Impact Factor
ISSN: 1449-1907
Int J Med Sci 2019; 16(6):882-892. doi:10.7150/ijms.30082 This issue Cite
Research Paper
Microarray and proteome array in an atherosclerosis mouse model for identification of biomarkers in whole blood
1. Department of Biomedical Laboratory Science, College of Health Science, Eulji University, Seongnam-si, Gyeonggi-do, 13135, Republic of Korea
2. Department of Senior Healthcare, BK21 plus Program, Graduated School, Eulji University, Daejeon, 34824, Republic of Korea
3. Department of Biomedical Laboratory Science, College of Health Sciences, Yonsei University at Wonju, Wonju, Gangwon-do 26493
Received 2018-9-20; Accepted 2019-5-2; Published 2019-6-2
Abstract
Cardiovascular disease (CVD) is highly fatal, and 80 percent of the mortality is attributed to heart attack and stroke. Atherosclerosis is a disease that increases a patient's risk to CVD and is characterized by atheroma formed by immune cells, lipids, and smooth muscle cells. When an atherosclerotic lesion grows and blocks blood vessels or when an atheroma ruptures and blocks blood vessels by embolism, sudden angina, or stroke can occur. It is therefore important to diagnose atherosclerosis early and prevent its progression to more severe disease. Although myeloperoxidase, plasma fibrinogen, cardiac troponin-I, and C-reactive protein have been considered as diagnostic markers for multiple cardiac risks, specific biomarkers for atherosclerosis have not been clearly determined yet. Particularly, reliable biomarkers for the diagnosis of atherosclerosis using whole blood are not yet available. In this study, we screened potential biomarker genes and proteins from whole blood of apolipoprotein E knockout (ApoE-/-) mice maintained on a Western diet, by comparing them to ApoE+/+ mice. We used whole blood for microarray and proteome array. Candidate genes and proteins identified from each method were confirmed with quantitative real-time PCR and ELISA. Based on our data, we speculate that Lilrb4a, n-R5s136, and IL-5 are potential targets that can be developed into novel biomarkers of atherosclerosis. Our study contributes to the diagnosis of atherosclerosis using whole blood in clinical settings.
Keywords: Atherosclerosis, ApoE knockout, microarray, proteome array, biomarker
Introduction
The WHO reports cardiovascular disease (CVD) to be the most fatal disease in the world. Approximately 80% of that mortality is caused by heart attack and stroke. Although CVD is manifested suddenly, people with symptoms such as atherosclerosis or hyperlipidemia are at higher risk of the disease [1]. Atherosclerosis is known to be a major underlying pathology of CVD. Age, hypertension, smoking, hyperlipidemia, obesity and metabolic syndrome, and diabetes are the major risk factors for atherosclerosis [2]. Regardless of the cause, atherosclerosis is usually accompanied by a chronic inflammatory reaction and thickening of the endothelium, which limits blood flow. It is characterized by rupture of the atheroma generated from the intima of endothelium, or by the formation of thrombus in the blood vessel, resulting in a sharp narrowing and blocking of the blood vessel. Atherosclerosis does not tend to have symptoms at first and most people are unaware that they have the disease, but as the disease progresses symptoms, such as chest pain are manifested. Because symptoms do not appear until late stages of the disease, it is imperative to diagnose atherosclerosis in early stages in order to prevent severe symptoms or CVD.
To study atherosclerosis, many animal models, including knockouts have been developed. The apolipoprotein E knockout (ApoE-/-) and low-density lipoprotein receptor deficient (LDLR-/-) C57BL/6 mice are the most frequently used [3, 4]. ApoE-/- mice develop atherosclerotic lesions, like humans, when maintained on normal chew for several months. However, the LDLR-/- mice require more than a year to develop atherosclerotic lesions [5, 6]. The predominant plasma lipoproteins in LDLR-/- mice are very-low-density lipoprotein (VLDL) and low-density lipoprotein (LDL), whereas ApoE-/- mice have cholesterol with lipoprotein, like the apolipoprotein B48 [7]. Unlike LDLR-/- mice, the ApoE-/- mice are not affected by natural killer T-cells [8], and it is also known that the amount of VLDL does not correlate with atherosclerosis of the aortic root in ApoE-/- mice. In addition to mice, animal models for atherosclerosis have also been developed in rat, rabbit, and pig [9, 10].
Several studies have used the ApoE-/- mice for atherosclerosis [9-15]. Mice lacking the ApoE gene show similar growth as healthy C57BL/6 mice [3]. ApoE-/- mice fed a diet of normal chew for 8-9 months, show lipid accumulation and foam cell deposition in the aorta. However, when ApoE-/- mice were maintained on a Western diet, lipid accumulation was found in the aorta after 10 weeks [5, 13], and lipid staining of the aorta showed the presence of atherosclerotic lesions [16].
Using cDNA filter array, mRNA extracted from the aorta of ApoE+/+ and ApoE-/- mice were compared, and transcript levels of vascular cell adhesion molecule (VCAM), intercellular adhesion molecule (ICAM), nerve growth factor (NGF), hepatocyte growth factor (HGF), monocyte chemotactic protein-3 (MCP3), cellular retinoic acid binding protein 2 (CRABP-II), and selectin P (SELP) were found to be elevated in ApoE-/- mice [17]. The proteins VCAM, ICAM, and P-selectin play a role in the formation of foam cells. They are expressed on endothelial cells, where they play a role in holding leukocytes and rolling them. Other studies have shown that transcripts of CD44, lymphocyte function-associated antigen 1 (LFA-1), cathepsin B, and cyclooxygenase-2 (COX-2), in addition to VCAM and ICAM, are also increased in the aorta of ApoE-/- mice [18]. Furthermore, the elevated levels of VCAM, ICAM, cathepsin B proteins in the aorta were confirmed. In addition, a bioinformatics analysis of microarray data obtained from mRNA of ApoE-/- and ApoE+/+ mice identified positive regulation of B-cell activation, chemotaxis, antigen binding, and lipid-related pathways to be associated with atherosclerosis [19]. Analysis of serum protein and RNA of aorta found elevated levels of the chemokine (C-C motif) ligand (CCL) proteins CCL2, CCL19, and CCL21 along with their corresponding transcripts [20]. Additionally, analysis of proteins from the aorta and plasma of ApoE-/- mice found increased levels of immunoglobulins or CD5 antigens in both [21].
Multiple molecules have been reported to be associated with atherosclerosis. Cytokines, such as tumor necrosis factor alpha (TNFα) and interleukin 1 (IL-1), nitric oxide synthase (NOS) involved in the production of nitric oxide (NO), selectin, and membrane proteins VCAM and ICAM activated during the progress of atherosclerosis, have been identified to influence the development of atherosclerosis [22]. The effects of TNFα and endothelial NOS (eNOS) knock outs in ApoE-/- mice have also been verified. The ApoE/TNFα double knockout mice showed lower plasma cholesterol levels and weaker atherosclerotic lesions than the ApoE-/- mice [23]. The double knockout of eNOS and ApoE confirmed an increase in atherosclerosis [24], suggesting that eNOS plays a protective role against atherosclerosis. In addition, studies on double knockout of selectin, cyclooxygenase, scavenger receptor class B, interleukin-10, fractalkine (CXC3CL1), retinoid X receptor, or Fcγ receptor with ApoE were also performed, but their effects on atherosclerosis remain unknown [25].
Several diagnostic studies for atherosclerosis are underway. Myeloperoxidase, plasma fibrinogen, and cardiac troponin-I have been reported as biomarkers for cardiovascular risk [26]. In addition, clinicians use high-sensitivity C-reactive protein (hs-CRP) levels along with family history and other risk factors, including atherosclerosis, for CVD diagnosis [27]. However, hs-CRP is used broadly as a marker of systemic inflammatory disease. A high hs-CRP count increases the probability of being at risk of atherosclerosis but also increases the likelihood that it is a different CVD. Current diagnostic methods for atherosclerosis include ultrasound, computed tomography (CT), magnetic resonance imaging (MRI), and angiography [28, 29]. However, these methods are costly and require professionals for interpretation. In addition, angiography can cause an allergic reaction to the catheter, caused by contrast media or vascular injury. Atherosclerosis is a complex disease that cannot be represented by a single biomarker at a time.
Some studies have extracted monocyte and macrophage from blood and atherosclerotic plaque of atherosclerosis patients and found the Finkel-Biskis-Jinkins osteosarcoma (FOS) gene to be elevated. Based on this observation, the analysis of circulating cells was suggested to be useful for atherosclerosis diagnosis [30]. However, most animal experiments have analyzed aortic tissue and/or serum or plasma. Microarray or proteome array studies of atherosclerosis are usually performed using aortic tissue. As a result, applying these methods to a patient requires the collection of aorta tissue, and acquiring atherosclerotic lesions is burdensome to the patient. In biomarker studies of atherosclerosis, serum or plasma has been used to confirm the results of aortic tissue. In this study, we used whole blood rather than serum or plasma to examine differential gene expression levels in ApoE-/- and ApoE+/+ mice and find biomarkers using microarray experiments. Furthermore, we used whole blood in proteome array studies to examine the differential expression of proteins.
Materials and Methods
Animals
Animals were purchased from GHBio (Daejeon, Korea). The planning, management, and experimentation of the animal study was approved by the Eulji University Institutional Animal Care and Use Committee (approval No. EUIACUC16-24, approval date 12 December 2016). Male C57BL/6 ApoE+/+ mice (6-8 weeks old, n = 15) and ApoE-/- mice (6-8 weeks old, n = 15) were fed Western diet containing 21% fat (Research Diets, USA) and provided free access to drinking water. Experiments were performed three times independently using 5 mice per group. Each independent experiment has been described as a batch in this manuscript. After 10 weeks, blood and aorta were collected from the mice. An aliquot of the whole blood was stored in a PAXgene tube (PreAnalytiX, Hombrechtikon, Switzerland) for microarray analysis. The remaining whole blood was stored with ethylenediaminetetraacetic acid (EDTA) in an Eppendorf tube at -80 °C until RNA and protein extraction. The aorta were fixed in 4% paraformaldehyde for 24 hours at 4 °C and stored at 4 °C until further use.
Oil red O stain by the en face method
The fixed aorta were transferred into 78% methanol in an Eppendorf tube for 5 min and this step was repeated twice. The aorta were moved into fresh Oil red O solution (filtered 0.2% Oil red O in 100% methanol) and incubated for 1 h on a rocker at room temperature. Then the aorta were washed twice in 78% methanol for 5 min. The stained aorta were stored in PBS at 4 °C. Using fine forceps, the stained aorta was placed on black paper in a petri dish. Under the stereomicroscope, the aorta was cut longitudinally using spring scissors. In the dark room, pictures of the stained aorta were taken with a digital camera attached to the stereomicroscope [16]. The ImageJ software (National Institutes of Health, USA) was used to quantify surface area of lesions and to count the number of spots [31]. The percentage of lesion area was calculated by dividing it by the total aortic area.
RNA extraction, cDNA synthesis, and quantitative real-time (qRT-) PCR
Total RNA was prepared using the QIAamp RNA blood mini kit (Qiagen, Valencia, CA, USA) according to manufacturer's instructions. The cDNA was synthesized from 1 µg of total RNA using the SensiFAST cDNA synthesis kit (Bioline, Taunton, MA, USA), and qRT-PCR was performed on an ABI StepOnePlus system (Applied Biosystems, Foster City, CA, USA). The following primer sequences were used for Lilrb4a, 5'-CCATGCTCACAGTGCTGCTA-3' and 5'-CCAGATGATGGGCTTTGGGA-3'; Cybb, 5'-CTGAAGGGGGCCTGTATGTG-3' and 5'-ATGGCAAGGCCGATGAAGAA-3' [32]; n-R5s136, 5'-GTCTACGGCCATACCACCCT-3' and 5'-AAAGCCTACAGCACCCGGTAT-3'; Pf4, 5'-CCTCAAGGTAGAACTTTACTCACTA-3' and 5'-GGATCCCAGAGGAGATGGTCT-3'; IFNγ, 5'-GGATGCATTCATGAGTATTGC-3' and 5'-CCTTTTCCGCTTCCTGAGG-3' [33]; IL-5, 5'-CGCTCACCGAGCTCTGTTG-3' and 5'-CCAATGCATAGCTGGTGATTTTT-3' [33]; TNFα, 5'-CTCCAGGCGGTGCCTATGT-3' and 5'-GAAGAGCGTGGTGGCCC-3' [33]; and GAPDH, 5'-AAGGTCATCCCAGAGCTGAA-3' and 5'-CTGCTTCACCACCTTCTTGA-3' [34]. GAPDH was used as the housekeeping gene to normalize expression levels of target genes, which was calculated using the 2-ΔΔCT method [35]. As for the reduced value, however, the negative reciprocal was taken for convenience.
Microarray
The whole blood collected in the PAXgene tube was used for RNA extraction, and the purity and integrity of the RNA was measured using the 260/280 optical density ratio on the Agilent 2100 Bioanalyzer (Agilent Technologies, Palo Alto, CA, USA) according to the manufacturer's protocol. Experiments were performed three times independently. The microarray was analyzed using a GeneChip Mouse Gene 2.0 ST Array in Macrogen Co. (Seoul, Korea). The data were summarized and normalized using a robust multi-average (RMA) method implemented in Affymetrix® Power Tools (APT). We exported the results of gene-level RMA analysis and conducted an analysis of the differentially expressed genes (DEGs). Statistical significance of the expression data was determined using independent t-test and fold changes, in which the null hypothesis was that no difference exists among the groups. Therefore, to analyze the difference between the two groups, the following formula was used to obtain the fold change (FC) value: FC = 2^(mean value of ApoE-/- group - mean value of ApoE+/+ group). However, for the reduced value, the negative reciprocal was considered for convenience. Only the values with P < 0.05 and 2.5-fold difference were used for the analysis.
Proteome array
The mouse atherosclerosis antibody array (Abcam, Cambridge, UK) was used according to the manufacturer's instruction. The whole blood with EDTA stored at -80 °C was thawed once and 50 μL of blood was tested. Experiments were performed three times independently. The HLImage software (Western Vision Software, Salt Lake City, UT, USA) was used to analyze the spot density.
Enzyme-linked immunosorbent assay (ELISA)
The Mouse IFNγ, IL-5, and TNFα ELISA kit (Abcam, Cambridge, UK) was used for the analysis of IFNγ, IL-5, and TNFα. We used 50 µL of whole blood with EDTA according to the manufacturer's instructions. The intensity of the color was measured at 450 nm using Infinite M200 PRO Multimode Microplate Reader (Tecan, Switzerland).
Statistical analysis
To compare the two groups, the Student's t-test was used in Excel software (Microsoft, Redmond, WA, USA). Statistical significance was analyzed based on P < 0.05.
Results
Mouse model of atherosclerosis
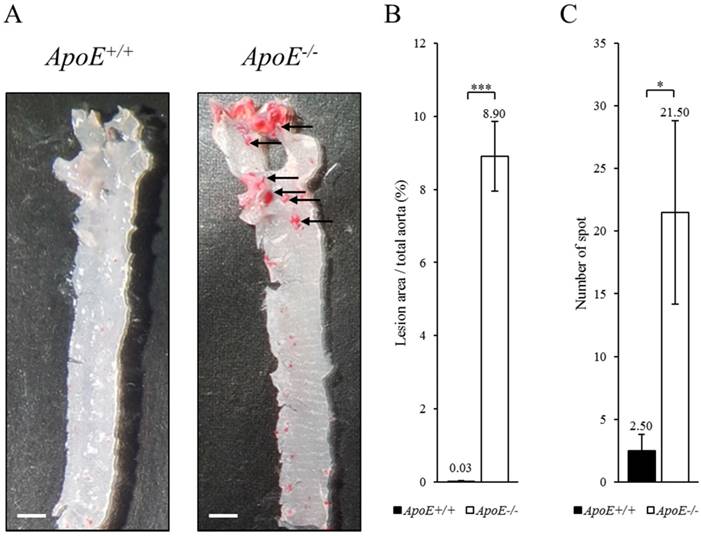
To identify biomarker candidates of atherosclerosis, we compared ApoE-/- and ApoE+/+ mice fed with Western diet for 10 weeks. The mice were euthanized, and whole blood and tissue were collected under animal care guidelines of Eulji University. The aorta of ApoE+/+ and ApoE-/- mice were stained with Oil red O using the en face method (Figure 1). The atherosclerotic lesion in the aorta root of ApoE-/- mice were stained red (arrow). The dyed area is concentrated in the root of the aorta in ApoE-/- mice, suggesting that atherosclerosis was progressing in the ApoE-/- mice, unlike the ApoE+/+ mice.
Histological analysis of the aorta from ApoE+/+ and ApoE-/- mice. (A) The oil red O stain of aorta in ApoE+/+ (left panel) and ApoE-/- (right panel) mice. Arrow indicates atherosclerotic lesions stained by oil red O. Scale bars = 1 mm. (B) Lesion area quantification. (C) Number of stained spots. Data represent mean ± S.E.M. Experiments were performed three times independently. *P < 0.05, ***P < 0.001.
Gene expression in blood of ApoE+/+ and ApoE-/- mice. Increased genes
| No. | Gene symbol | Relative fold change (batch 1) | Relative fold change (batch 2) | Relative fold change (batch 3) | Avg. | P-value | ApoE+/+ S.E.M. | ApoE-/- S.E.M. |
|---|---|---|---|---|---|---|---|---|
| 1 | Lilrb4a | 4.5 | 2.7 | 6.9 | 4.7 | 0.041 | 0.5 | 0.1 |
| 2 | Sirpb1b | 4.5 | 2.9 | 5.1 | 4.2 | 0.017 | 0.4 | 0.3 |
| 3 | Tlr7 | 3.7 | 5.8 | 2.7 | 4.1 | 0.014 | 0.2 | 0.4 |
| 4 | Cybb | 4.3 | 3.9 | 3.7 | 4.0 | 0.003 | 0.2 | 0.2 |
| 5 | Itgam | 3.6 | 2.8 | 4.3 | 3.6 | 0.019 | 0.4 | 0.2 |
| 6 | Fn1 | 3.5 | 2.8 | 4.3 | 3.5 | 0.001 | 0.1 | 0.1 |
| 7 | Rassf4 | 4.1 | 4.0 | 2.3 | 3.5 | 0.025 | 0.1 | 0.3 |
| 8 | Cers6 | 4.2 | 3.4 | 2.5 | 3.4 | 0.032 | 0.2 | 0.4 |
| 9 | Pld4 | 3.3 | 3.9 | 2.8 | 3.3 | 0.005 | 0.1 | 0.2 |
| 10 | Ctss | 2.8 | 3.6 | 3.5 | 3.3 | 0.028 | 0.4 | 0.3 |
| 11 | C3 | 4.0 | 2.7 | 2.8 | 3.2 | 0.001 | 0.1 | 0.1 |
| 12 | Ctsc | 3.0 | 3.5 | 2.8 | 3.1 | 0.002 | 0.1 | 0.2 |
| 13 | Il13ra1 | 3.8 | 2.5 | 2.8 | 3.1 | 0.016 | 0.2 | 0.3 |
| 14 | Cd68 | 3.3 | 3.1 | 2.6 | 3.0 | 0.011 | 0.2 | 0.3 |
| 15 | Ddx5 | 2.3 | 3.8 | 2.9 | 3.0 | 0.018 | 0.3 | 0.2 |
| 16 | Serpinb10 | 2.7 | 3.4 | 2.5 | 2.9 | 0.038 | 0.3 | 0.4 |
| 17 | Tnfrsf1b | 3.1 | 2.8 | 2.8 | 2.9 | 0.040 | 0.3 | 0.4 |
| 18 | Bmpr2 | 2.9 | 2.7 | 2.8 | 2.8 | 0.001 | 0.1 | 0.1 |
| 19 | Psap | 3.1 | 2.7 | 2.6 | 2.8 | 0.012 | 0.2 | 0.3 |
| 20 | Scd2 | 2.0 | 4.4 | 1.9 | 2.8 | 0.014 | 0.2 | 0.2 |
| 21 | Ifi30 | 2.4 | 3.5 | 2.1 | 2.7 | 0.006 | 0.1 | 0.2 |
| 22 | Naip6 | 3.0 | 2.2 | 3.0 | 2.7 | 0.012 | 0.2 | 0.2 |
| 23 | Soat1 | 1.8 | 3.3 | 2.9 | 2.7 | 0.019 | 0.3 | 0.1 |
| 24 | Hsp90b1 | 1.9 | 3.3 | 2.5 | 2.6 | 0.029 | 0.3 | 0.2 |
| 25 | Oas2 | 2.6 | 2.6 | 2.3 | 2.5 | 0.006 | 0.1 | 0.2 |
| 26 | Atp2b1 | 2.2 | 2.7 | 2.6 | 2.5 | 0.009 | 0.2 | 0.1 |
| 27 | Fgr | 2.4 | 2.3 | 2.8 | 2.5 | 0.013 | 0.2 | 0.2 |
| 28 | Rel | 2.7 | 3.0 | 1.9 | 2.5 | 0.020 | 0.1 | 0.2 |
| 29 | Ifngr1 | 2.2 | 2.7 | 2.7 | 2.5 | 0.050 | 0.4 | 0.3 |
Gene expression in blood of ApoE+/+ and ApoE-/- mice. Decreased genes
| No. | Gene symbol | Relative fold change (batch 1) | Relative fold change (batch 2) | Relative fold change (batch 3) | Avg. | P-value | ApoE+/+ S.E.M. | ApoE-/- S.E.M. |
|---|---|---|---|---|---|---|---|---|
| 1 | n-R5s136 | -3.8 | -2.2 | -2.7 | -2.9 | 0.024 | 0.0 | 0.3 |
| 2 | Thbs1 | -2.2 | -3.9 | -2.7 | -2.9 | 0.030 | 0.4 | 0.2 |
| 3 | Slc6a4 | -2.1 | -4.2 | -2.5 | -2.9 | 0.035 | 0.3 | 0.4 |
| 4 | Pf4 | -2.2 | -3.4 | -2.4 | -2.7 | 0.001 | 0.1 | 0.1 |
| 5 | Pde5a | -2.1 | -3.8 | -2.3 | -2.7 | 0.012 | 0.2 | 0.1 |
| 6 | Cd226 | -2.5 | -3.1 | -2.3 | -2.6 | 0.001 | 0.1 | 0.1 |
| 7 | Gp6 | -2.3 | -3.4 | -2.2 | -2.6 | 0.002 | 0.1 | 0.1 |
| 8 | Itgb3 | -2.4 | -3.0 | -2.5 | -2.6 | 0.012 | 0.2 | 0.2 |
| 9 | Mpl | -2.2 | -3.3 | -2.3 | -2.6 | 0.041 | 0.4 | 0.3 |
| 10 | Gp5 | -1.9 | -3.3 | -2.2 | -2.5 | 0.005 | 0.2 | 0.1 |
| 11 | Angpt1 | -2.3 | -3.1 | -2.2 | -2.5 | 0.006 | 0.2 | 0.2 |
| 12 | Trpc6 | -2.6 | -2.7 | -2.2 | -2.5 | 0.006 | 0.2 | 0.2 |
| 13 | Parvb | -2.6 | -2.9 | -2.0 | -2.5 | 0.009 | 0.2 | 0.2 |
| 14 | Alox12 | -2.2 | -3.1 | -2.2 | -2.5 | 0.017 | 0.2 | 0.2 |
| 15 | Arhgap10 | -2.2 | -3.1 | -2.2 | -2.5 | 0.018 | 0.2 | 0.2 |
Gene expression profiling
The gene expression profile in whole blood of ApoE+/+ and ApoE-/- mice was analyzed using microarray. The expression of 44 genes were altered by more than 2.5-fold in the blood mRNA of ApoE-/- mice compared to ApoE+/+ mice. Of these the expression of 29 genes were upregulated (Table 1A) and 15 genes were downregulated in the ApoE-/- mice (Table 1B). Four genes, Lilrb4a, Sirpb1b, Tlr7, and Cybb, were upregulated by more than 4-fold. Lilrb4a was the most upregulated gene (P < 0.041), and Cybb was most significantly upregulated gene (P < 0.003). Five genes, n-R5s136, Thbs1, Slc6a4, Pf4, and Pde5a were downregulated by more than 2.7-fold. n-R5s136 was the most downregulated gene (P < 0.024), and Pf4 was most significantly downregulated gene (P < 0.024). The most significantly upregulated genes were Cybb, fibronectin 1 (Fn1), complement 3 (C3), cathepsin C (Ctsc), and bone morphogenetic protein receptor type 2 (Bmpr2) (Table 1A). These genes were upregulated by 4.0-, 3.5-, 3.2-, 3.1-, and 2.8-fold, respectively (P < 0.003, P < 0.001, P < 0.001, P < 0.002, and P < 0.001). The most significantly downregulated genes (Table 1B) were Pf4, cluster of differentiation 226, platelet and T-cell activation antigen 1 (Cd226), glycoprotein VI (Gp6), and glycoprotein V (Gp5). They were downregulated by 2.7-, 2.6-, 2.6-, and 2.5-fold, respectively (P < 0.001, P < 0.001, P < 0.002, and P < 0.005).
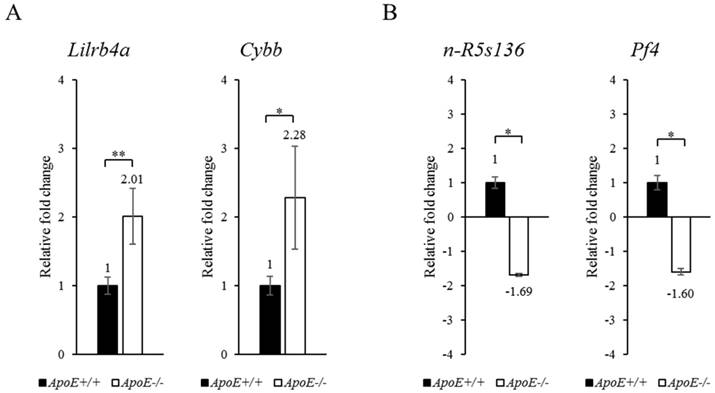
To confirm the microarray results, we performed qRT-PCR (Figure 2). The microarray data was confirmed based on the most upregulated (Lilrb4a) and the most significantly upregulated (Cybb) genes. In the qRT-PCR analysis, Lilrb4a and Cybb were upregulated by 2.01- and 2.28-fold, respectively (P < 0.01 and P < 0.05), in the ApoE-/- mice compared to the ApoE+/+ mice. Table 1B shows the most downregulated (n-R5s136) and the most significantly downregulated (Pf4) genes. n-R5s136 and Pf4 were downregulated by 1.69- and 1.60-fold (each P < 0.05) in the ApoE-/- mice compared to the ApoE+/+ mice. The qRT-PCR results confirm the microarray results.
Protein expression profiling
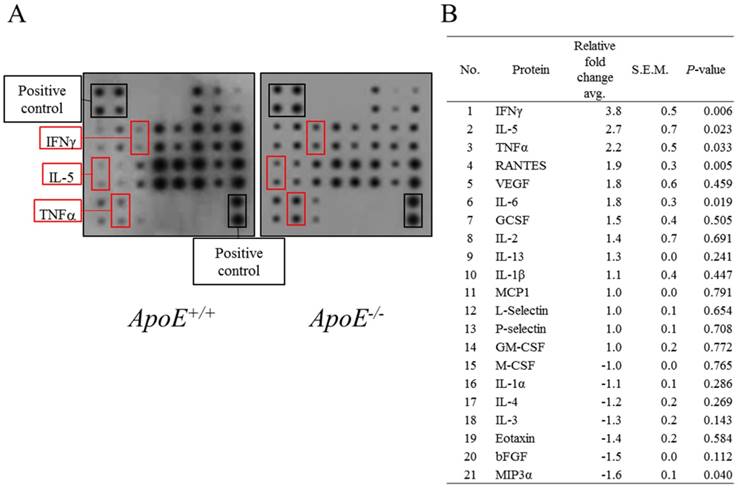
Proteome array was performed using whole blood of ApoE+/+ and ApoE-/- mice (Figure 3). Three proteins, IFNγ, IL-5, and TNFα, were 2-times more abundant in ApoE-/- mice compared to ApoE+/+ mice. In addition, IL-1β, IL-13, IL-2, granulocyte colony-stimulating factor (GCSF), IL-6, vascular endothelial growth factor (VEGF), and regulated upon activation normal T cell expressed and secreted (RANTES) tended to increase in each batch but did not increase more than 2-fold. In contrast, the abundance of macrophage colony-stimulating factor (M-CSF), IL-1α, IL-4, IL-3, Eotaxin, basic fibroblast growth factor (bFGF) and macrophage inflammatory protein-3a (MIP3a) proteins tended to decrease but did not decrease more than 2-fold. In addition, the levels of the four proteins MCP1, L-Selectin, P-selectin, and granulocyte-macrophage colony-stimulating factor (GM-CSF), showed no change between ApoE+/+ and ApoE-/- mice.
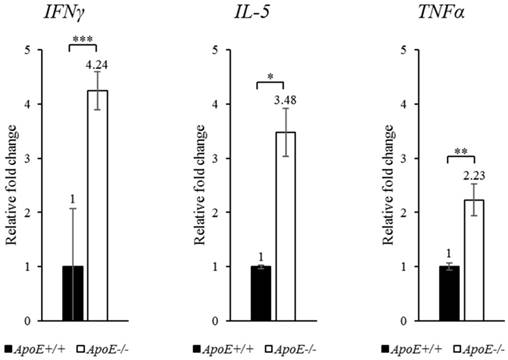
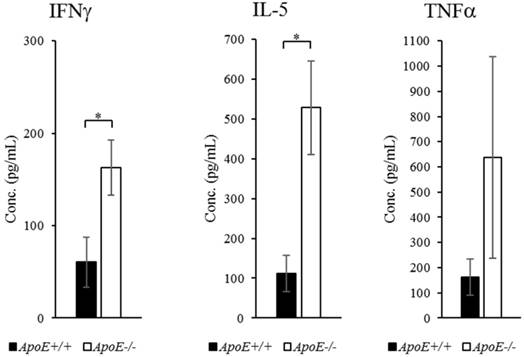
Proteins, which showed more than 2-fold change in abundance, were selected for qRT-PCR confirmation (Figure 3). The transcripts of IFNγ, IL-5, and TNFα were upregulated by 4.24-, 3.48-, and 2.23-fold (P-values < 0.001, < 0.05, and < 0.01, respectively) in ApoE-/- mice compared to ApoE+/+ mice (Figure 4). To quantify each protein separately, we used an ELISA kit to confirm the change in protein abundance. ELISA results reflected the proteome array results (Figure 5), and both IFNγ and IL-5 levels were more than 2-fold (P < 0.05) in the blood of ApoE-/- mice.
qRT-PCR analysis of target genes in blood of ApoE+/+ and ApoE-/- mice. (A) Upregulated and (B) downregulated target genes are shown. GAPDH was used as the housekeeping gene. Data represent mean ± S.E.M. Experiments were performed three times independently. *P < 0.05, **P < 0.01.
Protein levels in blood of ApoE+/+ and ApoE-/- mice. (A) Representative proteome array panel. (B) Fold change in spot density of (A). Relative fold change corresponds to the density of spot in ApoE-/- compared with control spot. Density was normalized with density of blanks, negative, and positive controls. Data represent mean ± S.E.M. Experiments were performed three times independently.
qRT-PCR analysis of target genes in ApoE+/+ and ApoE-/- mice. The expression of each gene was confirmed using specific primers. GAPDH was used as the housekeeping gene. Data represent mean ± S.E.M. Experiments were performed three times independently. *P < 0.05, **P < 0.01, *** P < 0.001.
Protein expression in ApoE+/+ and ApoE-/- mice measured using ELISA. Data represent mean ± S.E.M. Experiments were performed three times independently. *P < 0.05.
Discussion
Atherosclerosis is a disease that forms atheroma in the blood vessels, which if left untreated can cause fatal complications. In our microarray analysis, the expression of Lilrb4a, Sirpb1b, Tlr7 and Cybb were upregulated, while the expression of n-R5s136, Thbs1, Slc6a4 and Pf4 were downregulated. Both transcript and protein levels of TNFα were increased in the ApoE-/- mice, whereas protein levels of IFNγ and IL-5 were increased but not their corresponding transcripts in the microarray data. The discrepancy in the results obtained from the microarray and proteome array experiments might be the result of differing techniques and sensitivity between the two methods. However, through qRT-PCR also the upregulation in expression of these transcripts were confirmed. Microarray experiments were conducted to find novel biomarkers for atherosclerosis, and proteome array experiments were employed to determine the progression of the disease. Using ELISA, we further confirmed the proteome array results.
In the microarray data and by qRT-PCR, the expression of Lilrb4a was found to be upregulated by over 2-fold. The leukocyte immunoglobulin-like receptor, subfamily B, member 4A (Lilrb4a) gene encodes glycoprotein 49B (Gp49b), which is a member of the transmembrane gp49 family. This gene is expressed in immune cells that can bind to MHC class I for capturing or presenting antigen. In other words, the immune response can be modulated through the expression of this gene and its various isoforms [36]. No association of Lilrb4a in atherosclerosis has been previously reported. However, the expression of Lilrb4a in dendritic cells for the inhibition of excessive activation of T-cells and lowering cellular activity has been reported [37]. In animal models of allergic pulmonary inflammation, the expression of Lilrb4a has been shown to reduce the activity of dendritic cells. When the inhalation of ovalbumin and lipopolysaccharide (LPS) was compared in control and Lilrb4-/- mice, the secretion of IL-4 and IL-5 was increased in Lilrb4-/- mice together with increased Th2 lung pathology [38]. On the other hand, the transcripts of Lilrb1, Lilrb2, and Lilrb3 were upregulated in patients with acute myocardial infarction, but Lilrb4a levels did not change [39]. Consequently, Lilrb4a is a promising biomarker candidate of atherosclerosis, which allows distinction from acute myocardial infarction.
The most significantly upregulated transcript, Cybb has previously been studied in atherosclerosis. The cytochrome b-245 beta chain (Cybb or gp91phox) gene encodes the subunit constituting cytochrome b-245 and is better known as NADPH oxidase 2 (Nox2). It is primarily expressed in endothelial cells, smooth muscle cells (SMC), and adventitia [40]. Cybb, along with cytochrome b-245 alpha chain (Cyba), forms a protein that is essential for the activation of NADPH oxidase. NADPH oxidase is a major enzyme in the phagocyte that digests bacteria and fungi. Cybb deficiency causes chronic granulomatous disease, in which the activity of phagocytic NADPH oxidase is reduced and neutrophils do not completely remove bacteria even when digested [41]. There is also considerable research in understanding the association between Cybb expression and atherosclerosis. Reduced Cybb expression in ApoE-/- mice, resulted in reduced atherosclerotic lesions [42]. Decreased in vivo reactive oxygen species (ROS) production, increased NO bioavailability and reduced atherosclerotic plaque formation have been reported in ApoE/gp91phox double knockout mice compared to ApoE-/- mice [43], suggesting that Cybb deficiency reduces atherosclerosis by limiting superoxidase in the macrophage and vessel wall. Atherosclerosis was also attenuated in ApoE/p47phox double knockout mice, where p47phox is a subunit of Nox2 [44]. However, the role of Cybb in atherosclerosis remains unclear as studies with no prevention effect have also been published [45], while microarray of atherosclerosis rat model shows upregulation of Cybb [46], and knockdown of Cybb decreases restenosis [47].
On the other hand, studies on atherosclerosis and n-R5s136, the most downregulated gene in microarray, are lacking. In addition, there are not many studies on n-R5s136 itself. The nuclear encoded rRNA 5S 136 (n-R5s136) gene encodes components that make up the ribosome. The human 5S rRNA gene was published in 1991 as a repetitive sequence gene containing a pseudogene [48]. This property of 5S rRNA is also maintained in mice [49]. However, much research is still needed to deduce its function. So far, its involvement in the interaction of ribosomes has been reported [50]. In recent studies, a relationship between atherosclerosis and micro RNA and its application in diagnosis has been reported [51-53]. However, further studies to confirm the relationship between atherosclerosis and n-R5s136, and to determine the mechanism of atherosclerosis in relation to the ribosome are needed.
The most significantly downregulated gene, Pf4 has been studied in relation to atherosclerosis. Pf4 or CXCL4 encodes platelet factor 4 (PF4), a member of the CXC chemokine family. PF4 is secreted from alpha granules of platelet and assists the aggregation of platelets. It also inhibits hematopoiesis and angiogenesis. However, the role of platelets in atherosclerosis has not been elucidated. PF4 has been reported to inhibit the process of elimination of oxidized LDL in vitro [54]. Studies have also shown that removal of PF4 from platelets in ApoE-/- mice results in a reduction in atherosclerotic plaque burden compared to ApoE-/- mice [55]. The reported studies, use artificial addition or removal of PF4, which does not explain the mechanism by which the transcription of Pf4 changes. Therefore, the role of Pf4 transcriptional downregulation in our experiments is unclear and needs further investigation.
Atherosclerosis is a complex disease, however, our microarray data presented a small number of mRNAs, which are listed in Table 1A and B. In general, mRNA expression profiling with blood presented significantly less differentially expressed genes (DEGs) than using that using aortic tissue. If aortic tissues were used, more DEGs might be obtained, including the genes involved in cell proliferation. However, obtaining vascular tissues from a patient is more difficult and more dangerous than whole blood.
As atherosclerosis is a chronic inflammatory disease, the increase in IFNγ and TNFα levels is expected. IFNγ is an immunoregulatory factor secreted by lymphocytes that has antiviral and antitumor effect. It is a soluble cytokine belonging to the type II interferon class, which is associated with both innate and adaptive immune responses and is primarily activated in response to viral and bacterial infection. ApoE/IFNγ double knockout mice have been reported to have reduced lesion size compared to ApoE-/- mice [56], and ApoE-/- mice injected with IFNγ in the peritoneal cavity [57]. Similarly, TNFα is a proinflammatory cytokine and a member of the tumor necrosis factor superfamily with various functions. TNFα primarily secreted by macrophages is involved in various pathways, such as proliferation, differentiation, apoptosis, and lipid metabolism. ApoE/TNFα double knockout mice have been reported to have similar levels of serum cholesterol, but smaller plaques compared to ApoE-/- mice, in addition to reduced transcripts of ICAM, VCAM, and MCP1 [58]. However, because both IFNγ and TNFα are cytokines that enhance inflammatory responses, they are not of interest in application as a specific biomarker of atherosclerosis.
The transcript and protein levels of IL-5 were also elevated in the ApoE-/- mice. IL-5 is a cytokine required for the growth and differentiation of B-cells and eosinophils. Studies have reported elevated levels of IL-5 through cytokine assay in the serum of ApoE-/- mice [59]. On the other hand, when macrophage-specific IL-5 is overexpressed in LDLR1-/- mice, IL-5 secreted by the transplanted macrophages inhibits phagocytosis of LDL, thereby weakening the disease [60]. Other studies have also reported that IL-5 is antiatherogenic [61]. Although IL-5 may be useful in the early treatment of atherosclerosis, its mechanism of action remains unknown [62], and the role of increased IL-5 in reducing atherosclerosis needs further investigation.
Regarding the use of whole blood for the biomarker study, whole blood samples were used to identify biomarkers for acute allograft rejection in cardiac transplantation patient. Accordingly, 12 genes were suggested as biomarker with 83% sensitivity and 100% specificity [63]. In a breast cancer study, mass-spectrometry was performed on whole blood to report differential DNA methylation as a marker of breast cancer [64]. Other studies have reported that plasma and test results are not different between whole blood proteins. There was a positive correlation between the amount of sCD25 detected in whole blood and the detected amount of plasma in Alzheimer's disease [65]. In addition, a positive correlation was found between three representative markers of myocardial infarction (cTnl, CK-MB, and myoglobin) when comparing whole blood and plasma [66]. Therefore, using whole blood might not be inappropriate for a biomarker study.
In this study, potential candidate biomarkers for atherosclerosis were investigated using whole blood of animal models. The association of atherosclerosis with Lilrb4a, n-R5s136 and IL-5 had not been previously reported. The roles of Cybb and Pf4 transcriptional changes in atherosclerosis, also need to be further explored. Future efforts should validate the current results using blood of atherosclerosis patients by comparing gene expression and protein levels at various stages of atherosclerosis progression to identify early diagnostic markers in blood. The results in this study contribute to the development of diagnosis of atherosclerosis using whole blood.
Abbreviations
CVD: cardiovascular disease; ApoE-/- mice: apolipoprotein E deficiency mice; LDLR-/- mice: low density lipoprotein receptor deficiency mice; VLDL: very low-density lipoprotein; VCAM: vascular cell adhesion protein; ICAM: intercellular adhesion molecule; NGF: nerve growth factor; HGF: hepatocyte growth factor; MCP: monocyte-chemotactic protein; CRABP II: cellular retinoic acid binding protein 2; SELP: selectin P; CD: cluster of differentiation; LFA-1: lymphocyte function-associated antigen 1; COX-2: cyclooxygenase-2; CCL: chemokine (C-C motif) ligand; TNFα: tumor necrosis factor alpha; IL: interleukin; NOS: nitric oxide synthase; NO: nitric oxide; eNOS: endothelial NOS; hs-CRP: high-sensitivity C-reactive protein; CT: computerized tomography; MRI: magnetic resonance imaging; EDTA: ethylenediaminetetraacetic acid; GAPDH: glyceraldehyde 3-phosphate dehydrogenase; Fn1: fibronectin 1; C3: complement 3; Ctsc: cathepsin C; CD226: platelet and T-cell activation antigen 1; Gp6: glycoprotein VI; Gp5: glycoprotein V; IFN: interferon; GCSF: granulocyte colony-stimulating factor; VEGF: vascular endothelial growth factor; RANTES: regulated upon activation normal T cell expressed and secreted; M-CSF: macrophage colony-stimulating factor; bFGF: basic fibroblast growth factor; MIP3a: macrophage inflammatory protein-3a; GM-CSF: granulocyte-macrophage colony-stimulating factor; Lilrb4a: leukocyte immunoglobulin-like receptor; subfamily B: member 4A; Gp49b: glycoprotein 49B; Cybb: cytochrome b-245 beta chain; Nox2: NADPH oxidase 2; SMC: smooth muscle cells; Cyba: cytochrome b-245 alpha chain; ROS: reactive oxygen species; n-R5s136: nuclear encoded rRNA 5S 136; PF4: platelet factor 4.
Acknowledgements
This research was supported by the Bio & Medical Technology Development Program of the National Research Foundation (NRF) & funded by the Korean government (MSIP&MOHW) (No. 2016M3A9B6904244).
Authors' Contributions
Sun-Yeong Gwon and Ho Joong Sung conceived and designed the experiments; Sun-Yeong Gwon performed the experiments; Sun-Yeong Gwon, Hae Min Lee, Ki-Jong Rhee and Ho Joong Sung analyzed the data; Ho Joong Sung contributed all reagents/materials/analysis tools; Sun-Yeong Gwon, and Ho Joong Sung wrote the paper.
Competing Interests
The authors have declared that no competing interest exists.
References
1. Herrington W, Lacey B, Sherliker P, Armitage J, Lewington S. Epidemiology of Atherosclerosis and the Potential to Reduce the Global Burden of Atherothrombotic Disease. Circulation research. 2016;118:535-46
2. Rafieian-Kopaei M, Setorki M, Doudi M, Baradaran A, Nasri H. Atherosclerosis: Process, Indicators, Risk Factors and New Hopes. International Journal of Preventive Medicine. 2014;5:927-46
3. Plump AS, Smith JD, Hayek T, Aalto-Setala K, Walsh A, Verstuyft JG. et al. Severe hypercholesterolemia and atherosclerosis in apolipoprotein E-deficient mice created by homologous recombination in ES cells. Cell. 1992;71:343-53
4. Ishibashi S, Brown MS, Goldstein JL, Gerard RD, Hammer RE, Herz J. Hypercholesterolemia in low density lipoprotein receptor knockout mice and its reversal by adenovirus-mediated gene delivery. The Journal of clinical investigation. 1993;92:883-93
5. Nakashima Y, Plump AS, Raines EW, Breslow JL, Ross R. ApoE-deficient mice develop lesions of all phases of atherosclerosis throughout the arterial tree. Arteriosclerosis and thrombosis: a journal of vascular biology. 1994;14:133-40
6. Ishibashi S, Goldstein JL, Brown MS, Herz J, Burns DK. Massive xanthomatosis and atherosclerosis in cholesterol-fed low density lipoprotein receptor-negative mice. The Journal of clinical investigation. 1994;93:1885-93
7. Wouters K, Shiri-Sverdlov R, van Gorp PJ, van Bilsen M, Hofker MH. Understanding hyperlipidemia and atherosclerosis: lessons from genetically modified apoe and ldlr mice. Clinical chemistry and laboratory medicine: CCLM / FESCC. 2005;43:470-9
8. Getz GS, Vanderlaan PA, Reardon CA. Natural killer T cells in lipoprotein metabolism and atherosclerosis. Thrombosis and haemostasis. 2011;106:814-9
9. Getz GS, Reardon CA. Animal models of atherosclerosis. Arteriosclerosis, thrombosis, and vascular biology. 2012;32:1104-15
10. Liao J, Huang W, Liu G. Animal models of coronary heart disease. Journal of biomedical research. 2015:30
11. Piedrahita JA, Zhang SH, Hagaman JR, Oliver PM, Maeda N. Generation of mice carrying a mutant apolipoprotein E gene inactivated by gene targeting in embryonic stem cells. Proceedings of the National Academy of Sciences of the United States of America. 1992;89:4471-5
12. Reddick RL, Zhang SH, Maeda N. Atherosclerosis in mice lacking apo E. Evaluation of lesional development and progression. Arteriosclerosis, thrombosis, and vascular biology. 1994;14:141-7
13. Jawien J, Nastalek P, Korbut R. Mouse models of experimental atherosclerosis. Journal of physiology and pharmacology: an official journal of the Polish Physiological Society. 2004;55:503-17
14. Meir KS, Leitersdorf E. Atherosclerosis in the apolipoprotein-E-deficient mouse: a decade of progress. Arteriosclerosis, thrombosis, and vascular biology. 2004;24:1006-14
15. Zaragoza C, Gomez-Guerrero C, Martin-Ventura JL, Blanco-Colio L, Lavin B. et al. Animal Models of Cardiovascular Diseases. Journal of Biomedicine and Biotechnology. 2011;2011:13
16. Andres-Manzano MJ, Andres V, Dorado B. Oil Red O and Hematoxylin and Eosin Staining for Quantification of Atherosclerosis Burden in Mouse Aorta and Aortic Root. Methods Mol Biol. 2015;1339:85-99
17. Wuttge DM, Sirsjö A, Eriksson P, Stemme S. Gene expression in atherosclerotic lesion of ApoE deficient mice. Molecular Medicine. 2001;7:383-92
18. Ma Y, Malbon CC, Williams DL, Thorngate FE. Altered gene expression in early atherosclerosis is blocked by low level apolipoprotein E. PloS one. 2008;3:e2503
19. Papadodima O, Chatziioanou A, Sirsjo A, Kolisis FN. Bioinformatic transcriptomic analysis of ApoE deficient mice suggests Alterations in atherosclerosis related molecular mechanisms. Proceedings of the 10th IEEE International Conference on Information Technology and Applications in Biomedicine. 2010 p. 1-4
20. Tabibiazar R, Wagner RA, Deng A, Tsao PS, Quertermous T. Proteomic profiles of serum inflammatory markers accurately predict atherosclerosis in mice. Physiological genomics. 2006;25:194-202
21. Hanzawa H, Sakamoto T, Kaneko A, Manri N, Zhao Y, Zhao S. et al. Combined Plasma and Tissue Proteomic Study of Atherogenic Model Mouse: Approach To Elucidate Molecular Determinants in Atherosclerosis Development. J Proteome Res. 2015;14:4257-69
22. Cybulsky MI, Gimbrone MA Jr. Endothelial expression of a mononuclear leukocyte adhesion molecule during atherogenesis. Science. 1991;251:788-91
23. Branen L, Hovgaard L, Nitulescu M, Bengtsson E, Nilsson J, Jovinge S. Inhibition of tumor necrosis factor-alpha reduces atherosclerosis in apolipoprotein E knockout mice. Arteriosclerosis, thrombosis, and vascular biology. 2004;24:2137-42
24. Kuhlencordt PJ, Gyurko R, Han F, Scherrer-Crosbie M, Aretz TH, Hajjar R. et al. Accelerated atherosclerosis, aortic aneurysm formation, and ischemic heart disease in apolipoprotein E/endothelial nitric oxide synthase double-knockout mice. Circulation. 2001;104:448-54
25. Kolovou G, Anagnostopoulou K, Mikhailidis DP, Cokkinos DV. Apolipoprotein E knockout models. Current pharmaceutical design. 2008;14:338-51
26. Brown TM, Bittner V. Biomarkers of Atherosclerosis: Clinical Applications. Current cardiology reports. 2008;10:497-504
27. Vlachopoulos C, Xaplanteris P, Aboyans V, Brodmann M, Cifkova R, Cosentino F. et al. The role of vascular biomarkers for primary and secondary prevention. A position paper from the European Society of Cardiology Working Group on peripheral circulation: Endorsed by the Association for Research into Arterial Structure and Physiology (ARTERY) Society. Atherosclerosis. 2015;241:507-32
28. Anderson JD, Kramer CM. MRI of Atherosclerosis: Diagnosis and Monitoring Therapy. Expert review of cardiovascular therapy. 2007;5:69-80
29. Ibanez B, Badimon JJ, Garcia MJ. Diagnosis of atherosclerosis by imaging. The American journal of medicine. 2009;122:S15-25
30. Patino WD, Mian OY, Kang J-G, Matoba S, Bartlett LD, Holbrook B. et al. Circulating transcriptome reveals markers of atherosclerosis. Proceedings of the National Academy of Sciences of the United States of America. 2005;102:3423-8
31. Chen PY, Qin L, Baeyens N, Li G, Afolabi T, Budatha M. et al. Endothelial-to-mesenchymal transition drives atherosclerosis progression. The Journal of clinical investigation. 2015;125:4514-28
32. Jung YS, Lee S-W, Park JH, Seo HB, Choi BT, Shin HK. Electroacupuncture preconditioning reduces ROS generation with NOX4 down-regulation and ameliorates blood-brain barrier disruption after ischemic stroke. Journal of Biomedical Science. 2016;23:32
33. Amsen D, de Visser KE, Town T. Approaches to Determine Expression of Inflammatory Cytokines. Methods in molecular biology (Clifton, NJ). 2009;511:107-42
34. Wilkinson RDA, Young A, Burden RE, Williams R, Scott CJ. A bioavailable cathepsin S nitrile inhibitor abrogates tumor development. Molecular cancer. 2016;15:1-11
35. Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001;25:402-8
36. Fanger NA, Borges L, Cosman D. The leukocyte immunoglobulin-like receptors (LIRs): a new family of immune regulators. J Leukoc Biol. 1999;66:231-6
37. Kasai S, Inui M, Nakamura K, Kakizaki Y, Endo S, Nakamura A. et al. A novel regulatory role of gp49B on dendritic cells in T-cell priming. European journal of immunology. 2008;38:2426-37
38. Fanning LB, Buckley CC, Xing W, Breslow RG, Katz HR. Downregulation of key early events in the mobilization of antigen-bearing dendritic cells by leukocyte immunoglobulin-like Receptor B4 in a mouse model of allergic pulmonary inflammation. PloS one. 2013;8:e57007
39. Yan W, Song H, Jiang J, Xu W, Gong Z, Duan Q. et al. Characteristics of B cellassociated gene expression in patients with coronary artery disease. Mol Med Rep. 2016;13:4113-21
40. Csanyi G, Taylor WR, Pagano PJ. NOX and inflammation in the vascular adventitia. Free radical biology & medicine. 2009;47:1254-66
41. Violi F, Carnevale R, Loffredo L, Pignatelli P, Gallin JI. NADPH Oxidase-2 and Atherothrombosis: Insight From Chronic Granulomatous Disease. Arteriosclerosis, thrombosis, and vascular biology. 2017;37:218-25
42. Vendrov AE, Hakim ZS, Madamanchi NR, Rojas M, Madamanchi C, Runge MS. Atherosclerosis is attenuated by limiting superoxide generation in both macrophages and vessel wall cells. Arteriosclerosis, thrombosis, and vascular biology. 2007;27:2714-21
43. Judkins CP, Diep H, Broughton BR, Mast AE, Hooker EU, Miller AA. et al. Direct evidence of a role for Nox2 in superoxide production, reduced nitric oxide bioavailability, and early atherosclerotic plaque formation in ApoE-/- mice. American journal of physiology Heart and circulatory physiology. 2010;298:H24-32
44. Barry-Lane PA, Patterson C, van der Merwe M, Hu Z, Holland SM, Yeh ET. et al. p47phox is required for atherosclerotic lesion progression in ApoE(-/-) mice. The Journal of clinical investigation. 2001;108:1513-22
45. Hsich E, Segal BH, Pagano PJ, Rey FE, Paigen B, Deleonardis J. et al. Vascular effects following homozygous disruption of p47(phox): An essential component of NADPH oxidase. Circulation. 2000;101:1234-6
46. Li JM, Zhang X, Nelson PR, Odgren PR, Nelson JD, Vasiliu C. et al. Temporal evolution of gene expression in rat carotid artery following balloon angioplasty. Journal of cellular biochemistry. 2007;101:399-410
47. Li JM, Newburger PE, Gounis MJ, Dargon P, Zhang X, Messina LM. Local arterial nanoparticle delivery of siRNA for NOX2 knockdown to prevent restenosis in an atherosclerotic rat model. Gene therapy. 2010;17:1279-87
48. Sørensen PD, Frederiksen S. Characterization of human 5S rRNA genes. Nucleic acids research. 1991;19:4147-51
49. Hallenberg C, Nederby Nielsen J, Frederiksen S. Characterization of 5S rRNA genes from mouse. Gene. 1994;142:291-5
50. Dinman JD. 5S rRNA: Structure and Function from Head to Toe. International journal of biomedical science: IJBS. 2005;1:2-7
51. Gardner PP, Daub J, Tate JG, Nawrocki EP, Kolbe DL, Lindgreen S. et al. Rfam: updates to the RNA families database. Nucleic acids research. 2009;37:D136-40
52. Maitrias P, Metzinger-Le Meuth V, Massy ZA, M'Baya-Moutoula E, Reix T, Caus T. et al. MicroRNA deregulation in symptomatic carotid plaque. Journal of vascular surgery. 2015;62:1245-50.e1
53. Holdt LM, Stahringer A, Sass K, Pichler G, Kulak NA, Wilfert W. et al. Circular non-coding RNA ANRIL modulates ribosomal RNA maturation and atherosclerosis in humans. Nat Commun. 2016;7:12429
54. Nassar T, Sachais BS, Akkawi S, Kowalska MA, Bdeir K, Leitersdorf E. et al. Platelet factor 4 enhances the binding of oxidized low-density lipoprotein to vascular wall cells. The Journal of biological chemistry. 2003;278:6187-93
55. Sachais BS, Turrentine T, Dawicki McKenna JM, Rux AH, Rader D, Kowalska MA. Elimination of platelet factor 4 (PF4) from platelets reduces atherosclerosis in C57Bl/6 and apoE-/- mice. Thrombosis and haemostasis. 2007;98:1108-13
56. Gupta S, Pablo AM, Jiang X, Wang N, Tall AR, Schindler C. IFN-gamma potentiates atherosclerosis in ApoE knock-out mice. The Journal of clinical investigation. 1997;99:2752-61
57. Whitman SC, Ravisankar P, Elam H, Daugherty A. Exogenous interferon-gamma enhances atherosclerosis in apolipoprotein E-/- mice. The American journal of pathology. 2000;157:1819-24
58. Ohta H, Wada H, Niwa T, Kirii H, Iwamoto N, Fujii H. et al. Disruption of tumor necrosis factor-alpha gene diminishes the development of atherosclerosis in ApoE-deficient mice. Atherosclerosis. 2005;180:11-7
59. Smith E, Prasad K-MR, Butcher M, Dobrian A, Kolls JK, Ley K. et al. Blockade of IL-17A results in reduced atherosclerosis in Apoe-deficient mice. Circulation. 2010;121:1746-55
60. Zhao W, Lei T, Li H, Sun D, Mo X, Wang Z. et al. Macrophage-specific overexpression of interleukin-5 attenuates atherosclerosis in LDL receptor-deficient mice. Gene therapy. 2015;22:645-52
61. Fatkhullina AR, Peshkova IO, Koltsova EK. The Role of Cytokines in the Development of Atherosclerosis. Biochemistry Biokhimiia. 2016;81:1358-70
62. Silveira A, McLeod O, Strawbridge RJ, Gertow K, Sennblad B, Baldassarre D. et al. Plasma IL-5 concentration and subclinical carotid atherosclerosis. Atherosclerosis. 2015;239:125-30
63. Lin D, Hollander Z, Ng RT, Imai C, Ignaszewski A, Balshaw R. et al. Whole blood genomic biomarkers of acute cardiac allograft rejection. The Journal of heart and lung transplantation: the official publication of the International Society for Heart Transplantation. 2009;28:927-35
64. Loke SY, Lee ASG. The future of blood-based biomarkers for the early detection of breast cancer. Eur J Cancer. 2018;92:54-68
65. May JE, Pemberton RM, Hart JP, McLeod J, Wilcock G, Doran O. Use of whole blood for analysis of disease-associated biomarkers. Analytical biochemistry. 2013;437:59-61
66. Pettersson K, Katajamaki T, Irjala K, Leppanen V, Majamaa-Voltti K, Laitinen P. Time-resolved fluorometry (TRF)-based immunoassay concept for rapid and quantitative determination of biochemical myocardial infarction markers from whole blood, serum and plasma. Luminescence: the journal of biological and chemical luminescence. 2000;15:399-407
Author contact
![]() Corresponding author: Tel.: +82-31-740-7108; Fax: +82-31-740-7425; E-mail: hjsungac.kr
Corresponding author: Tel.: +82-31-740-7108; Fax: +82-31-740-7425; E-mail: hjsungac.kr

 Global reach, higher impact
Global reach, higher impact