentially expressed. The CNC network was composed of 289 nodes and 713 edges. There were 512 positive and 201 negative interactions within the network (Fig. 3). The lncRNA uc009ewo.1 is correlated with 138 mRNAs, ENSMUST00000132340 is correlated with 132 mRNAs, uc029sdr.1 is correlated with 152 mRNAs, ENSMUST00000180519 is correlated with 127 mRNAs, AK148935 is correlated with 76 mRNAs, while uc007mds.1 and AK030988 are correlated with 49 and 39 mRNAs respectively. According to the networks, the most relevant mRNAs are B430306N03Rik, C-type lectin receptor 4e (Clec4e) and paired immunoglobulin-like receptor A6 (Pira6), all correlated with 6 lncRNAs. To predict the functions of the lncRNAs, GO analyses and KEGG pathway analyses of those interacted mRNAs which were differentially expressed were conducted based on the results of the co-expression network. GO analysis showed that the most enriched GO terms of the targeted genes were immune system process (ontology: biological process, GO: 0002376), plasma membrane (ontology: cellular component, GO: 0005886) and protein binding (ontology: molecular function, GO: 0005515) (Fig. 4A). KEGG pathway analysis showed those targeted mRNAs were enriched in chemokine signaling pathway, osteoclast differentiation and cytokine-cytokine receptor interaction (Fig. 4B).
The GO and KEGG analyses of altered mRNAs. The GO analyses of up- (A) and downregulated (B) mRNAs. KEGG pathway analysis of altered mRNAs indicated: the top 10 significant enriched pathways of the upregulated genes (C), and the top 5 significant enriched pathways of the downregulated genes (D).
CNC networks by validated lncRNAs. The co-expression network profiles of lncRNAs and mRNAs based on 7 validated lncRNAs and correlated mRNAs which were differentially expressed. This co-expression network composed of 713 edges and 289 nodes. The diamond nodes represent lncRNAs, in which yellow denotes upregulated lncRNAs and blue denotes downregulated lncRNAs. The round nodes represent mRNAs, in which green denotes upregulated mRNAs and red denotes downregulated mRNAs. Continuous and dotted lines indicate positive and negative interactions between lncRNAs and mRNAs, respectively.
Discussion
Despite several studies reported important roles of particular lncRNAs such as ZNF503-AS1 and BANCR in RPE cells through in vitro studies [22, 23], none of them have investigated the role of lncRNAs in CNV. The present study profiled lncRNA and mRNA expression in the mouse model of laser-induced CNV by integrated microarray analysis. We identified 821 significantly altered mRNAs (588 upregulated; 233 downregulated) and 716 differentially expressed lncRNAs (442 upregulated; 274 downregulated) in the RPE-choroid-sclera complexes from CNV mice compared to control mice. Validation of seven randomly chosen lncRNAs by qRT-PCR further confirmed the reliability of microarray analysis. The profiles of the lncRNAs and mRNAs in CNV mice provide novel insights into our understanding of the pathogenesis of CNV.
A number of studies have reported that different cytokines, such as IL-10, IL-12, IFN-γ, IL-17, IL-18 and IL-33, may play a role in the pathogenesis of CNV [33-37]. The pro- and anti-angiogenic effects of the cytokines relevant to Th1, Th2 and Th17 cells, participate in a complicated immunological network [38]. It has been reported that the number of macrophages increased significantly after laser photocoagulation [24], and M1-M2 polarization of macrophages have diverse distributions and functions in laser-induced CNV as well as in wet AMD [39-41]. Pro-inflammatory M1 macrophages increased in the site of CNVs [39], suggesting that inflammation might play a key role in the pathogenesis of CNV and AMD. Our GO analyses showed that the most upregulated genes participate in immune system process, whereas the downregulated genes participate in plasma membrane region. Moreover, the KEGG pathway analysis revealed that the functions of altered genes are enriched in the chemokine signaling pathway, cytokine-cytokine receptor interaction and the GABAergic synapse. Similarly, GO and KEGG pathway analyses of the lncRNA-interacted mRNAs showed that the most enriched GO terms are associated with immune system process and immune response, and the most enriched KEGG pathway is also chemokine signaling pathway. These analyses demonstrated that CNV might be mainly immune-regulated by cytokines and chemokines.
The GO and KEGG analyses of interacted and altered mRNAs in CNC networks. GO analysis of the CNC-associated altered mRNAs (A). KEGG pathway analysis of the CNC-associated altered mRNAs (B).
Macrophage inducible Ca2+-dependent lectin receptor (Mincle) is a member of the C-type lectin family of immune receptors, which encoded by the gene of Clec4e, and Clec4e is also a type 2 transmembrane receptor [42]. As the CNC network analysis showed, Clec4e is a significantly altered gene correlated with 6 validated lncRNAs. Lv et al. reported that clec4e is induced specifically on M1 macrophages, and it plays an essential role in maintaining the phenotype of M1 macrophage in acute renal inflammation [43]. Another study showed that that Clec4e enhanced proinflammatory phenotype of macrophages through activation of the unfolded protein response [44]. Our previous study demonstrated that M1 macrophages mainly distribute around the site of CNVs, and inflammatory M1 macrophage-associated cytokines increased in RPE and choroid in laser-induced CNV mouse model [39]. Thus Clec4e, as well as macrophage polarization are possibly involved in mRNA-lncRNA network.
Nevertheless, the relevance of lncRNAs and cytokines or macrophages in intraocular neovascularization is still unclear. Our GO analysis suggested that several altered lncRNAs are involved in the regulation of cytokines and immunological networks in the pathogenesis of CNV, however, further studies should be guaranteed to investigate the exact functions and mechanisms of the dysregulated lncRNAs. Chemokines are a group of small chemoattractant cytokines, which play great roles in inflammation and regulating angiogenesis as well as macrophage polarization [45, 46]. Studies have shown that chemokine (C-X-C motif) ligand 8 (CXCL8) and monocyte chemoattractant protein-1 (MCP-1), and chemokine receptors such as CXC chemokine receptor 3 (CXCR3) have been reported to be involved in CNV and AMD pathogenesis [47, 48]. A study showed that miR-539-5p attenuates experimental CNV through targeting CXC chemokine receptor 7 (CXCL7) [49], indicating that non-coding RNAs are involved in the functional effect of chemokines and their receptors that contribute to the formation of CNV. In the present study, GO and KEGG analyses showed that altered genes enriched in immune system process, immune response, chemokine signaling pathway and cytokine-cytokine receptor interaction, indicating that lncRNAs may regulate chemokines, cytokines and their receptors in the pathogenesis of CNV formation through their target genes. Thus, it is worth further investigate the mechanisms of involvement of lncRNAs in chemokines and chemokine receptors in CNV and AMD.
Zhu et al. presented identified lncRNAs involved in early AMD [21]. In that study, GO and KEGG analyses showed that lncRNA-related genes enriched different functions, such as visual perception, sensory perception of light stimuli, and phototransduction pathway, but not immunological regulations. It might because that study focused on early-stage AMD patients without geographic atrophy and CNV, and our present study investigated the pathogenesis of CNV, which demonstrated different roles of lncRNAs in AMD.
We recently demonstrated that both mRNAs and lncRNAs have differential expression profiles in the oxygen-induced retinopathy (OIR) mouse model [20], which is used for investigation of retinal neovascular diseases. We recognized 539 altered mRNAs and 373 altered lncRNAs in OIR retinas (FC≥2.0, P<0.05) [20]. And we tried to check the intersection of altered RNAs in both models, and identified 161 mRNAs and 53 lncRNAs which altered in OIR as well as CNV models, and the majority of the RNAs have the same trend of alteration. For instance, ENSMUST00000165968 and CD68 were up-regulated in both models, while ENSMUST00000144657 and cpa2 were down-regulated in both models. These double-dysregulated mRNAs and lncRNAs might play more essential roles in angiogenesis, and this suggested that lncRNAs could have potential roles in the pathogenesis of ocular neovascular diseases.
In conclusion, the present study reveals that lncRNAs and mRNAs are significantly altered in CNV mice through microarray analysis. Further, GO and pathway analyses indicate that altered lncRNAs are not only involved in biological processes of immune mechanisms and inflammation but also are involved in the related pathways which might contribute to CNV and AMD pathogenesis. In particular, immunological networks, including cytokines, chemokines and their receptors are mainly involved and relevant to the effect of lncRNAs in CNV. The limitation of this study is the lack of functional assessment of the identified lncRNA and mRNAs. Therefore, further studies are required to investigate the potential roles and exact mechanisms of the altered lncRNAs and mRNAs in CNV.
Acknowledgements
This work was supported by National Natural Science Foundation of China (No. 81500746 and 81800855), Natural Science Foundation of Hunan Province (No. 2018JJ3765), and Department of Science and Technology, Hunan (No.2015TP2007). The authors declare that they have no competing interests.
Competing Interests
The authors have declared that no competing interest exists.
References
1. Klein R, Peto T, Bird A, Vannewkirk MR. The epidemiology of age-related macular degeneration. American journal of ophthalmology. 2004;137:486-95
2. Ambati J, Ambati BK, Yoo SH, Ianchulev S, Adamis AP. Age-related macular degeneration: etiology, pathogenesis, and therapeutic strategies. Survey of ophthalmology. 2003;48:257-93
3. van Wijngaarden P, Qureshi SH. Inhibitors of vascular endothelial growth factor (VEGF) in the management of neovascular age-related macular degeneration: a review of current practice. Clin Exp Optom. 2008;91:427-37
4. Ferrara N. Vascular endothelial growth factor and age-related macular degeneration: from basic science to therapy. Nat Med. 2010;16:1107-11
5. Kovach JL, Schwartz SG, Flynn HW Jr, Scott IU. Anti-VEGF Treatment Strategies for Wet AMD. J Ophthalmol. 2012;2012:786870
6. Ferrara N, Adamis AP. Ten years of anti-vascular endothelial growth factor therapy. Nat Rev Drug Discov. 2016;15:385-403
7. Park YG, Rhu HW, Kang S, Roh YJ. New Approach of Anti-VEGF Agents for Age-Related Macular Degeneration. Journal of Ophthalmology. 2012
8. Eghoj MS, Sorensen TL. Tachyphylaxis during treatment of exudative age-related macular degeneration with ranibizumab. Br J Ophthalmol. 2012;96:21-3
9. Schaal S, Kaplan HJ, Tezel TH. Is There Tachyphylaxis to Intravitreal Anti-Vascular Endothelial Growth Factor Pharmacotherapy in Age-Related Macular Degeneration? Ophthalmology. 2008;115:2199-205
10. Gasperini JL, Fawzi AA, Khondkaryan A, Lam L, Chong LP, Eliott D. et al. Bevacizumab and ranibizumab tachyphylaxis in the treatment of choroidal neovascularisation. Brit J Ophthalmol. 2012;96:14-20
11. Mercer TR, Dinger ME, Mattick JS. Long non-coding RNAs: insights into functions. Nat Rev Genet. 2009;10:155-9
12. Yang L, Lin C, Jin C, Yang JC, Tanasa B, Li W. et al. lncRNA-dependent mechanisms of androgen-receptor-regulated gene activation programs. Nature. 2013;500:598-602
13. Sun X, Yuan Y, Xiao Y, Lu Q, Yang L, Chen C. et al. Long non-coding RNA, Bmcob, regulates osteoblastic differentiation of bone marrow mesenchymal stem cells. Biochem Biophys Res Commun. 2018
14. Liu K, Yao H, Wen Y, Zhao H, Zhou N, Lei S. et al. Functional role of a long non-coding RNA LIFR-AS1/miR-29a/TNFAIP3 axis in colorectal cancer resistance to pohotodynamic therapy. Biochim Biophys Acta Mol Basis Dis. 2018;1864:2871-80
15. Jia J, Zhang M, Li Q, Zhou Q, Jiang Y. Long noncoding ribonucleic acid NKILA induces the endoplasmic reticulum stress/autophagy pathway and inhibits the nuclear factor-k-gene binding pathway in rats after intracerebral hemorrhage. J Cell Physiol. 2018;233:8839-49
16. Yan BA, Yao J, Liu JY, Li XM, Wang XQ, Li YJ. et al. lncRNA-MIAT Regulates Microvascular Dysfunction by Functioning as a Competing Endogenous RNA. Circ Res. 2015;116:1143 -+
17. Gong QY, Su GF. Roles of miRNAs and long noncoding RNAs in the progression of diabetic retinopathy. Biosci Rep. 2017:37
18. Xu XD, Li KR, Li XM, Yao J, Qin J, Yan B. Long non-coding RNAs: new players in ocular neovascularization. Mol Biol Rep. 2014;41:4493-505
19. Huang J, Yang Y, Fang F, Liu K. MALAT1 modulates the autophagy of retinoblastoma cell through miR-124-mediated stx17 regulation. J Cell Biochem. 2018;119:3853-63
20. Zhang L, Fu X, Zeng H, Wang JH, Peng Y, Zhao H. et al. Microarray Analysis of Long Non-Coding RNAs and Messenger RNAs in a Mouse Model of Oxygen-Induced Retinopathy. Int J Med Sci. 2019;16:537-47
21. Zhu W, Meng YF, Xing Q, Tao JJ, Lu J, Wu Y. Identification of lncRNAs involved in biological regulation in early age-related macular degeneration. Int J Nanomed. 2017;12:7589-602
22. Chen X, Jiang C, Qin B, Liu GH, Ji JD, Sun XT. et al. LncRNA ZNF503-AS1 promotes RPE differentiation by downregulating ZNF503 expression. Cell Death Dis. 2017:8
23. Kutty RK, Samuel W, Duncan T, Postnikova O, Jaworski C, Nagineni CN. et al. Proinflammatory cytokine interferon-gamma increases the expression of BANCR, a long non-coding RNA, in retinal pigment epithelial cells. Cytokine. 2018;104:147-50
24. Lambert V, Lecomte J, Hansen S, Blacher S, Gonzalez MLA, Struman I. et al. Laser-induced choroidal neovascularization model to study age-related macular degeneration in mice. Nat Protoc. 2013;8:2197-211
25. Gu LP, Chen H, Tuo JS, Gao XW, Chen L. Inhibition of experimental choroidal neovascularization in mice by anti-VEGFA/VEGFR2 or non-specific siRNA. Exp Eye Res. 2010;91:433-9
26. Kobayashi Y, Yoshida S, Zhou Y, Nakama T, Ishikawa K, Kubo Y. et al. Tenascin-C secreted by transdifferentiated retinal pigment epithelial cells promotes choroidal neovascularization via integrin alpha V. Lab Invest. 2016;96:1178-88
27. Nakama T, Yoshida S, Ishikawa K, Kobayashi Y, Zhou Y, Nakao S. et al. Inhibition of choroidal fibrovascular membrane formation by new class of RNA interference therapeutic agent targeting periostin. Gene Ther. 2015;22:127-37
28. Zhang L, Liu S, Wang JH, Zou J, Zeng H, Zhao H. et al. Differential Expressions of microRNAs and Transfer RNA-derived Small RNAs: Potential Targets of Choroidal Neovascularization. Curr Eye Res. 2019;44:1226-35
29. Zhang H, Yang Y, Takeda A, Yoshimura T, Oshima Y, Sonoda KH. et al. A Novel Platelet-Activating Factor Receptor Antagonist Inhibits Choroidal Neovascularization and Subretinal Fibrosis. PloS one. 2013;8:e68173
30. Cui HJ, Tao L, Li PF, Ali Y, Zhou HJ, Luo JK. et al. Altered Long Noncoding RNA and Messenger RNA Expression in Experimental Intracerebral Hemorrhage - a Preliminary Study. Cell Physiol Biochem. 2018;45:1284-301
31. Bustin SA. Absolute quantification of mRNA using real-time reverse transcription polymerase chain reaction assays. J Mol Endocrinol. 2000;25:169-93
32. Yang B, Xia ZA, Zhong B, Xiong X, Sheng C, Wang Y. et al. Distinct Hippocampal Expression Profiles of Long Non-coding RNAs in an Alzheimer's Disease Model. Mol Neurobiol. 2017;54:4833-46
33. Doyle SL, Lopez FJ, Celkova L, Brennan K, Mulfaul K, Ozaki E. et al. IL-18 Immunotherapy for Neovascular AMD: Tolerability and Efficacy in Nonhuman Primates. Invest Ophthalmol Vis Sci. 2015;56:5424-30
34. Nakamura R, Sene A, Santeford A, Gdoura A, Kubota S, Zapata N. et al. IL10-driven STAT3 signalling in senescent macrophages promotes pathological eye angiogenesis. Nat Commun. 2015;6:7847
35. Zhu Y, Tan W, Demetriades AM, Cai Y, Gao Y, Sui A. et al. Interleukin-17A neutralization alleviated ocular neovascularization by promoting M2 and mitigating M1 macrophage polarization. Immunology. 2016;147:414-28
36. Theodoropoulou S, Copland DA, Liu J, Wu J, Gardner PJ, Ozaki E. et al. Interleukin-33 regulates tissue remodelling and inhibits angiogenesis in the eye. J Pathol. 2017;241:45-56
37. Kleinman ME, Yamada K, Takeda A, Chandrasekaran V, Nozaki M, Baffi JZ. et al. Sequence- and target-independent angiogenesis suppression by siRNA via TLR3. Nature. 2008;452:591-7
38. Kwee BJ, Budina E, Najibi AJ, Mooney DJ. CD4 T-cells regulate angiogenesis and myogenesis. Biomaterials. 2018;178:109-21
39. Zhou Y, Yoshida S, Kubo Y, Yoshimura T, Kobayashi Y, Nakama T. et al. Different distributions of M1 and M2 macrophages in a mouse model of laser-induced choroidal neovascularization. Mol Med Rep. 2017;15:3949-56
40. Zandi S, Nakao S, Chun KH, Fiorina P, Sun D, Arita R. et al. ROCK-isoform-specific polarization of macrophages associated with age-related macular degeneration. Cell reports. 2015;10:1173-86
41. Zhou YD, Yoshida S, Peng YQ, Kobayashi Y, Zhang LS, Tang LS. Diverse roles of macrophages in intraocular neovascular diseases: a review. Int J Ophthalmol-Chi. 2017;10:1902-8
42. Lu XY, Nagata M, Yamasaki S. Mincle: 20 years of a versatile sensor of insults. Int Immunol. 2018;30:233-9
43. Lv LL, Tang PMK, Li CJ, You YK, Li JH, Huang XR. et al. The pattern recognition receptor, Mincle, is essential for maintaining the M1 macrophage phenotype in acute renal inflammation. Kidney Int. 2017;91:587-602
44. Clement M, Basatemur G, Masters L, Baker L, Bruneval P, Iwawaki T. et al. Necrotic Cell Sensor Clec4e Promotes a Proatherogenic Macrophage Phenotype Through Activation of the Unfolded Protein Response. Circulation. 2016;134:1039-51
45. Dimberg A. Chemokines in angiogenesis. Curr Top Microbiol Immunol. 2010;341:59-80
46. Owen JL, Mohamadzadeh M. Macrophages and chemokines as mediators of angiogenesis. Front Physiol. 2013:4
47. Fujimura S, Takahashi H, Yuda K, Ueta T, Iriyama A, Inoue T. et al. Angiostatic Effect of CXCR3 Expressed on Choroidal Neovascularization. Invest Ophthalmol Vis Sci. 2012;53:1999-2006
48. Lechner J, Chen M, Hogg RE, Toth L, Silvestri G, Chakravarthy U. et al. Peripheral blood mononuclear cells from neovascular age-related macular degeneration patients produce higher levels of chemokines CCL2 (MCP-1) and CXCL8 (IL-8). J Neuroinflammation. 2017:14
49. Feng YF, Wang J, Yuan YZ, Zhang X, Shen MQ, Yuan F. miR-539-5p inhibits experimental choroidal neovascularization by targeting CXCR7. FASEB J. 2018;32:1626-39
Author contact
![]() Corresponding author: Yedi Zhou, MD, PhD, Department of Ophthalmology, The Second Xiangya Hospital, Central South University, Changsha, Hunan 410011, China. Telephone: +86-731-85292175. E-mail: zhouyediedu.cn
Corresponding author: Yedi Zhou, MD, PhD, Department of Ophthalmology, The Second Xiangya Hospital, Central South University, Changsha, Hunan 410011, China. Telephone: +86-731-85292175. E-mail: zhouyediedu.cn

 Global reach, higher impact
Global reach, higher impact