3.2
Impact Factor
ISSN: 1449-1907
Int J Med Sci 2020; 17(1):45-52. doi:10.7150/ijms.35173 This issue Cite
Research Paper
Molecular identification of isolates of the Trichophyton mentagrophytes complex
1. Research Unit, High Speciality Regional Hospital of Ixtapaluca. Ixtapaluca, Edo. Mexico.
2. Dermatological Center of the Southeast “Dr. Fernando Latapí”, Mérida, Yucatán, México.
3. Efficiency, quality and costs in Health Services Research Group (EFISALUD), Galicia Sur Health Research Institute (IIS Galicia Sur). SERGAS-UVIGO.
4. Mycoloy Service, Hospital Manuel Gea González, Mexico City, Mexico.
5. Dermatology Department, Hospital do Meixoeiro and University of Vigo, Vigo, Spain.
6. European Women's Dermatologic and Venereologic Society (EWDVS), Tui, Spain.
7. Psychodermatology task force of the Ibero-Latin American College of Dermatology (CILAD).
Received 2019-3-24; Accepted 2019-12-10; Published 2020-1-1
Abstract
Background: The Trichophyton mentagrophytes complex is the second most common causal agent of dermatophytosis. It comprises five species—T. mentagrophytes, T. interdigitale, T. erinacei, T quinckeanum, and T. benhamie, as well as nine different genotypes of T. mentagrophytes / T. interdigitale—which are morphologically similar; however, their susceptibility to antifungal agents may differ. For targeted therapy and better prognosis, it is important to identify these species at a molecular level. However, since many hospitals lack molecular methods, the actual aetiology of dermatophytosis caused by this complex remains unknown.
Objective: To characterize 55 anthropophilic isolates of the T. mentagrophytes complex recovered from a dermatological centre in Yucatán, Mexico.
Material and methods: Fifty-five isolates of the T. mentagrophytes complex were obtained from patients with tinea capitis, tinea pedis, tinea corporis, tinea barbae, and tinea unguium. They were characterized by their colonial and microscopic morphology on Sabouraud dextrose agar (SDA) and through the sequencing of a fragment from the region ITS1-5.8S-ITS2.
Results: All colonies grown on SDA were white. Forty-six isolates formed colonies with a powdery texture, while nine isolates formed colonies with a velvety texture. The micromorphological features were typical of the T. mentagrophytes complex. The molecular analysis revealed that 55 isolates were microorganisms that belonged to the T. mentagrophytes complex, that 46 formed powdery colonies representing T. mentagrophytes, and that the other nine isolates that formed velvety colonies represented T. interdigitale. The latter nine isolates were obtained from patients with tinea pedis, tinea corporis, and tinea unguium.
Conclusions: The colony morphology on SDA led to the identification of 46 isolates as T. mentagrophytes and nine isolates as T. interdigitale. At a molecular level, the species identified by their morphology were identified only as T. mentagrophytes complex.
Keywords: Trichophyton mentagrophytes complex, Trichophyton mentagrophytes, Trichophyton interdigitale, Sabouraud dextrose agar, Internal transcribed spacer
Introduction
Dermatophytes are a group of fungi that are closely related to each other and have the enzyme keratinase; thus, they can cause infections in the skin, hair, and nails in both humans and animals1. Among the dermatophytes, Trichophyton mentagrophytes stands out as the second most common causative agent of dermatophytosis2,3, after T. rubrum. This fungus is characterized morphologically based on the development of macro and microconidia with smooth walls. The macroconidia originate laterally in the hyphae or in short pedicles of thin or thick walls and are club-shaped or fusiform, with a size that varies from 4-8 to 8-50 μm. The microconidia are abundant, spherical, pyriform, or irregularly shaped, with sizes varying from 2-3 to 2-4 μm. The most consistent feature of T. mentagrophytes is the production of globose micro-aleuriospores arranged in groups (like a bunch of grapes)3. The taxonomy of T. mentagrophytes is complex due to the changes it has undergone in recent years. Until 2017, T. mentagrophytes-series included seven species: T. tonsurans, T. mentagrophytes, T. interdigitale, T, equinum, T. quinckeanum, T. schoenleinii, and T. simii characterized through ecological data, morphological characteristics, mating type studies, and molecular analysis4.
However, nowadays, only five species are considered—T. mentagrophytes, T. interdigitale, T. erinacei, T quinckeanum, and T. benhamie—as well as nine different genotypes of T. mentagrophytes / T. interdigitale associated with the geographical origin and the source of infection5. These species differ with regards to their ecological preferences; for example, T. interdigitale is anthropophilic and produces dispersal aerial mycelium with numerous conidia, while T. mentagrophytes is zoophilic and produces powdery colonies6. Conventionally, T. mentagrophytes (sensu lato) is identified based on its macro and microscopic features, and sometimes, for its physiological characteristics (hair perforation and urease activity), particularly in the case of atypical isolates7,8; however, the results are usually uncertain due to phenotypic variations among isolates, such as the mycelial growth rate, the colour (white or beige) and appearance of colonies (powdery or velvety), the number of microconidia, the presence or absence of spiral filaments, etc9. In clinical practice, it is common for the anthropophilic velvety isolates obtained from the human foot to be named as T. interdigitale, while the powdery isolates, regardless of their origin, are categorised as T. mentagrophytes9. However, this strategy is insufficient and unsuitable to differentiate T. mentagrophytes from the rest of the species that can cause infection in humans; additionally, like T. interdigitale, it also produces velvety colonies. Therefore, to ensure the accuracy of the identification, the use of molecular methods is recommended in combination with morphological analyses2,6,10,11.
The treatment used for tinea caused by T. mentagrophytes sensu lato is, in general, effective against all members of the complex; however, an increase in the number of cases caused by T. interdigitale resistant to terbinafine, which is the treatment of choice, has been reported12-14. Thus, in order to achieve a targeted therapy and better prognosis, and for epidemiological purposes, it is important to perform the identification of the fungus at a species level using molecular methods11.
In Mexico, many hospitals lack the infrastructure to identify the species of the T. mentagrophytes complex at a molecular level. Thus, conventional methods are used habitually, which leads to a lack of awareness of the actual aetiology of the dermatophytosis.
The present study aimed to characterize the anthropophilic isolates of the T. mentagrophytes complex recovered from a dermatological centre in the Yucatán Peninsula, Mexico, through the conventional method (cultivation on Sabouraud dextrose agar (SDA)) and the molecular method (sequencing of a fragment of the internal transcribed spacer (ITS) region).
Materials and methods
Isolates
In total, 55 patients (22 men and 33 women) with an age range of 1-80 years, a clinical suspicion of infection with the T. mentagrophytes complex, and a clinical presentation of tinea capitis, tinea corporis, tinea unguium, tinea pedis, and tinea barbae that were referred to the Central Mycology Laboratory of Dermatological Center "Dr. Fernando Latapí" in Mérida, Yucatán, Mexico, from 2006 to 2016, were studied for 11 years. The fungi had been identified through the morphological characteristics observed via direct examination with lactophenol cotton blue and were preserved in tubes with SDA (Bioxón, CDMX, MX) and actidione at 4°C.
Morphological characterization
The isolates were inoculated on SDA and were incubated at 28°C for 7 days. The macroscopic and microscopic characteristics of the colonies were observed through microcultures15.
Molecular characterization
DNA Extraction
From the culture of each isolate on SDA plates, a block of approximately 1 cm3 was cut out and transferred into flasks that contained Malt Extract Broth (Bioxon), followed by incubation at 25°C under conditions of agitation for 7-10 days. The mycelium of each isolate was filtered, lyophilized, and preserved at 4°C. The total fungal DNA was extracted from the mycelium using the Animal and Fungi DNA Preparation Kit (Jena Bioscience Gmbh, Jena, TH, DE), following the manufacturer's instructions. The DNA obtained was analysed using a Nanodrop (Thermo Scientific, Waltham, MA, USA) at 260 nm.
PCR and sequencing
The amplification reactions of a fragment of the ITS1-5.8S-ITS2 region were carried out using the oligonucleotides ITS1 (5′-tccgtaggtgaacctgcgg-3′) and ITS4 (5′-tcctccgcttattgatatgc-3′) (Sigma Aldrich Co Ltd., Missouri, USA), which produced a 600 to 750-bp-long fragment, according to the process described by Ziółkowska et al.16. The amplification products were analysed by electrophoresis on a 1.5% agarose gel, followed by staining with GelRed™ 3x (Biotium, Fremont, CA, USA) in TAE buffer (Tris-Acetate-EDTA) 1X. A 100-bp DNA ladder (Fermentas Life Sciences, Waltham, MA, USA) was used as the molecular-weight marker. The images from the gels were visualized and documented with the Gel Doc™ EZ Gel Documentation System (Bio-Rad Laboratories Inc., Hercules, CA, USA).
The amplicons were purified with the PCR Purification Kit (Jena Bioscience GmbH) and sequenced in both directions (Langebio, Guanajuato, Mexico). The final determination of the species was based on the comparison of the sequences of the isolates with the reference sequences (MH865908.1, MH864960.1, MH859073.1, MH865946.1, MH865915.1, and MH859166.1) in the GenBank database, using the BLAST algorithm (https://blast.ncbi.nlm.nih.gov/Blast)17.
To determine the phylogenetic position of the isolates of the T. mentagrophytes complex, a Maximum Likelihood tree was built. This analysis was carried out using RAxML v.8.0.0 software. To obtain 1000 bootstrap replicates, the tree-bisection-reconnection method was used and the GTR+G evolutionary model obtained with the JModeltest 2.1.10 program using the Bayesian Information Criterion (BIC) was employed.
Results
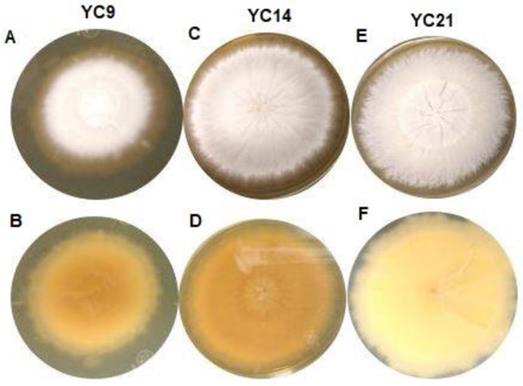
All the studied isolates formed white colonies on SDA; however, 46 isolates formed powdery colonies with a brown-yellowish pigmentation on the back, while nine isolates formed colonies with a velvety surface and yellowish pigmentation on the reverse (Figure 1), suggestive of T. mentagrophytes and T. interdigitale, respectively.
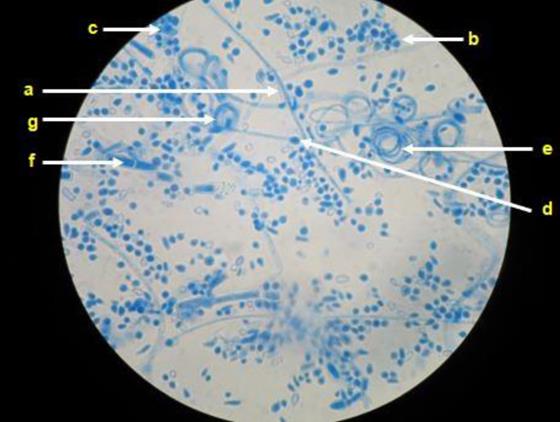
Upon the micromorphological analysis of the 55 isolates, the presence of branched and septate hyaline hyphae was observed; in case of eight isolates, it was possible to observe the formation of spiral hyphae, particularly in fungi that developed velvety colonies. Spherical and/or pyriform microconidia were found in case of all the isolates but were abundant in those that formed powdery colonies, where they were observed to be alternating through the length of the hyphae (Cross of Lorraine). Additionally, club-shaped, multiseptate macroconidia were visualized in case of 20 fungal isolates that formed powdery colonies (Figure 2).
Macroscopic view of the isolates from the Trichophyton mentagrophytes complex cultured for 10 to 15 days on Sabouraud dextrose agar at 25-30°C. A: Isolate YC9, obverse view showing white-coloured, limited-growth colony with velvety texture. B: Isolate YC9, reverse view showing yellow-coloured colony with no diffusible pigment. C: Isolate YC14, obverse view showing white-coloured colony with radial, unlimited growth and powdery texture. D: Isolate YC14, reverse view showing yellow-coloured colony with no diffusible pigment. E: Isolate YC21, obverse view showing white-coloured colony with unlimited growth and granular texture. F) Isolate YC14, reverse view showing beige-coloured colony with no diffusible pigment.
Representative microscopy image. Hyaline, septate, and branched hyphae (a) can be observed in Trichophyton mentagrophytes complex, as well as abundant spherical or semi-spherical microconidia (b) that resemble clusters of grapes (c), spherical chlamyconidia (d), spiral hyphae (e), macroconidia (f) and nodular bodies (g).
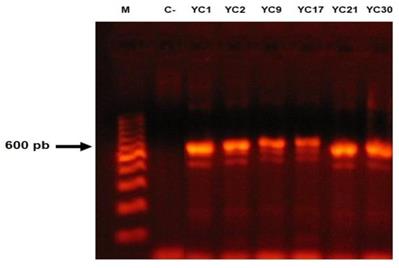
For all isolates, a fragment approximately 600- to 750-bp long was amplified (Figure 3). High similarity (100% identity and 100% coverage) was found by BLASTn among the sequences of the 46 isolates studied, when compared with the sequences of the T. mentagrophytes strains CBS (gbǀMH866908.1, gbǀMH864960.1 and gbǀMH859073.1) deposited in the GenBank database, while sequences of the nine isolates identified as the T. interdigitale by morphological features showed a high similarity with T. interdigitale strains CBS (gbǀMH865946.1, gbǀMH865915.1 and gbǀMH859166.1) deposited in the GenBank database. Importantly, the 46 sequences of T. mentagrophytes showed 98% identity with T. interdigitale and the nine sequences of T. interdigitale showed 99% identity with T. mentagrophytes strains reported in GenBank. The sequences of the 55 studied isolates were deposited in the GenBank database (Access No.: MK045530-MK045584). No isolates of T. erinacei, T quinckeanum or T. benhamie were found (Table 1).
To determine the phylogenetic position of the isolates of the T. mentagrophytes complex, a Maximum Likelihood tree was built, using Nannizzia gypsea (Accession Number KT155807.1) as an outgroup. The phylogenetic tree formed four groups. Group I included the reference sequence for Trichophyton erinacei (Accession Number KT155878.1). Group II included the reference sequence Trichophyton quinckeanum (Accession Number KY680503.1), with a 86% bootstrap. Group III included the reference sequence for Athroderma benhamiae (Accession Number AF506034.1) and Trichophyton mentagrophytes (Accession Number KT155726.1) with an 84% bootstrap. Finally, group IV was composed of 55 isolates from the study that were associated with the reference sequence of Trichophyton interdigitale (Accession Number KC595991.1) with an 85% bootstrap.
Amplification reactions of a fragment of the ITS1-5.8S-ITS2 region, from the fungal isolates of the Trichophyton mentagrophytes complex. Molecular marker (M), negative control (C-).
Conventional and molecular characterization of the anthropophilic isolates of the Trichophyton mentagrophytes complex examined in this study
| Isolate | Initial identification/molecular characterization species | Accession number | Sex/Age (years) | Area | Source/Conventional characterization | Colonies on SDA |
|---|---|---|---|---|---|---|
| YC1 | T. mentagrophytes/T. mentagrophytes | MK045530 | Male/8 | Urban | Hairy skin / tinea capitis | Granular |
| YC2 | T. mentagrophytes/T. mentagrophytes | MK045531 | Female/10 | Rural | Hairy skin / tinea capitis | Granular |
| YC3 | T. mentagrophytes/T. mentagrophytes | MK045532 | Male/3 | Urban | Hairy skin / tinea capitis | Granular |
| YC4 | T. mentagrophytes/T. mentagrophytes | MK045533 | Male/7 | Rural | Hairy skin / tinea capitis | Powdery |
| YC5 | T. mentagrophytes/T. mentagrophytes | MK045534 | Female/4 | Rural | Hairy skin / tinea capitis | Granular |
| YC6 | T. mentagrophytes/T. mentagrophytes | MK045535 | Female/13 | Rural | Hairy skin / tinea capitis | Granular |
| YC7 | T. mentagrophytes/T. mentagrophytes | MK045536 | Female/12 | Rural | Hairy skin / tinea capitis | Granular |
| YC8 | T. mentagrophytes/T. mentagrophytes | MK045537 | Female/4 | Urban | Hairy skin / tinea capitis | Granular |
| YC9 | T. mentagrophytes/T. interdigitale | MK045538 | Female/48 | Rural | Nail / tinea unguium | Velvety |
| YC10 | T. mentagrophytes/T. interdigitale | MK045539 | Male/62 | Rural | Nail / tinea unguium | Velvety |
| YC11 | T. interdigitale/T. interdigitale | MK045540 | Female/69 | Rural | Skin /tinea pedis | Velvety |
| YC12 | T. mentagrophytes/T. interdigitale | MK045541 | Female/54 | Urban | Skin /tinea pedis | Velvety |
| YC13 | T. mentagrophytes/T. mentagrophytes | MK045542 | Male/3 | Rural | Hairy skin / tinea capitis | Granular |
| YC14 | T. mentagrophytes/T. mentagrophytes | MK045543 | Male/5 | Rural | Hairy skin / tinea capitis | Powdery |
| YC15 | T. mentagrophytes/T. interdigitale | MK045544 | Male/38 | Urban | Hairless skin / tinea corporis | Velvety |
| YC16 | T. mentagrophytes/T. mentagrophytes | MK045545 | Female/4 | Urban | Hairy skin / tinea capitis | Granular |
| YC17 | T. mentagrophytes/T. mentagrophytes | MK045546 | Male/9 | Rural | Hairy skin / tinea capitis | Granular |
| YC18 | T. mentagrophytes/T. mentagrophytes | MK045547 | Male/8 | Rural | Hairy skin / tinea capitis | Granular |
| YC19 | T. mentagrophytes/T. mentagrophytes | MK045548 | Female/2 | Rural | Hairy skin / tinea capitis | Granular |
| YC20 | T. mentagrophytes/T. interdigitale | MK045549 | Male/33 | Rural | Hairless skin / tinea corporis | Velvety |
| YC21 | T. mentagrophytes/T. mentagrophytes | MK045550 | Female/5 | Urban | Hairy skin / tinea capitis | Granular |
| YC22 | T. mentagrophytes/T. interdigitale | MK045551 | Female/11 | Rural | Hairless skin / tinea corporis | Velvety |
| YC23 | T. mentagrophytes/T. interdigitale | MK045552 | Female/80 | Urban | Hairless skin / tinea corporis | Velvety |
| YC24 | T. mentagrophytes/T. interdigitale | MK045553 | Female/51 | Urban | Hairless skin / tinea corporis | Velvety |
| YC25 | T. mentagrophytes/T. mentagrophytes | MK045554 | Male/6 | Urban | Hairy skin / tinea capitis | Granular |
| YC26 | T. mentagrophytes/T. mentagrophytes | MK045555 | Male/2 | Rural | Hairy skin / tinea capitis | Granular |
| YC27 | T. mentagrophytes/T. mentagrophytes | MK045556 | Female/10 | Urban | Hairy skin / tinea capitis | Granular |
| YC28 | T. mentagrophytes/T. mentagrophytes | MK045557 | Male/5 | Rural | Hairy skin / tinea capitis | Powdery |
| YC29 | T. mentagrophytes/T. mentagrophytes | MK045558 | Female/8 | Rural | Hairy skin / tinea capitis | Granular |
| YC30 | T. mentagrophytes/T. mentagrophytes | MK045559 | Female/4 | Rural | Hairy skin / tinea capitis | Granular |
| YC31 | T. mentagrophytes/T. mentagrophytes | MK045560 | Female/4 | Rural | Hairy skin / tinea capitis | Granular |
| YC32 | T. mentagrophytes/T. mentagrophytes | MK045561 | Female/1 | Urban | Hairy skin / tinea capitis | Granular |
| YC33 | T. mentagrophytes/T. mentagrophytes | MK045562 | Female/11 | Rural | Hairy skin / tinea capitis | Granular |
| YC34 | T. mentagrophytes/T. mentagrophytes | MK045563 | Female/9 | Urban | Hairy skin / tinea capitis | Granular |
| YC35 | T. mentagrophytes/T. mentagrophytes | MK045564 | Female/4 | Rural | Hairy skin / tinea capitis | Granular |
| YC36 | T. mentagrophytes/T. mentagrophytes | MK045565 | Male/5 | Rural | Hairy skin / tinea capitis | Granular |
| YC37 | T. mentagrophytes/T. mentagrophytes | MK045566 | Male/7 | Urban | Hairy skin / tinea capitis | Granular |
| YC38 | T. mentagrophytes/T. mentagrophytes | MK045567 | Female/5 | Rural | Hairy skin / tinea capitis | Granular |
| YC39 | T. mentagrophytes/T. mentagrophytes | MK045568 | Female/6 | Rural | Hairy skin / tinea capitis | Granular |
| YC40 | T. mentagrophytes/T. mentagrophytes | MK045569 | Male/3 | Rural | Hairy skin / tinea capitis | Granular |
| YC41 | T. mentagrophytes/T. mentagrophytes | MK045570 | Male/62 | Rural | Hairless skin / tinea corporis | Granular |
| YC42 | T. mentagrophytes/T. mentagrophytes | MK045571 | Female/37 | Urban | Hairless skin / tinea corporis | Granular |
| YC43 | T. mentagrophytes/T. mentagrophytes | MK045572 | Female/16 | Rural | Hairless skin / tinea corporis | Granular |
| YC44 | T. mentagrophytes/T. mentagrophytes | MK045573 | Female/32 | Urban | Hairless skin / tinea corporis | Granular |
| YC45 | T. mentagrophytes/T. mentagrophytes | MK045574 | Female/10 | Rural | Hairless skin / tinea corporis | Granular |
| YC46 | T. mentagrophytes/T. mentagrophytes | MK045575 | Female/9 | Urban | Hairless skin / tinea corporis | Powdery |
| YC47 | T. mentagrophytes/T. mentagrophytes | MK045576 | Female/35 | Rural | Hairless skin / tinea corporis | Granular |
| YC48 | T. mentagrophytes/T. mentagrophytes | MK045577 | Male/24 | Urban | Hairless skin / tinea corporis | Granular |
| YC49 | T. mentagrophytes/T. mentagrophytes | MK045578 | Female/40 | Rural | Hairless skin / tinea corporis | Granular |
| YC50 | T. mentagrophytes/T. mentagrophytes | MK045579 | Female/38 | Rural | Hairless skin / tinea corporis | Granular |
| YC51 | T. mentagrophytes/T. mentagrophytes | MK045580 | Male/74 | Urban | Hairless skin / tinea corporis | Granular |
| YC52 | T. mentagrophytes/T. mentagrophytes | MK045581 | Female/39 | Urban | Hairless skin / tinea corporis | Granular |
| YC53 | T. mentagrophytes/T. mentagrophytes | MK045582 | Male/64 | Urban | Hairless skin / tinea corporis | Granular |
| YC54 | T. mentagrophytes/T. mentagrophytes | MK045583 | Male/63 | Urban | Nail / tinea unguium | Powdery |
| YC55 | T. mentagrophytes/T. mentagrophytes | MK045584 | Male/40 | Rural | Hairy skin / tinea barbae | Granular |
Discussion
In clinical practice, the name T. mentagrophytes is still used incorrectly to name dermatophytes that form powdery or cottony-white colonies with yellowish pigmentation on the back, and show multiseptate macroconidia, pyriform microconidia grouped in clusters, and spiral hyphae3. This error occurs for two reasons: first, dermatologists do not consider the possibility that all species comprising the T. mentagrophytes complex, including zoophilic fungi, can cause infection in humans6, and second, the identification of T. mentagrophytes is performed through the correlation of the clinical manifestations of the infection with a microscopic examination of the morphology, and eventually, in combination with physiological tests10, since these methods do not allow the differentiation between the four species of the T. mentagrophytes complex.
The microscopic characteristics of these species are practically identical, while macroscopically, their growths on SDA have been reported to be indicative of the identification of the most frequent species, particularly, T. mentagrophytes and T. interdigitale18,19. However, the variability that the fungus can present in its morphologic features with subcultures, prevents these characteristics from being considered as specific markers of each species; thus, the use of more specific methods, such as molecular techniques, are necessary for their proper identification11. In this study, the macromorphological findings pertaining to the growth of the fungi on SDA were oriented towards the presence of two types of fungal isolates, since colonies with two types of textures, i.e. powdery and velvety, were observed, which have been associated with T. mentagrophytes and T. interdigitale, respectively18. Additionally, although all the isolates of the complex present the typical micromorphology of T. mentagrophytes sensu lato, different characteristics among the species were observed: the presence of macroconidia and/or abundance of microconidia was observed in 100% of the T. mentagrophytes isolates, while in 88.9% of the T. interdigitale isolates, the presence of spiral hyphae20 stood out.
Concerning the molecular data, the Blastn analysis showed that the sequences of the STIS1-5.8S-ITS2 marker of T. mentagrophytes and T. interdigitale are very similar. This echoes the data of de Hoog et al.4, who were only able to identify groups at the genus level with this marker. The same situation occurred in the present study, and because T. mentagrophytes and T. interdigitale are very close, they were included within the same group in the phylogenetic tree. Therefore, from a multilocus phylogeny, Hoog et al.4 proposed better-resolved clades in which the T mentagrophytes series falls within the clade A-1 that includes seven species (T. mentagrophytes, T. interdigitale, T. simii, T. tonsurans, T. equinum, T. quinckeanum and Trichophyton schoenleinii)4. Considering that in this study the sequencing was only performed with the ITS marker, it was confirmed at the molecular level that the morphological identification of the fungi was the genus Trichophyton.
Maximum likelihood Phylogenetic tree internal transcribed sequence rDNA, (RAxML v.8.0.0), sequences of Trichophyton interdigitale and Trichophyton mentagrophytes using GTR+G as model substitution, with 1000 bootstrap replications, shown when >70%, Nannizzia gypsea (Accession Number KT155807.1) was selected as outgroup and as reference strains Trichophyton mentagrophytes (Accession Number KT155726.1), Trichophyton interdigitale (Accession Number KC595991.1), Trichophyton quinckeanum (Accession Number KY680503.1), Trichophyton erinacei (Accession Number KT155878.1) and Athroderma benhamiae (Accession Number AF506034.1).

Nennoff et al.5 proposed a new classification for the T. mentagrophytes complex according to the data reported by Heidemann et al.21 and the results obtained by them in a study performed with population from India using the molecular marker ITS, in which they assure that this marker is good for the differentiation between the species T. mentagrophytes and T. interdigitales. The authors proposed nine genotypes based on the phylogenetic comparison of different sequences obtained from these two species of fungus isolated from different regions of the world. In the present study, the problematic sequences were evaluated by comparing them with the sequences used by Nennoff et al2. They all grouped in the same branch without distinction. Thus, it may be considered that the ITS marker is not resolute and that these fungi may belong to ITS type I, ITS Type III or both.
However, these results do not justify the use of the culture on SDA for a presumptive identification of these species, since the five species of the T. mentagrophytes complex can cause infection in humans8,18,22,23; however, some fungi, such as T. erinacei, are rare6. Furthermore, some fungi may be resistant to first-line antifungal drugs14. The isolates identified as T. interdigitale were obtained from two patients with tinea pedis, five with tinea corporis, and two with tinea unguium, which coincides with findings of other studies, in which this fungus has increasingly been identified as the causal agent of tinea with different clinical manifestations8,11,16,18. Conducting molecular studies on a regular basis is recommended to gain a better understanding of the epidemiology of the infections caused by the T. mentagrophytes complex.
Conclusions
In conclusion, considering the difficulties associated with the identification of dermatophytes and the relatedness of the taxa in our study group, the amplification reactions of a fragment of the ITS1-5.8S-ITS2 region did not provide a powerful alternative for the identification of T. interdigitale. At the molecular level, the species could only be identified as T. mentagrophytes complex. Since the morphological analysis of colonies in SDA led to their preliminary identification, it should not be ruled out for the identification of species in this complex.
Therefore, it is necessary to use a polyphasic approach of classification, using the ITS marker along with other markers or genes during the molecular analysis, to identify the T. mentagrophytes complex at a species level.
Acknowledgements
MSc. Tania Vite Garin from the “Universidad Nacional Autónoma de México (UNAM)” for her assistance in the Molecular systematics assessment.
Funding
No funding was received.
Availability of data and materials
The datasets used and/or analysed during the present study are available from the corresponding author on reasonable request.
Ethics approval and consent to participate
The authors declare that the procedures followed were in accordance with the ethical standards of the responsible committee on human experimentation (institutional and national) and with the Helsinki Declaration of 1975, as revised in 2000.
Patient consent for publication
Written informed consent was obtained from the patient regarding the publication of the case details and any accompanying images.
Authors' contributions
CR-C, MGF-D-L, CEA-D followed up the patient, wrote the manuscript and designed the study. Moreover, they contributed to addressing all questions related to the accuracy and integrity of this study. RA and EM-H provided the analyses of the isolates and the molecular identification results. All the authors have read and approved the final version of the manuscript.
Competing Interests
The authors have declared that no competing interest exists.
References
1. Mercer DK, Stewart CS. Keratin hydrolysis by dermatophytes. Med Mycol. 2019;57(1):13-22 doi: 10.1093/mmy/myx160
2. Nenoff P, Herrmann J, Gräser Y. Trichophyton mentagrophytes sive interdigitale? A dermatophyte in the course of time. J Dtsch Dermatol Ges. 2007;5(3):198-202 DOI: 10.1111/j.1610-0387.2007.06180.x
3. Rivas L. Trichophyton mentagrophytes complex. Rev Chilena Infectol. 2015;32(3):319-320 DOI:10.4067/S0716-10182015000400009
4. de Hoog GS, Dukik K, Monod M. et al. Toward a novel multilocus phylogenetic taxonomy for the dermatophytes. Mycopathologia. 2017;182(1-2):5-31 doi: 10.1007/s11046-016-0073-9
5. Nenoff P, Verma SB, Vasani R. et al. The current Indian epidemic of superficial dermatophytosis due to Trichophyton mentagrophytes-A molecular study. Mycoses. 2019;62(4):336-56 doi: 10.1111/myc.12878
6. Kim J, Tsuchihashi H, Hiruma M. et al. Tinea Corporis due to Trichophyton erinacei probably transmitted from a hedgehog. Med Mycol. 2018;59:E77-E79 doi: 10.3314/mmj.18-00006
7. Robert R, Pihet M. Conventional methods for the diagnosis of dermatophytosis. Mycopathologia. 2008;166(5-6):295-306 doi: 10.1007/s11046-008-9106-3
8. Tartabini ML, Bonino GS, Raccab L. et al. Taxonomic study of clinic isolates of Trichophyton in Rosario, Argentina. Rev Argent Microbiol. 2013;45(4):248-253
9. Symoens F, Jousson O, Planard C. et al. Molecular analysis and mating behavior of the Trichophyton mentagrophytes species complex. Int J Med Microbiol. 2011;301(3):260-266 doi: 10.1016/j.ijmm.2010.06.001
10. Fréalle E, Rodrigue M, Gantois N. et al. Phylogenetic analysis of Trichophyton mentagrophytes human and animal isolates based on MnSOD and ITS sequence comparison. Microbiology. 2007;153(Pt10):3466-3477.DOI 10.1099/mic.0.2006/004929-0
11. Packeu A, Hendrickx M, Beguin H. et al. Identification of the Trichophyton mentagrophytes complex species using MALDI-TOF mass spectrometry. Med Mycol. 2013;51(6):580-585 doi: 10.3109/13693786.2013.770605
12. Avelar Pires CA, Ferreira Santos da Cruz N, Monteiro Lobato A. et al. Clinical, epidemiological, and therapeutic profile of dermatophytosis. An Bras Dermatol. 2014;89(2):259-265 DOI:10.1590/abd1806-4841.20142569
13. Lammoglia-Ordiales L, Martínez-Herrera E, Toussaint-Caire S. et al. Mexican case of tinea incognito and granuloma de Majochi acquired from a hedgehog. Rev Chilena Infectol. 2018;35(2):204-206 doi: 10.4067/s0716-10182018000200204
14. Singh A, Masih A, Khurana A. et al. High terbinafine resistance in Trichophyton interdigitale isolates in Delhi, India harbouring mutations in the squalene epoxidase gene. Mycoses. 2018;61(7):477-484 doi: 10.1111/myc.12772
15. Ridell R. Permanent stained mycological preparations obtained by slide culture. Mycology. 1950;42(2):265-270 DOI: 10.2307/3755439
16. Ziółkowska G, Nowakiewicz A, Gnat S. et al. Molecular identification and classification of Trichophyton mentagrophytes complex strains isolated from humans and selected animal species. Mycoses. 2015;58(3):119-126 doi: 10.1111/myc.12284
17. NCBI. National Center for Biotechnology Information [Internet]. Bethesda (MD): National Library of Medicine; c1988-2018. BLAST Basic Local Alignment Search Tool. 2018 Sep 27 [Revised 15 February 2019]. https://blast.ncbi.nlm.nih.gov/Blast
18. Dhib I, Khammari I, Yaacoub A. et al. Relationship between phenotypic and genotypic characteristics of Trichophyton mentagrophytes strains isolated from patients with dermatophytosis. Mycopathologia. 2017;182(5-6):487-493 doi: 10.1007/s11046-017-0110-3
19. Zhan P, Liu W. The changing face of dermatophytic infections worldwide. 2017; 182 (1-2): 77-86. doi: 10.1007/s11046-016-0082-8.
20. Ramaraj V, Vijayaraman RS, Hemanth V. et al. Molecular strain typing of Trichophyton mentagrophytes (T. mentagrophytes var. interdigitale) using non-transcribed spacer region as a molecular marker. Indian J Med Res. 2017;146(5):636-341 doi: 10.4103/ijmr.IJMR_51_15
21. Heidemann S, Monod M, Gräser Y. Signature polymorphisms in the internal transcribed spacer region relevant for the differentiation of zoophilic and anthropophilic strains of Trichophyton interdigitale and other species of T. mentagrophytes sensu lato. Br J Dermatol. 2010;162(2):282-295 doi: 10.1111/j.1365-2133.2009.09494.x
22. Beguin H, Goens K, Hendrickx M. et al. Is Trichophyton simii endemic to the Indian subcontinent? Med Mycol. 2013;51(4):444-448 doi: 10.3109/13693786.2012.737032
23. Drira I, Neji S, Hadrich I. et al. Tinea manuum due to Trichophyton erinacei from Tunisia. J Mycol Med. 2015;25(3):200-3 doi: 10.1016/j.mycmed.2015.05.001
Author contact
![]() Corresponding author: Carmen Rodriguez-Cerdeira; Meixoeiro Hospital,CHUVI, C/Meixoeiro S/N 36200, Vigo, Spain. Tel: 0034986814517/0034600536114, Fax: 0034986276416, E-mail: carmencerdeira33com.
Corresponding author: Carmen Rodriguez-Cerdeira; Meixoeiro Hospital,CHUVI, C/Meixoeiro S/N 36200, Vigo, Spain. Tel: 0034986814517/0034600536114, Fax: 0034986276416, E-mail: carmencerdeira33com.

 Global reach, higher impact
Global reach, higher impact