3.2
Impact Factor
ISSN: 1449-1907
Int J Med Sci 2020; 17(1):112-124. doi:10.7150/ijms.35133 This issue Cite
Research Paper
Comparative Gene Expression Profiling of Tobacco-Associated HPV-Positive versus Negative Oral Squamous Carcinoma Cell Lines
1. Laboratory of Pre-Clinical and Translational Research, IRCCS, Referral Cancer Center of Basilicata, Rionero in Vulture, Italy;
2. Medical Oncology Unit, Department of Medical and Surgical Sciences, University of Foggia, Italy;
3. Medical Statistics Unit, University of Campania Luigi Vanvitelli, Naples, Italy;
4. Anatomic Pathology Unit, Department of Clinic and Experimental Medicine; University of Foggia, Italy;
5. Section of Clinic Pathology, Riuniti Hospital, Foggia, Italy
Received 2019-3-22; Accepted 2019-11-5; Published 2020-1-1
Abstract
Background: HPV-positive oral squamous cell carcinomas (OSCCs) are specific biological and clinical entities, characterized by a more favorable prognosis compared to HPV-negative OSCCs and occurring generally in non-smoking and non-drinking younger individuals. However, poor information is available on the molecular and the clinical behavior of HPV-positive oral cancers occurring in smoking/drinking subjects. Thus, this study was designed to compare, at molecular level, two OSCC cell lines, both derived from drinking and smoking individuals and differing for presence/absence of HPV infection.
Methods: HPV-negative UPCI-SCC-131 and HPV16-positive UPCI-SCC-154 cell lines were compared by whole genome gene expression profiling and subsequently studied for activation of Wnt/βCatenin signaling pathway by the expression of several Wnt-target genes, βCatenin intracellular localization, stem cell features and miRNA let-7e. Gene expression data were validated in head and neck squamous cell carcinoma (HNSCC) public datasets.
Results: Gene expression analysis identified Wnt/βCatenin pathway as the unique signaling pathway more active in HPV-negative compared to HPV-positive OSCC cells and this observation was confirmed upon evaluation of several Wnt-target genes (i.e., Cyclin D1, Cdh1, Cdkn2a, Cd44, Axin2, c-Myc and Tcf1). Interestingly, HPV-negative OSCC cells showed higher levels of total βCatenin and its active form, increase of its nuclear accumulation and more prominent stem cell traits. Furthermore, miRNA let-7e was identified as potential upstream regulator responsible for the downregulation of Wnt/βCatenin signaling cascade since its silencing in UPCI-SCC-154 cell resulted in upregulation of Wnt-target genes. Finally, the analysis of two independent gene expression public datasets of human HNSCC cell lines and tumors confirmed that Wnt/βCatenin pathway is more active in HPV-negative compared to HPV-positive tumors derived from individuals with smoking habit.
Conclusions: These data suggest that lack of HPV infection is associated with more prominent activation of Wnt/βCatenin signaling pathway and gain of stem-like traits in tobacco-related OSCCs.
Keywords: oral squamous carcinoma, Wnt/βCatenin pathway, stemness, tobacco
Background
Oral squamous cell carcinoma (OSCC) is responsible for 1.8% of cancer mortality worldwide with 300.000 new cases every years and is associated to tobacco use, unhealthy diet, excessive alcohol consumption and HPV infection [1]. Recent evidences suggest that HPV-positive OSCCs are characterized by a different clinical, molecular and biological behavior compared to HPV-negative cancers. Indeed, the majority of HPV-positive OSCCs have better prognosis and occur in non-smoking, non-drinking and younger individuals than HPV-negative OSCCs. The reason of this better survival is likely related to the different population-affected profile and lack of field cancerization [2]. However, a subset of HPV-related OSCCs, up to one-third of cases, occurs in smoking and drinking subjects, this representing a subgroup of tumors with unclear clinical and biological characters [3]. Indeed, recent evidence suggests that cigarette smoking changes the clinical behavior of HPV-positive OSCCs, being responsible for reduced responsiveness to therapies and worsening of their prognosis [4].
Several molecular and epidemiological studies showed relevant differences in terms of genome-wide gene expression profiles between HPV-positive and HPV-negative OSCCs, these likely influencing clinical outcomes [5-6]. Indeed, Holzinger et al. identified a different protein expression pattern between HPV-positive and HPV-negative OSCCs, with HPV-positive tumors characterized by high p16 expression, lower levels of pRb and Cyclin D1 and normal p53 level [7]. In addition, a number of studies identified DNA replication, cell cycle regulation and DNA repair as prominent cell functions associated to genes differentially expressed in HPV-positive versus HPV-negative cancers [8-9]. Since i) a broad understanding of the molecular differences between different subtypes of OSCCs represents an important step in the development of personalized treatments and ii) poor information is available on the role of HPV infection in tobacco-related oral squamous carcinomas, this study was designed to evaluate the gene expression profile of two OSCCs cell lines derived from smoking and drinking patients and differing for presence/absence of HPV infection and to validate these profiles in public datasets. Our data suggest that lack of HPV infection is associated with a more prominent activation of Wnt/βCatenin pathway and gain of stem-like traits.
Materials and Methods
Cell lines and siRNA transfection
OSCC UPCI-SCC-131 and UPCI-SCC-154 cell lines (DSMZ, Braunschweig Germany) were cultured at 37 °C in a humidified atmosphere of 5% CO2 in DMEM (Gibco, Life Technologies, Carlsbad, CA, USA) supplemented with 10% (v/v) fetal bovine serum (FBS, Gibco, Life Technologies, Carlsbad, CA, USA) and 1% (v/v) penicillin/streptomycin (Gibco, Life Technologies, Carlsbad, CA, USA).
miRNA let-7e was silenced using 60 nM mirVana miRNA Let-7e-5p inhibitor (Ambion, Thermo Fisher Scientific, Waltham, Massachusetts, USA, Catalog. 4464084, ID: MH12304). Negative mirVana inhibitor (Ambion, Thermo Fisher Scientific, Waltham, Massachusetts, USA, Catalog. 464076) was used as negative control. Transfection was performed with Lipofectamine RNAiMAX (Invitrogen, Thermo Fisher Scientific, Waltham, Massachusetts, USA,) according to the manufacturer's instructions.
Immunoblot analysis
Cells were lysed with RIPA buffer containing 25mM Tris HCl pH 7.4, 150mM NaCl, 1% (v/v) NP-40, 1% (w/v) sodium deoxycholate, 0.1% (w/v) SDS for 30 min on ice. Cell lysates were centrifuged at 1200 rpm for 10 minute at 4°C and supernatants assayed for protein concentration by Bradford method. Thirty micrograms of total proteins were loaded onto ready-to-use 4-20% polyacrylamide gels (Bio-Rad, Hercules, CA, USA), separated by electrophoresis and transferred onto nitrocellulose membranes (Trans-Blot, Bio-Rad, Hercules, CA, USA). The following primary antibodies were used: mouse monoclonal anti-active-βCatenin (cod.8814), rabbit polyclonal anti-βCatenin (cod.9562), rabbit monoclonal anti-Cyclin D1 (cod.92G2), rabbit monoclonal anti-cMyc (D84C12)XP (cod. 5605), rabbit monoclonal anti-Tcf-1 (C63D9) (cod.2203) from Cell Signaling Technology (Cell Signaling, Boston, MA, USA); mouse monoclonal anti-E-Cadherin (cod.610181) from BD (Becton Dickinson, BD, Franklin, NJ, USA); rabbit monoclonal anti-Axin-2 (cod. ab109307) from Abcam (Abcam, Cambridge, UK); rabbit polyclonal anti-Lgr5 (H-76) (cod. sc-135238), rabbit polyclonal anti-GAPDH (cod. sc-47724) and rabbit polyclonal anti-βActin (cod. sc-47778) from Santa Cruz Biotechnology. Specific proteins were reveled using the enhanced chemiluminescence (ECL) reagent (Bio-Rad, Hercules, CA, USA). GAPDH or β-Actin were detected as loading controls.
Clonogenic assay
Single cell suspension of 1000 cells were seeded on 60 mm tissue culture dishes and cultured for 15 days with medium changes every 3 days. Colonies were fixed with 4% paraformaldehyde at room temperature for 10 minutes and, after removing the fixation solution, stained with crystal violet solution for 20 minutes, rinsed with H2O and air-dried at room temperature. Colonies were counted under a light microscope and numbers used as index for clonogenicity.
Flow-cytometry
Non-confluent cell cultures were trypsinized into single cell suspension, washed with phosphate buffered saline (PBS) and counted. 1.0x106 cells were incubated with APC-conjugated mouse monoclonal Anti-Human CD44 (Becton Dickinson, BD, Franklin, NJ, USA) or mouse IgG2b κ Isotype Control (Becton Dickinson, BD, Franklin, NJ, USA), used as negative control. After washing twice with PBS, samples were analyzed by FACSb Calibur (Becton Dickinson, San Jose, CA, USA).
Immunocytochemistry
Cells were grown on glass slides at a density of 5x103 cells, rinsed with PBS, fixed in alcohol, rehydrated in graded ethanol solution and finally washed for 5 minutes with distilled water. p16 expression was assessed by standard linked streptavidin-biotin horseradish peroxidase technique (LSAB-HRP) using a specific mouse monoclonal primary antibody produced against the p16 INK4a protein (CINtec p16 Histology, clone E6H4, Ventana) delivered by the Benchmark XT autostainer (Ventana Medical Systems Inc, Tucson, AZ) in combination with Ventana detection kits and accessories. Immunocytochemical staining was evaluated using the Olympus BX41 microscope.
Immunofluorescence and confocal microscopy analysis
15x103 cells were seeded on chamber slides and cultured for 48 hours. Cells were fixed with 4% (w/v) paraformaldehyde in PBS at room temperature for 15 minutes and permeabilized with 0.1% (v/v) Triton-X100 (Sigma-Aldrich, Milan, Italy) for 10 minutes at room temperature. Non-specific binding sites were blocked by incubation in 1% (w/v) bovine serum albumin (BSA) in PBS for 30 minutes at room temperature. Cells were incubated with mouse monoclonal anti-active-βCatenin (1:400, Cell Signaling Technology, Boston, MA, USA) overnight at 4°C and subsequently with FITC-anti-mouse IgG antibody (1:1000, Sigma-Aldrich, Milan, Italy) for 1 hour at room temperature. Nuclei were counterstained with 4',6-diamidino-2-phenylindole (DAPI) using VECTASHIELD® Mounting Medium (Biorad, Hercules, CA, USA) and viewed under a confocal microscope Nikon Eclipse Ti-E microscope equipped with a unique Perfect Focus System (PFS) and coupled to Laser scanning confocal microscope C2 (Nikon Instruments S.p.A, Florence, Italy). Specimens were viewed through a 60X Plan APO oil immersion objective. Digital images were processed using the Nis Elements AR software.
Multiplex real-time PCR
DNA was extracted by the DNA mini kit (Qiagen, Hilden, Germany) and HPV genotype evaluated by Anyplex II HPV28 assay, according to manufacturer's recommendations (Seegene, Seoul, South Korea). Data recording and interpretation were automated with the Seegene viewer software.
RNA Extraction and Reverse Transcriptase PCR analysis
Total RNA was extracted using the TRIzol Reagent (Life Technologies, Carlsbad, CA, USA). For first strand synthesis of cDNA, 1 µg of RNA was used in a 20 µl reaction mixture utilizing a Transcriptor First Strand cDNA Synthesis Kit (Roche, Mannheim, Germany). For Real-time PCR analysis, 0.5 ng of cDNA samples were amplified using the LightCycler 480 SYBR Green I Master in a LightCycler 480 (Roche, Basel, Switzerland). The following primers were used: c-Myc forward 5'-TTCGGGTAGTGGAAAACCAG-3', reverse 5'-CAGCAGCTCGAATTTCTTCC-3' (PCR product 203 bp); Cyclin D1 forward 5'-CTACTACCGCCTCACACGCTT-3', reverse 5'-AGCCCTGGAGTCAAGCC-3' (PCR product 198 bp); Axin2 forward 5'-AGGTTCTGGCTATGTCTTTG-3', reverse 5'-AAATGAGGTAGAGACACTTGG-3' (PCR product 201 bp). PCR reaction conditions were as follows: pre-incubation at 95°C for 5 min, followed by 45 cycles of 10 s at 95°C, 10 s at 60°C, 10 s at 72°C. β-Actin was chosen as an internal control.
For miRNA let-7e quantification, RT-PCR was performed in UPCI-SCC-131 and UPCI-SCC-154 cell lines and in UPCI-SCC-154 upon miRNA let-7e silencing. cDNA was synthetized by reverse transcription using the TaqMan™ MicroRNA reverse transcription kit (Applied Biosystems, Monza, Italy), according to the manufacturer's instructions. Real-time PCR analysis was performed using the TaqMan™ MicroRNA assay with specific primer sets (probe has-miR-let-7e-5p, ID 002406, Applied Biosystems, Monza, Italy). The expression of RNU6B was used as internal control (probe RNU6B, ID 001093, Applied Biosystems, Monza, Italy) and the results were calculated using the ∆∆CT (where CT is threshold cycle) method.
Microarray experiments and data analysis
RNAs were isolated by Trizol method, using three replicates for each cell lines, and evaluated for quality and integrity by the 2100 Bioanalyzer (Agilent Technologies, Waldbronn, Germany). Gene expression analysis was performed using the Illumina TotalPrep RNA (Ambion, Thermo Fisher Scientific, Waltham, Massachusetts, USA) for amplification, labeling and generation of cRNA, while hybridization by Microarray HumanHT-12 v4 Expression BeadChip (Illumina Inc., San Diego, CA, USA) according to the manufacturer's instructions. BeadChip was dried and scanned with an Illumina HiScanSQ system (Illumina Inc., San Diego, CA, USA). Raw data were obtained using Genome Studio Software (Illumina Inc., San Diego, CA, USA). Briefly, quantile normalization algorithm was applied on the data set to correct systematic errors: values below a detection score of 0.05 were filtered out and missing values were imputed. Differential expression analyses were conducted using GenomeStudio Software (Illumina Inc., San Diego, CA, USA) and differently expressed genes (DEGs) were selected with differential score (DiffScore) cutoff set at ±18 (p<0.01). Microarray data were submitted to Array Express under accession number E-MTAB-6152.
Differentially expressed transcripts were displayed using a classic Heatmap, with chromatic variations that reflect the different levels of gene expression between the two cell lines. The heatmap was generated using normalized gene expression values with -1≤log≥1 fold change. The DEGs list was used to evaluate the functional behavior in terms of Biological Processes, performing an enrichment analysis with IPA software (Ingenuity Systems, Redwood City, CA; http://www.ingenuity.com).
MTT assay
Cell viability was evaluated using the dimethylthiazoldiphenyltetrazoliumbromide (MTT, Sigma-Aldrich, Milan, Italy) dye assay as previously described [10]. Briefly, cells were seeded into 24-well plates (1x104 cells/well) and grown up to 144 hours. Cells were then incubated in presence of 500 µM MTT solution for 3 hours at 37°C to allow MTT metabolism into formazan crystals. The formazan crystals were finally solubilized by adding 200 µl of 0.04 N HCl in isopropanol to each microplate well. Adsorbance at 570 nm was measured using a Bio-Tek microplate reader (model EL-340; BioMetallics, Priceston, NJ). Wells containing only DMEM, FBS and MTT were used as controls. In specific experiments, cell viability was evaluated upon exposure to cisplatin (1mg/ml, Accord Healthcare Italia S.r.l.). Cells were treated with cisplatin (1, 10, and 20 μM) for 48 hours and, then, assayed by MTT, as descripted above.
Public dataset analysis
The GEO (Gene Expression Omnibus) portal was used to expand gene expression analysis. Public datasets were selected using 'head and neck cancer' as keyword and results were filtered by selecting only those reporting data about HPV status of cell lines and tumors and the smoking habit of patients. Among all, only two datasets with sufficient information were selected (GSE65858 and GSE52088), the first reporting data on human head and neck squamous cell carcinomas (HNSCCs) [11] and the other on HNSCC cell lines. Geo2R application was used to compare groups by calculating the logFC and the p-value. We defined DEGs genes resulting with -0.26 > logFC>0.26 and p-value <0.05. To assess if Wnt/βCatenin pathway is active in HPV-negative versus HPV-positive tumors/cell lines, a Hypergeometric test was performed on all 39 mSigDB collections [12] reporting the word “wnt” into their denomination; the test was performed on previously obtained up-/down-regulated genes of Wnt/βCatenin pathway and was considered significantly enriched if the adjusted p-value was < 0.05 with fdr correction [13]. Pathways significantly enriched in tumors or cell lines datasets were further investigated in the respective non-significant dataset with a more relaxed threshold (p-value <0.05, no correction) to assess, at least, a possible common trend. All the analysis were performed in R [14].
Statistical analysis
Data were analyzed using Prism5 (GraphPad Software Inc., La Jolla CA). Statistical evaluation of data was performed by t-test. Significant differences were assigned to p value <0.05. Data were presented as means ± standard deviation. All experiments were repeated at least three times.
Results
Gene expression profiles of OSCC cell lines
In order to address the biological relevance of HPV infection in OSCCs occurring in smoking and drinking individuals, UPCI-SCC-131 and UPCI-SCC-154 cell lines, both derived from Caucasian male patients affected by a squamous cell carcinoma of the oral cavity and with a clinical history of long-time tobacco smoking and excessive alcohol drinking, were selected for this study. Both cell lines are p53 wild type, UPCI-SCC-131 cells positive for the amplification of chromosomal band 11q13, UPCI-SCC-154 cells positive for HPV infection (Table 1).
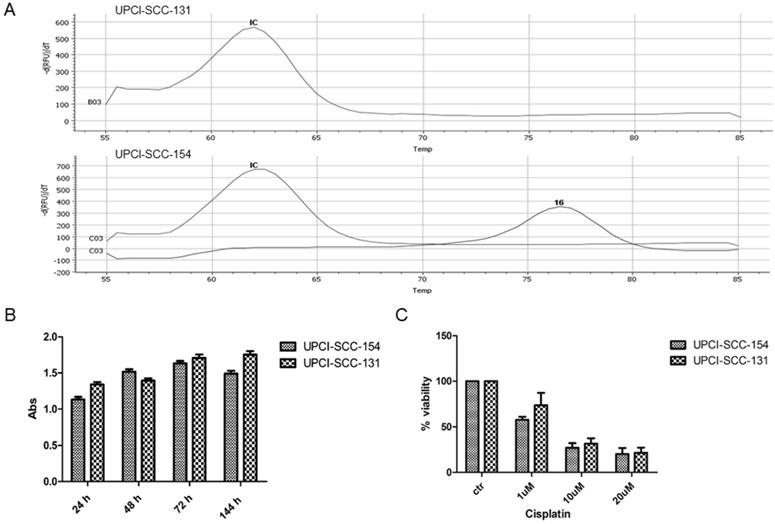
In preliminary experiments, RT-PCR analysis confirmed the positivity of UPCI-SCC-154 cells for HPV strain 16 infection (Figure 1A). In addition, since HPV-positive OSCC cell lines are generally more resistant to cisplatin cytotoxicity [15], UPCI-SCC-154 and UPCI-SCC-131 cell lines were compared for proliferation rates and response to cisplatin. As reported in Figure 1B-C, no major differences were observed between the two OSCC cell lines in terms of growth rate and sensitivity to cisplatin, this suggesting that UPCI-SCC-154 cells, besides being HPV-positive, behave similarly to HPV-negative UPCI-SCC-131 cells. Thus, in further experiments, UPCI-SCC-154 cells were used as a model of tobacco-related HPV-positive cell line and UPCI-SCC-131 as the HPV-negative counterpart.
In order to identify genes differentially expressed between UPCI-SCC-154 and UPCI-SCC-131 cells, we performed a full genome gene expression profiling analysis by Illumina technology. Four hundred seventy-two genes were shown to be differentially expressed (230 up- and 242 downregulated), as reported in Supplementary Table S1 and Figure 2A. IPA was used to predict main signaling pathways differentially modulated between HPV-positive and HPV-negative OSCC cell lines and the top five canonical pathways are reported in Figure 2B. Interestingly, Wnt/βCatenin pathway was identified as the unique signaling pathway unambiguously less active in HPV-positive compared to HPV-negative OSCC cells, with a negative z-score (Figure 2B). Consistently, 13 genes encoding for regulators or target genes of Wnt/βCatenin pathway were differentially expressed between HPV-positive compared to HPV-negative OSCC cells, 8 of them with a log ratio ≥ ±1.5 (Figure 2C). Noteworthy, the expression profile of the vast majority of these genes is consistent with a reduced activity of Wnt/βCatenin signaling pathway in UPCI-SCC-154 compared to UPCI-SCC-131 cells.
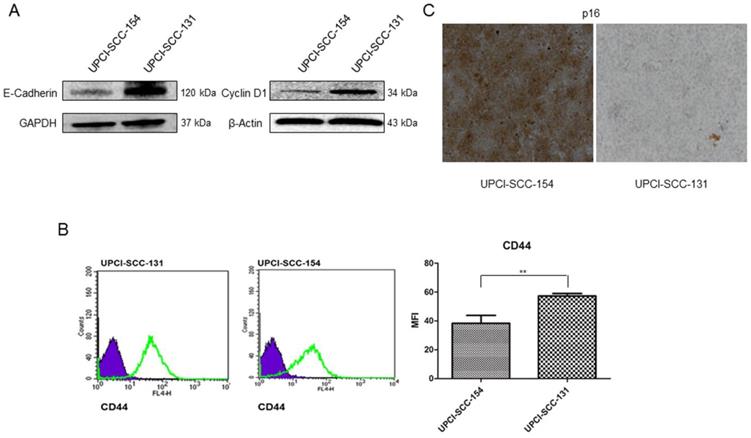
To further validate microarray data, the expression of CCND1 (Cyclin D1) and CDH1 (E-Cadherin) was analyzed by immunoblot, CDKN2A (p16) by immunocytochemistry and CD44 by flow-cytometry in both cell lines. As shown in Figure 3, UPCI-SCC-131 cells exhibited higher expression of Cyclin D1 and E-Cadherin (Figure 3A) and CD44 (Figure 3B) compared to UPCI-SCC-154 cells. Furthermore, HPV-positive OSCC cells showed the upregulation of p16 (Figure 3C), a surrogate marker of HPV infection in oropharyngeal cancer [16-17].
Clinicopathological features of cell lines
| Cell Line | UPCI-SCC-154 | UPCI-SCC-131 |
|---|---|---|
| Gender | M | M |
| Age at diagnosis (years) | 54 | 73 |
| Ethnicity | Caucasian | Caucasian |
| Smoking | Yes | Yes |
| Alcohol | Yes | Yes |
| Origin | n.p. | n.p. |
| Primary tumor size | Tongue | FOM |
| Grading | 3 | 1 |
| TNM | T4N2 | T2N2 |
| TP53 | wt | wt |
| 11q13 amplification | No | Yes |
| p16 | Positive | Negative |
| HPV+ | Positive | Negative |
M, male; n.p., new primary; FOM, floor of mouth; wt, wild type
Characterization of UPCI-SCC-131 and UPCI-SCC-154 cell lines. (A) HPV-DNA detection in UPCI-SCC-131 and UPCI-SCC-154 cell lines by multiplex RT-PCR. (B-C) Comparative analysis of cell proliferation (B) and cell viability (C) between UPCI-SCC-131 and UPCI-SCC-154 cell lines grown up to 6 days (B) or exposed to increasing concentrations of cisplatin for 48 hours (C) and evaluated by MTT incorporation.
Differential gene expression profile between UPCI-SCC-131 and UPCI-SCC-154 cell lines. (A) Heatmap reporting normalized expression values (z score of ΔCT) of significant genes (adjusted p ≤ 0.05, n= 2) differentially expressed between UPCI-SCC-154 and UPCI-SCC-131 cells. The scale ranges from -3 = bright green to +3 = bright red. Each cell line is reported in triplicate. (B) List of top canonical pathways according to IPA. Bars represent -log (p-value). Colors represent z-score; in blue are reported pathways with decreased activity while in grey pathways with conflicting or missing data. (C) List of 13 Wnt-related/target differentially expressed genes between UPCI-SCC-154 and UPCI-SCC-131 cells. Bar Graphs represent gene expression log ratios.

Validation of Wnt-target genes differentially expressed between UPCI-SCC-131 and UPCI-SCC-154 cells. (A-C) Analysis of E-Cadherin and Cyclin D1, CD44 and p16 expression in UPCI-SCC-131 and UPCI-SCC-154 cells by, respectively, immunoblot analysis (A), immunocytochemistry (B) and flow cytometry (C). (B) LSAB-HRP, nuclear counterstaining with haematoxylin; original magnification 10X. (C) Histograms represent mean fluorescence intensity (MFI) ±SD. **p value=0.05 respect to UPCI-SCC-131 cells.
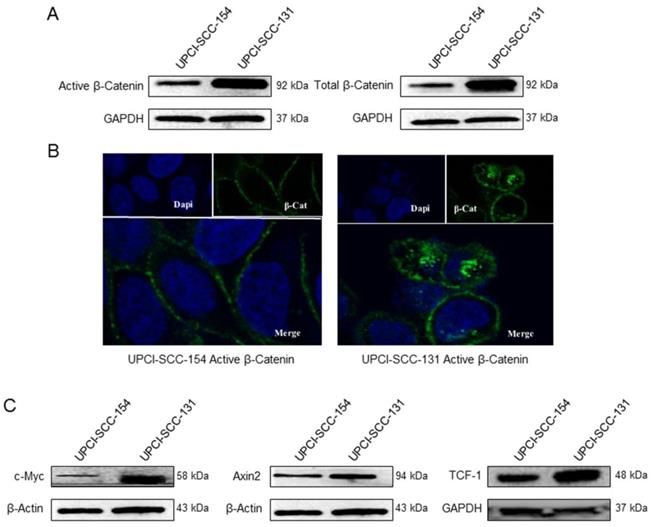
Wnt/βCatenin signaling pathway analysis in UPCI-SCC-131 and UPCI-SCC-154 cells. (A) Immunoblot analysis of total and active βCatenin protein levels in UPCI-SCC-131 and UPCI-SCC-154 cell lines. (B) Confocal microscopy analysis of active βCatenin intracellular distribution in UPCI-SCC-131 and UPCI-SCC-154 cell lines. Images were obtained with an original magnification of 60X. (C) Immunoblot analysis of cMyc, Axin-2 and Tcf-1 in UPCI-SCC-131 and UPCI-SCC-154 cell lines.
Wnt/βCatenin pathway is down-regulated in HPV-positive OSCC cells
βCatenin, a subunit of the cadherin protein complex, is an intracellular signal transducer in Wnt signaling pathway, being able to translocate to the nucleus and induce the transcription of several target genes [18]. The cellular level of βCatenin is mostly controlled by its phosphorylation, which drives the ubiquitination and the proteasomal degradation of the protein; whereas the activation of Wnt pathway results in increase of non-phosphorylated-βCatenin active form, thus enabling its transcriptional activity [19]. Thus, to further study the differential activity of Wnt/βCatenin pathway between HPV-positive/negative OSCC cell lines, the expression of βCatenin and its non-phosphorylated active form was evaluated in both cell lines by immunoblot analysis. Indeed, HPV-negative UPCI-SCC-131 cells showed higher levels of total βCatenin and its active form compared to HPV-positive UPCI-SCC-154 (Figure 4A). In addition, confocal microscopy analysis showed higher nuclear accumulation of active βCatenin in UPCI-SCC-131 cells compared to UPCI-SCC-154 cells (Figure 4B), consistently with a lower activation of Wnt/βCatenin pathway in the HPV-positive cell line. Finally, UPCI-SCC-131 cells showed higher levels of Axin-2, c-Myc and Tcf-1, three Wnt/βCatenin target genes [20] (Figure 4C). Altogether, these data suggest that Wnt/βCatenin signaling pathway is positively modulated in our HPV-negative OSCC cell line compared to the HPV-positive counterpart.
Higher activity of Wnt/βCatenin pathway correlates with stem cell traits
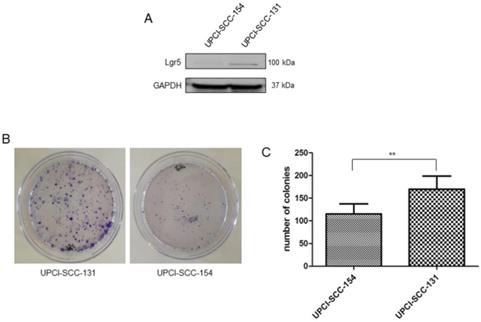
Since Wnt/βCatenin pathway is responsible for cancer stem cell maintenance and self-renewal [21-23], the correlation between modulation of Wnt/βCatenin pathway and stem-like features was further evaluated in our panel of OSCC cells lines. Consistently with the higher level of CD44, an established marker of stemness in OSCCs [24] (Figure 3B), UPCI-SCC-131 cells exhibited higher expression of the cancer stem cell marker, Lgr5 [25-26] compared to HPV-positive UPCI-SCC-154 cells (Figure 5A). In addition, colony forming assay showed higher number of colonies and increase of their size in UPCI-SCC-131 cells compared to UPCI-SCC-154 cells (Figure 5B-C). These results suggest that higher levels of activation of Wnt/βCatenin pathway correlate with more prominent stem-like traits in HPV-negative OSCC cells.
miRNAlet-7e is involved in suppression of Wnt/βCatenin pathway in HPV-positive OSCC
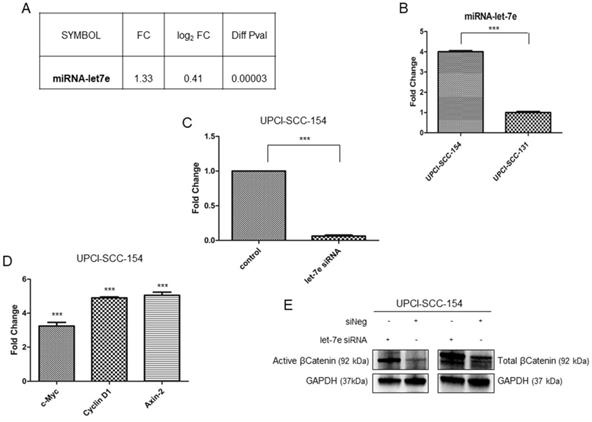
Microarray data suggest that multiple genes belonging to Wnt/βCatenin pathway are modulated in HPV-positive OSCC cells; thus, IPA software was used to identify potential upstream regulators responsible for modulation of Wnt/βCatenin signaling genes. Based on the list of Wnt/βCatenin pathway components or target genes modulated in our data set, IPA identified miRNA let-7e as potential upstream regulator. Indeed, miRNAlet-7e is known to act as a tumor suppressor by inhibiting the expression of βCatenin and stemness genes [27] and, thereby, is downregulated in oropharyngeal squamous carcinomas [28]. The analysis of our data set (Figure 6A) and the subsequent real time validation assay (Figure 6B) showed higher levels of miRNAlet-7e in UPCI-SCC-154 compared to UPCI-SCC-131 cells. Thus, to prove the causative role of miRNA let-7e in suppressing Wnt/βCatenin in UPCI-SCC-154 HPV-positive cells, the expression of total βCatenin and its active form as well as three Wnt/βCatenin-target genes (i.e., Axin-2, c-Myc and Cyclin D1) were evaluated upon silencing of miRNA let-7e in UPCI-SCC-154 cells. Of note, miRNA let-7e silencing (Figure 6C) resulted in increased levels of total and active βCatenin (Figure 6E) and the parallel upregulation of Axin-2, c-Myc and Cyclin D1 (Figure 6D), this suggesting that miRNAlet-7e is involved in suppression of Wnt/βCatenin pathway in HPV-positive OSCC cells.
Stem-like traits in UPCI-SCC-131 and UPCI-SCC-154 cell lines. (A-B) Immunoblot analysis of Lgr5 protein expression (A) and representative images of colony forming assay (B) in UPCI-SCC-131 and UPCI-SCC-154 cell lines. (C) Colony forming assay quantification in UPCI-SCC-131 and UPCI-SCC-154 cell lines. **p value<0.05 compared to UPCI-SCC-131 cells.
Wnt/βCatenin pathway is more active in HPV-negative HNSCC cell lines and tumors in two gene expression public datasets
The higher activation of Wnt/βCatenin pathway in HPV-negative versus HPV-positive HNSCCs was further investigated in two public datasets of HNSCC cell lines and tumors comparing the activation of Wnt/βCatenin pathway in HPV- positive versus HPV- negative tumors/cell lines both derived from patients with a smoking history. Two independent datasets were analyzed: i) GSE65858, which is a large dataset reporting gene expression data from 270 human HNSCCs and which allowed the comparison between 42 HPV16-positive versus 170 HPV-negative tumors, ii) GSE52088, which included gene expression data from 39 HNSCC cell lines and allowed the comparison between 2 HPV16 positive (i.e., SCC47 and SCC90 cells) and 11 HPV-negative (i.e., BIRC22, BIRC31, CAL27, DOK, H103, H357, H400, HN6, MSK921, OECM1 and SCC68 cells) cell lines. In both cases, only tumors/cell lines derived from patients with a smoking habit were analyzed, being the other cell lines/tumors uncertain for this issue. Clinicopathological and molecular characteristics of tumors from GSE65858 dataset and cell lines from the GSE52088 dataset are reported in, respectively, Supplementary Table S2 and Supplementary Table S3. The analysis of the GSE65858 dataset identified 482 differentially expressed genes (191 up- and 291 down-regulated) with -0.26>logFC>0.26, whereas the analysis of the GSE52088 dataset identified 1780 differentially-expressed genes (1091 up- and 689 down-regulated) with -0.26>logFC>0.26 (Supplementary Table S4). Interestingly, 16 up- and 36 down-regulated genes were coherently differentially expressed in both in house cell lines and HPV-positive/negative patients from GSE65858 dataset, as shown by the Venn diagram (Supplementary Figure S1 A-B). In addition, 23 up- and 26 down-regulated genes were coherently differentially expressed in both in house cell lines and HPV-positive/negative cell lines from GSE52088 dataset (Supplementary Figure S1 C-D).
Interestingly, the pathway analysis of gene expression data from both datasets identified the related Wnt/βCatenin gene set (Labbe WNT3a Targets) [29] as activated in HPV-negative tumors/cell lines (Table 2). More specifically, the analysis of gene expression data from HNSCC tumors identified 12/59 Wnt/βCatenin target genes significantly and coherently upregulated in HPV-negative tumors (adj. p-value 0.0036). By contrast, the analysis of HNSCC cell lines identified 9/110 Wnt/βCatenin target genes coherently downregulated in HPV-negative cell lines with a not-significant adjusted p-value. However, this difference was borderline significant (p-value 0.042) with a more relaxed threshold (Table 2). Altogether, these data provide a wider validation of our observation that Wnt/βCatenin pathway is more active in HPV-negative compared to HPV-positive HNSCC cells derived from smoking patients.
Discussion
Patients bearing HPV-positive OSCCs are predominantly younger, non-smokers and non-drinkers and with a better prognosis compared to HPV-negative OSCC patients, even though the basis for these clinical differences are still largely unknown. In such a context, clinical evidences showed that the smoking habit might modify the more favorable clinical behavior of HPV-positive OSCCs [4]. Indeed, several authors reported significant differences in survival rates between smoking and non-smoking HPV-positive OSCC patients, the latter being characterized by better outcome [5,30]. In such a context, Ang et al. grouped patients with oropharyngeal tumors into three subgroups according to their prognosis: HPV-positive non-smoking patients with a longer survival (low risk tumors), HPV-negative smoking patients with the worst survival (high risk tumors) and HPV-positive and smoking patients with an intermediate outcome (intermediate risk tumors) [5]. Therefore, patients with HPV-positivity and smoking habit may represent a distinct subgroup of oropharyngeal tumors, with a clinical phenotype that does not recapitulate classical HNSCCs positive for HPV infection. In such a perspective, a better understanding of the molecular pathobiology of this specific subgroup of oropharyngeal cancers is required.
This study addressed the relevance of HPV infection in the molecular profile of an oral squamous carcinoma cell line obtained from a smoking and drinking individual. Our data suggest that HPV-positive OSCC cells display a growth rate and a sensitivity to cisplatin comparable to HPV-negative cells. The gene expression analysis detected a differential activation of Wnt/βCatenin signaling pathway between the two cell lines, as shown by lower level of βCatenin and Wnt-target/related genes in HPV-positive compared to HPV-negative cells. Consistently, HPV-negative cells showed higher expression of stem cell antigens and increased clonogenic potential.
Enrichment analysis of Wnt/βCatenin related pathway.
| Gene Set | Data Set | Gene Ratio | p-value | Adj. p-value | Gene ID |
|---|---|---|---|---|---|
| Labbe Wnt3a Targets_Up | Tumors GSE65858 | 12/59 | 0.00010 | 0.0036 | Serpine1, Cav1, Ccnd1, Fabp5, Cyr61, Ctgf, Grem1, Cxcl1, Ahr, Ifit3, Nrp1, Ctsc |
| Labbe Wnt3a Targets_Down | Cell Lines GSE52088 | 9/110 | 0.042 | 0.77 | Cdh11, Prkcb, Gng4, Tgm3, Tnfrsf1, Berbb3, Hpgd, Slc27a2, Pter |
Wnt/βCatenin related pathway enrichment in HPV-negative gene expression data from HNSCC tumors/cell lines derived from smoking patients (from GSE65858 and GSE52088 datasets)
Role of miRNAlet-7e in suppression of Wnt/βCatenin pathway in HPV-positive OSCC UPCI-SCC-154 cells. (A-B). miRLet-7e expression in UPCI-SCC-154 compared to UPCI-SCC-131 cells, as obtained by gene expression (A) and real time PCR analysis (B) ***p value<0.0001 compared to UPCI-SCC-131 cells. (C-D) Expression of miRLet-7e (C) and c-Myc, Axin2 and Cyclin D1 genes (D) in UPCI-SCC-154 cells after transfection with control or miRNA Let-7e siRNA. ***p value<0.0001 compared to control. (E) Total and active βCatenin protein expression in UPCI-SCC-154 cells after transfection with control or Let-7e siRNA.
A major issue raised by these observations is their significance in the general context of the current literature about HPV-related oropharyngeal cancers. Indeed, the majority, but not all authors claim that Wnt/βCatenin pathway is up-regulated in patients with HPV-positive tumors [31-32]. More specifically, in vivo studies showed a synergy between HPV infection and activation of Wnt/βCatenin signaling cascade [33-34], as well as Felthaus et al. suggested a specific influence of Wnt/βCatenin signaling on CSC proliferation in HPV-positive tumors [35]. Thus, our observation that HPV-positive oral cancer cells are characterized by lower activation of Wnt/βCatenin signaling is apparently in contrast with the current view of the literature. However, it is important to note that the vast majority of studies that addressed the relationship between HPV infection, Wnt/βCatenin signaling modulation and stemness did not evaluated the relevance of the smoking habit in this process. Consistently with this interpretation, comparative gene expression analyses between HPV-positive and negative human oropharyngeal carcinomas, which did not consider the influence of other risk factors as the smoking habit, identified cell cycle regulation, DNA repair, apoptosis, DNA replication and recombination, but not Wnt/βCatenin signaling cascade, as major functions/pathways differentially active between HPV-positive and negative carcinomas [8-9]. Thus, our study, to our knowledge, is the first that specifically evaluated the relevance of HPV infection in the context of oral carcinoma cell lines derived from smoking individuals and that identified Wnt/βCatenin signaling as differentially active in such a specific cell context. It is important to highlight that a limitation of our study is the head-to-head comparison between only two tobacco-related cell lines differing for the HPV status and the lack of data from human samples. Thus, to support our conclusions we extended our analysis to two independent public datasets reporting comparative gene expression data from human HNSCC tumors and cell lines. This analysis validated and reinforced the hypothesis that Wnt/βCatenin pathway is more active in HPV-negative versus HPV-positive tumors derived from patients with a smoking history, further supporting the concept that HPV-positive oropharyngeal carcinomas from a smoking individual are likely characterized by a different molecular profile compared to classical HPV-positive oropharyngeal cancer cells.
Recent studies highlighted the relevance of Wnt/βCatenin pathway in tumorigenesis, metastatic process, epithelial-to-mesenchymal transition (EMT) and regulation of cancer stem cell (CSC) expansion and maintenance [36]. In fact, Wnt/βCatenin signaling cascade, among several other pathways, is a major driver in the intricate processes that govern CSC self-renewal [37-38] and EMT [39], this suggesting that the dysregulation of this pathway leads to cancer development [40]. In such a context, the abnormal activation of Wnt/βCatenin canonical pathway has been associated with an increase in CSC proliferation and self-renewal in HNSCC [41-43]. Thus, the correlation between higher activation of Wnt/βCatenin pathway and the expression of stem cell markers in our HPV-negative oral carcinoma cell line is consistent with the role of Wnt/βCatenin signaling in stemness maintenance in HNSCCs and other human malignancies [44]. Furthermore, it has been proposed that both HPV infection and the smoking habit enhance the transition toward a mesenchymal phenotype in HNSCC cells [45-46]. Indeed, HPV infection induces the repression of E-Cadherin and promotes EMT, this suggesting that HPV-related HNSCCs are likely to be characterized by a mesenchymal phenotype [47]. However, this hypothesis is apparently in contrast with our observation that HPV-positive UPCI-SCC-154 cells, besides being characterized by lower expression of E-Cadherin compared to the HPV-negative UPCI-SCC-131 cells, are also characterized by lower levels of N-Cadherin, Vimentin and Twist-1, all known biomarkers of epithelial phenotype [48]. This observation is consistent with a recent report suggesting that the association between HPV status and EMT in not statistically significant in a large cohort of 296 oropharyngeal carcinomas and this may be explained by the presence of mixed populations of HPV-positive patients with other associated risk factors (alcohol, tobacco) [49]. Thus, based on all this evidence and consistently with the observation that Wnt/βCatenin pathway is also less active in these tumors/cell lines, we can hypothesize that HPV-positive HNSCC cells derived from smoking patients behave differently from canonical HPV-positive tumors.
The observation that Wnt/βCatenin pathway may be relevant in oropharyngeal tumorigenesis provides preclinical implications for development of new therapeutic strategies in these human malignancies. Indeed, the treatment with Wnt-C59, an inhibitor of Wnt/βCatenin signaling, showed inhibitory activity toward CSCs in nasopharyngeal carcinoma cells [50]. Consistently, many studies reported reduced expression of βCatenin and suppression of CSC proliferation in HNSCC upon treatment with inhibitors of the Wnt/βCatenin canonical pathway, including secreted FRP4 (frizzled-related protein), ATRA (all-trans-retionic acid) and honokiol, an active natural compound [51-53]. Thus, future studies are needed to identify subgroups of HNSCCs with constitutive activation of Wnt/βCatenin signaling and amenable for selective targeting of CSCs through inhibition of Wnt/βCatenin pathway. This hypothesis is relevant taking into account that current treatments for locally advanced and metastatic HNSCC patients rely of radiotherapy and chemotherapy and both treatments are known to kill proliferating cancer cells and not CSCs. Thus, the identification and validation of novel molecular targets to inhibit specifically CSCs are relevant clinical needs in this deadly disease.
Finally, our analysis identified miRNAlet-7e as potential upstream regulator of Wnt/βCatenin pathway modulation in our cell lines. Indeed, miRNA let-7e is known to be functionally involved in regulating the differentiation of stem cells/progenitors and reprogramming cell fates [54-56], and modulating βCatenin expression and Wnt pathway [27]. Consistently with our observation, Vojtechova et al. reported that miRNA let-7e is down-regulated in HPV-negative HNSCC tumors [57]. Thus, our data support the hypothesis that the differential activation of Wnt/βCatenin signaling between our cell lines may be related to the differential expression of miRNA let7e and that the expression of miRNA let7e may represent a tool to select oropharyngeal tumors with activation of Wnt/βCatenin pathway and stem-like traits.
Conclusions
In conclusion, this is, to our knowledge, the first study that addressed, at molecular level, the significance of HPV infection in tobacco-related oropharyngeal cancer cell lines and showed that HPV-positive oral carcinoma cells from smoking patients are likely characterized by low activity of Wnt/βCatenin pathway compared to the HPV-negative counterpart. This unexpected molecular profile suggests that tobacco-associated HPV-positive OSCCs may represent a specific pathological entity with a molecular profile intermediate between classical HPV-positive and HPV-negative oropharyngeal cancers. Future studies are needed to confirm and validate this observation in selected subgroups of human oropharyngeal carcinomas and verify whether these molecular differences may explain the different outcome of tobacco-related HPV-positive tumors.
Abbreviations
BSA: bovine serum albumin; cDNA: complementary DNA; cRNA: complementary RNA; CSC: Cancer Stem Cells; DAPI: 4',6-diamidino-2-phenylindole; DEGs: differently expressed genes; DiffScore: differential score; FBS: Fetal bovine serum; HNSCC: Head and neck squamous cell carcinoma; HPV: Human Papilloma Virus; IPA: Ingenuity Pathway Analysis; MTT: 3-(4,5-Dimethylthiazol-2-yl)-2,5-Diphenyltetrazolium Bromide; OSCCs: oral squamous cell carcinomas; PBS: phosphate buffered saline; RT-PCR: real-time PCR.
Supplementary Material
Supplementary figures and tables.
Acknowledgements
This work was supported by AIRC Grant IG2015 Id. 16738 to ML.
Authors' contributions
Study concept and design: SL and ML; Acquisition of data: SL, VC, LS, AP, CP, IN, GL and MP; Analysis and interpretation of data: SL, PZ, VS and ML; Drafting of the manuscript: SL and ML; Critical revision of the manuscript: SL, GL and ML; Obtaining funding: ML; Study Supervision: SL and ML. All authors read and approved the final version of the manuscript.
Availability of data and materials
All data generated or analyzed during this study are included in this published article. The microarray dataset generated and analyzed during the current study is available to Array Express under accession number E-MTAB-6152.
Competing Interests
The authors have declared that no competing interest exists.
References
1. Ferlay J, Soerjomataram I, Dikshit R, Eser S, Mathers C, Rebelo M, Parkin DM, Forman D, Bray F. Cancer incidence and mortality worldwide: Sources, methods and major patterns in GLOBOCAN 2012. Int J Cancer. 2015;136:E359-86
2. Chai RC, Lambie D, Verma M, Punyadeera C. Current trends in the etiology and diagnosis of HPV-related head and neck cancers. Cancer Med. 2015;44:596-607
3. Hong AM, Martin A, Chatfield M, Jones D, Zhang M, Armstrong B, Lee CS, Harnett G, Milross C, Clark J, Elliott M, Smee R, Corry J, Liu C, Porceddu S, Rees G, Rose B. Human papillomavirus, smoking status and outcomes in tonsillar squamous cell carcinoma. Int J Cancer. 2013;132:2748-2754
4. Granata R, Miceli R, Orlandi E, Perrone F, Cortelazzi B, Franceschini M, Locati LD, Bossi P, Bergamini C, Mirabile A, Mariani L, Olmi P, Scaramellini G, Potepan P, Quattrone P, Ang KK, Licitra L. Tumor stage, human papillomavirus and smoking status affect the survival of patients with oropharyngeal cancer: an Italian validation study. Ann Oncol. 2012;23:1832-1837
5. Ang KK, Harris J, Wheeler R, Weber R, Rosenthal DI, Nguyen-Tân PF, Westra WH, Chung CH, Jordan RC, Lu C, Kim H, Axelrod R, Silverman CC, Redmond KP, Gillison ML. Human Papillomavirus and Survival of Patients with Oropharyngeal Cancer. N Engl J Med. 2010;363:24-35
6. Martinez I, Wang J, Hobson KF, Ferris RL, Khan SA. Identification of differentially expressed genes in HPV-positive and HPV-negative oropharyngeal squamous cell carcinomas. Eur J Cancer. 2007;43:415-432
7. Holzinger D, Flechtenmacher C, Henfling N, Kaden I, Grabe N, Lahrmann B, Schmitt M, Hess J, Pawlita M, Bosch FX. Identification of oropharyngeal squamous cell carcinomas with active HPV16 involvement by immunohistochemical analysis of the retinoblastoma protein pathway. Int J Cancer. 2013;133:1389-99
8. Lohavanichbutr P, Houck J, Fan W, Yueh B, Mendez E, Futran N, Doody DR, Upton MP, Farwell DG, Schwarts SM, Zhao LP, Chen C. Genomewide gene expression profiles of HPV-positive and HPV-negative oropharyngeal cancer: potential implications for treatment choices. Arch Otolaryngol Head Neck Surg. 2009;135:180-188
9. Suarez E, Gonzalez L, Perez-Mitchell C, Ortiz AP, Ramírez-Sola M, Acosta J, Bernabe-Dones RD, Gonzalez-Aquino C, Montes-Rodriguez I, Cadilla CL. Pathway Analysis using Gene-expression Profiles of HPV-positive and HPV-negative Oropharyngeal Cancer Patients in a Hispanic Population: Methodological Procedures. P R Health Sci J. 2016;35:3-8
10. Yedjou CG, Tchounwou PB. In vitro cytotoxic and genotoxic effects of arsenic trioxide on human leukemia (HL-60) cells using the MTT and alkaline single cell gel electrophoreis (comet) assays. Mol Cell Biochem. 2007;301:123-130
11. Wichmann G, Rosolowski M, Krohn K, Kreuz M. et al. The role of HPV RNA transcription, immune response-related gene expression and disruptive TP53 mutations in diagnostic and prognostic profiling of head and neck cancer. Int J Cancer. 2015;137(12):2846-57
12. Liberzon A, Birger C, Thorvaldsdóttir H, Ghandi M, Mesirov JP, Tamayo P. TheMolecular Signatures Database (MSigDB) hallmark gene set collection. Cell Syst. 2015;1(6):417-425
13. Benjamini Y, Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Statist Soc. B. 1995;57:289-300
14. R Core Team R. A language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria. 2016 URL https://www.R-project.org
15. Nagel R, Martens-de Kemp SR, Buijze M, Jacobs G, Braakhuis BJ, Brakenhoff RH. Treatment response of HPV-positive and HPV-negative head and neck squamous cell carcinoma cell lines. Oral Oncol. 2013;49:560-566
16. Kumar R, Ghosh SK, Verma AK, Talukdar A, Deka MK, Wagh M, Bahar HM, Tapkire R, Chakraborty KP, Kannan RR. p16 Expression as a Surrogate Marker for HPV Infection in Esophageal Squamous Cell Carcinoma can Predict Response to Neo-Adjuvant Chemotherapy. Asian Pac J Cancer Prev. 2015;16:7161-7165
17. Sritippho T, Pongsiriwet S, Lertprasertsuke N, Buddhachat K, Sastraruji T, Iamaroon A. p16 - a Possible Surrogate Marker for High-Risk Human Papillomaviruses in Oral Cancer? Asian Pac J Cancer Prev. 2016;17:4049-4057
18. Prakash S, Swaminathan U, Nagamalini BR, Krishnamurthy AB. Beta-catenin in disease. J Oral Maxillofac Pathol. 2016;20:289-299
19. Nusse R, Clevers H. Wnt/β-Catenin Signaling, Disease, and Emerging Therapeutic Modalities. Cell. 2017;169:985-999
20. Herbst A, Jurinovic V, Krebs S, Thieme SE, Blum H, Göke B, Kolligs FT. Comprehensive analysis of β-catenin target genes in colorectal carcinoma cell lines with deregulated Wnt/β-cateninsignaling. BMC Genomics. 2014;15:74
21. Ajani JA, Song S, Hochster HS, Steinberg IB. Cancer stem cells: the promise and the potential. Semin Oncol. 2015;42(Suppl 1):3-17
22. Sato N, Meijer L, Skaltsounis L, Greengard P, Brivanlou AH. Maintenance of pluripotency in human and mouse embryonic stem cells through activation of Wnt signaling by a pharmacological GSK-3-specific inhibitor. Nat Med. 2004;10:55-63
23. Ten Berge D, Kurek D, Blauwkamp T, Koole W, Maas A, Eroglu E, Siu RK, Nusse R. Embryonic stem cells require Wnt proteins to prevent differentiation to epiblast stem cells. Nat Cell Biol. 2011;13:1070-1075
24. Prince ME, Sivanandan R, Kaczorowski A, Wolf GT, Kaplan MJ, Dalerba P, Weissman IL, Clarke MF, Ailles Le. Identification of a subpopulation of cells with cancer stem cell properties in head and neck squamous cell carcinoma. Proc Natl Acad Sci U S A. 2007;104:973-978
25. Haegebarth A, Clevers H. Wnt signaling, Lgr5, and stem cells in the intestine and skin. Am J Pathol. 2009;174:715-721
26. Von Rahden BH, Kircher S, Lazariotou M, reiber C, Stuermer L, Otto C, Germer CT, Grimm M. LgR5 expression and cancer stem cell hypothesis: clue to define the true origin of esophageal adenocarcinomas with and without Barrett's esophagus? J Exp Clin Cancer Res. 2011;30:23
27. Vinas JL, Ventayol M, Brune B, Jung M, Sola A, Pi F, Mastore C, Hotter G. miRNA let-7e Modulates the Wnt Pathway and Early Nephrogenic Markers in Mouse Embryonic Stem Cell Differentiation. PloS One. 2013;8:e60937
28. Christensen BC, Moyer BJ, Avissar M, Ouellet LG, Plaza SL, McClean MD, Marsit CJ, Kelsey KT. A let-7 microRNA-binding site polymorphism in the KRAS 3' UTR is associated with reduced survival in oral cancers. Carcinogenesis. 2009;30:1003-1007
29. Labbé E, Lock L, Letamendia A, Gorska AE, Gryfe R, Gallinger S, Moses HL, Attisano L. Transcriptional cooperation between the transforming growth factor-beta and Wnt pathways in mammary and intestinal tumorigenesis. Cancer Res. 2007;67(1):75-84
30. Hafkamp HC, Manni JJ, Haesevoets A, Voogd AC, Schepers M, Bot FJ, Hopman AH, Ramaekers FC, Speel EJ. Marked differences in survival rate between smokers and nonsmokers with HPV 16-associated tonsillar carcinomas. Int J Cancer. 2008;122:2656-2664
31. Rampias T, Boutati E, Pectasides E, Sasaki C, Kountourakis P, Weinberger P, Psyrri A. Activation of Wnt signaling pathway by human papillomavirus E6 and E7 oncogenes in HPV16-positive oropharyngeal squamous carcinoma cells. Mol Cancer Res. 2010;8:433-43
32. Rampias T, Pectasides E, Prasad M, Sasaki C, Gouveris P, Dimou A, Kountourakis P, Perisanidis C, Burtness B, Zaramboukas T, Rimm D, Fountzilas G, Psyrri A. Molecular profile of head and neck squamous cell carcinomas bearing p16 high phenotype. Ann Oncol. 2013;24:2124-2131
33. Bonilla-Delgado J, Bulut G, Liu X, Cortés-Malagón EM, Schlegel R, Flores-Maldonado C, Contreras RG, Chung SH, Lambert PF, Uren A, Gariglio P. The E6 oncoprotein from HPV16 enhances the canonical Wnt/β-catenin pathway in skin epidermis in vivo. Mol Cancer Res. 2012;10:250-258
34. Bulut G, Fallen S, Beauchamp EM, Drebing LE, Sun J, Berry DL, Kallakury B, Crum CP, Toretsky JA, Schlegel R, Üren A. Beta-catenin accelerates human papilloma virus type-16 mediated cervical carcinogenesis in transgenic mice. PLoS One. 2011;6:e27243
35. Felthaus O, Ettl T, Gosau M, Driemel O, Brockhoff G, Reck A, Zeitler K, Hautmann M, Reichert TE, Schmalz G, Morsczeck C. Cancer stem cell-like cells from a single cell of oral squamous carcinoma cell linesBiochem. Biophys. Res. Commun. 2011;407:28-33
36. Jang GB, Kim JY, Cho SD, Park KS, Junk JY, Lee HY, Hong IS, Nam JS. Blockade of Wnt/β-catenin signaling suppresses breast cancer metastasis by inhibiting CSC-like phenotype. Sci Rep. 2015;5:12465
37. Katoh M, Katoh M. WNT signaling pathway and stem cell signaling network. Clin Cancer Res. 2007;13:4042-4045
38. Cheng Y, Cheung AK, Ko JM, Phoon YP, Chiu PM, Lo PH, Waterman ML, Lung ML. Physiological beta-catenin signaling controls self-renewal networks and generation of stem-like cells from nasopharyngeal carcinoma. BMC Cell Biol. 2013;14:44
39. Heuberger J, Birchmeier W. Interplay of Cadherin-Mediated Cell Adhesion and Canonical Wnt Signaling. Cold Spring Harb Perspect Biol. 2010;2(2):24
40. Serio RN. Wnt of the two horizons: putting stem cell self-renewal and cell fate determination into context. Stem Cells Dev. 2014;23:1975-1990
41. Aminuddin A, Py Ng. Promising Druggable Target in Head and Neck Squamous Cell Carcinoma: Wnt Signaling. Front Pharmacol. 2016;7:244
42. Vidal SJ, Rodriguez-Bravo V, Galsky M, Co rdon-Cardo C, Domingo-Domenech J. Targeting cancer stem cells to suppress acquired chemotherapy resistance. Oncogene. 2014;33:4451-4463
43. Reya T, Morrison SJ, Clarke MF, Weissman IL. Stem cells, cancer, and cancer stem cells. Nature. 2001;414:105-111
44. Anastas JN, Moon RT. WNT signalling pathways as therapeutic targets in cancer. Nat. Rev. Cancer. 2013;13:11-26
45. Jung YS, Kato I, Kim HR. A novel function of HPV16 E6/E7 in epithelial mesenchymal transition. Biochem Biophys Res Commun. 2013;435:339-344
46. Yu MA, Kiang A, Wang-Rodriguez J, Rahimy E, Haas M, Yu V, Ellies LG, Chen J, Fan JB, Brumund KT, Weisman RA, Ongkeko WM. Nicotine promotes acquisition of stem cell and epithelial-to-mesenchymal properties in head and neck squamous cell carcinoma. PLoS One. 2012;7(12 e):51967
47. Laurson J, Khan S, Chung R, Cross K, Raj K. Epigenetic repression of E-cadherin by human papillomavirus 16 E7 protein. Carcinogenesis. 2010;31(5):918-26
48. Liu P-F, Kang B-H, Wu Y-M, Sun J-H, Yen L-M, Fu T-Y, Lin Y-C, Liou H-H, Lin Y-S, Sie H-C, Hsieh I, Tseng Y-K, Shu C-W, Hsieh Y-D, Ger L-P. Vimentin is a potential prognostic factor for tongue squamous cell carcinoma among five epithelial-mesenchymal transition-related proteins. Plos One. 2017;12(6):e0178581
49. Lefevre M, Rousseau A, Rayon R, Dalstein V, Clavel C, Beby-Defaux A, Pretet J-L, Soussan P, Polette M, Saint Guily J L, Birembaut P, the Papillophar Study Group. Epithelial to mesenchymal transition and HPV infection in squamous cell oropharyngeal carcinomas: the papillophar study. Br J Cancer. 2017;116(3):362-369
50. Cheng Y, Phoon YP, Jin X, Chong SY, Ip JC, Wong BW, Lung M.L. Wnt-C59 arrests stemness and suppresses growth of nasopharyngeal carcinoma in mice by inhibiting the Wnt pathway in the tumor microenvironment. Oncotarget. 2015;6:14428-14439
51. Lim YC, Kang HJ, Kim YS, Choi EC. All-trans-retinoic acid inhibits growth of head and neck cancer stem cells by suppression of Wnt/β-catenin pathway. Eur J Cancer. 2012;48:3310-3318
52. Ya CJ, Lai GM, Yeh CT, Lai MT, Shih PH, Chao WJ, Whang-Peng J, Chuang SE, Lai TY. Honokiol eliminates human oral cancer stem-like cells accompanied with suppression of Wnt/βcatenin signaling and apoptosis induction. Evid Based Complement Alternat Med. 2013;2013:146136
53. Warrier S, Bhuvanalakshmi G, Arfuso F, Rajan G, Millward M, Dharmarajan A. Cancer stem-like cells from head and neck cancers are chemosensitized by the Wnt antagonist, sFRP4, by inducing apoptosis, decreasing stemness, drug resistance and epithelial to mesenchymal transition. Cancer Gene Ther. 2014;21:381-388
54. Bussing I, Slack FJ, Grosshans H. Let-7 microRNAs in development, stem cells and cancer. Trends Mol Med. 2008;14:400-409
55. Ortholan C, Puissegur MP, Ilie M, Barbry P, Mari B, Hofman P. MicroRNAs and lung cancer:New oncogenes and tumor suppressors, new prognostic factors and potential therapeutic targets. Curr Med Chem. 2009;16:1047-1061
56. Roush S, Slack FJ. The let-7 family of microRNAs. Trends Cell Biol. 2008;18:505-516
57. Vojtechova Z, Sabol I, Salakova M, Smahelova J, Zavaldi J, Turek L, Grega M, Klozar J, Procazka B, Tachezy R. Comparision of the miRNA profiles in HPV-positive and HPV-negative tonsillar Tumors and a model system of Human keratinocyte clones. BMC Cancer. 2016;16:382
Author contact
![]() Corresponding authors: Prof. Matteo Landriscina, Dipartimento di Scienze Mediche e Chirurgiche, Università degli Studi di Foggia, Viale Pinto, 1, 71100 Foggia, Italy. Tel.: +39 0881 736241; E-mail: matteo.landriscinait or Dr. Valentina Condelli, Laboratorio di Ricerca Preclinica e Traslazionale, IRCCS, Centro di Riferimento Oncologico della Basilicata, Via Padre Pio, 1, 85028 Rionero in Vulture, Italy. Tel.: +39 0971 726525; E-mail: valentina.condelliit
Corresponding authors: Prof. Matteo Landriscina, Dipartimento di Scienze Mediche e Chirurgiche, Università degli Studi di Foggia, Viale Pinto, 1, 71100 Foggia, Italy. Tel.: +39 0881 736241; E-mail: matteo.landriscinait or Dr. Valentina Condelli, Laboratorio di Ricerca Preclinica e Traslazionale, IRCCS, Centro di Riferimento Oncologico della Basilicata, Via Padre Pio, 1, 85028 Rionero in Vulture, Italy. Tel.: +39 0971 726525; E-mail: valentina.condelliit

 Global reach, higher impact
Global reach, higher impact