Impact Factor
ISSN: 1449-1907
Int J Med Sci 2019; 16(3):366-375. doi:10.7150/ijms.29946 This issue Cite
Research Paper
New Insight on Solute Carrier Family 27 Member 6 (SLC27A6) in Tumoral and Non-Tumoral Breast Cells
1. Department of Emergency Medicine, Kaohsiung Medical University Hospital, Kaohsiung Medical University, Kaohsiung 807, Taiwan;
2. Graduate Institute of Clinical Medicine, College of Medicine, Kaohsiung Medical University, Kaohsiung 807, Taiwan;
3. Graduate Institute of Medicine, College of Medicine, Kaohsiung Medical University, Kaohsiung 807, Taiwan;
4. Department of Breast Surgery, Kaohsiung Medical University Hospital, Kaohsiung Medical University, Kaohsiung 807, Taiwan.
Received 2018-9-14; Accepted 2018-12-17; Published 2019-1-24
Abstract
Long-chain fatty acids are the most abundant fatty acids and are essential for various physiological processes. Translocation of long-chain fatty acids across cell membrane is dependent on transport proteins. Solute carrier family 27 member 6 (SLC27A6) is a transport protein which mediates long-chain fatty acid uptake. The bioinformatic analysis revealed that the expression of SLC27A6 in non-tumoral breast tissue was higher than that in tumoral breast cancer in clinic samples. When SLC27A6 expression in non-tumorigenic cell H184B5F5/M10 was repressed, the fatty acids uptake capacity and cell proliferation was inhibited, and cell cycle was delayed. The protein expression of cell cycle regulators including cell division protein kinase 4 (CDK4), CDK6, and cyclin D1 was significantly decreased in SLC27A6-silenced H184B5F5/M10. By contrast, relatively low SLC27A6 expression in tumorigenic breast cancer cell Hs578T when compared to H184B5F5/M10. Repressing SLC27A6 expression did not affect these phenotypes in Hs578T. The interaction network of SLC27A6 was further investigated via STRING database. The function of these SLC27A6-associated proteins mainly involved in lipid biosynthesis, fatty acid metabolic process, and fatty acid transport. In conclusion, this study reveals inverse correlation between SLC27A6 expression and tumoral tissues and provides a new insight into SLC27A6-mediated cell growth and cell cycle regulation in non-tumorigenic breast cells.
Keywords: solute carrier family 27 member 6 (SLC27A6), fatty acid transport protein 6 (FATP6), very long-chain acyl-CoA synthetases member 2 (ACSVL2), fatty acid transport, breast, proliferation, cell cycle
Introduction
Dietary fat is one of important energy sources [1]. Triglyceride which composed of fatty acids, phospholipid, and cholesteryl esters is abundant in fat-diet [2]. In fasting condition, triglyceride which is stored in adipose tissue is hydrolyzed to free fatty acids and glycerol [3]. Based on carbon number of aliphatic tails, fatty acids categorized as short-chain (< 8 carbons), medium-chain (8-12 carbons), long-chain (16-22 carbons), or very-long-chain (>22 carbons) fatty acids [2]. In general, long-chain fatty acids (>16 carbons) are more abundant than short-chain and medium-chain fatty acids in animal tissues [4]. The transport of fatty acids across cell membrane could occur by passive diffusion, or be facilitated by proteins associated with fatty acid transport, including CD36 (also called fatty acid translocase), fatty acid binding protein (FABP), and a family of fatty acid transporter (SLC27, also called FATP) [5-7]. These long-chain fatty acids are important for various physiological processes, such as inflammation, synthesis of phospholipid and triglyceride [8, 9]. Therefore, these transporter proteins are usually associated with regulation of cell behaviors, including cancer cells.
Dysregulated metabolism is a hallmark of oncogenesis [10]. Emerging studies suggest that FABP5 is associated with poor survival and the FABP7-associated signaling pathway enhances cell survival and proliferation in triple-negative breast cancer [11, 12]. CD36 overexpression is associated with cell growth and metastasis in breast cancer cells [13, 14]. There are six members of SLC27 family in mammals (SLC27A1 through SLC27A6). According to the amino acid sequence of the conserved region, the SLC27 family proteins are proposed to bifunctional protein with long-chain fatty acids transport and acyl-CoA synthetase (ACS) activity [15, 16]. Therefore, SLC27 family proteins are also named very long-chain acyl-CoA synthetases (ACSVL) [15]. Currently, the association of SLC27 and tumor cells is not fully understood although the relationship between SLC27 proteins and some human diseases have been demonstrated.
SLC27A6 which also named FATP6 and ACSVL2 colocalizes with CD36 [17]. FATP6- 7 T>A polymorphism may protect from human cardio-metabolic diseases [18]. In human intrauterine growth restriction, increased protein expression of CD36 and SLC27A6 is observed in syncytiotrophoblast microvillous plasma membrane [19]. The association between SLC27A6 and malignant cells were not well-known. In our recent study, we found that the expression patterns of SLC27A family proteins were quite different in tumor samples when compared to non-tumor samples [20]. The SLC27A6 expression was the most significantly and inversely associated breast tumor samples in several public microarray datasets. Thus, the aim of the present study was to investigate whether SLC27A6 plays a role in human tumor progression. The function of SLC27A6 was evaluated in tumorigenic and non-tumorigenic breast cells.
Material and methods
Cell culture
Human mammary epithelial cell line H184B5F5/ M10 was obtained from Bioresource Collection and Research Center (BCRC Number: 60197) (Hsinchu, Taiwan). H184B5F5/M10 was cultured in alpha- Minimum Essential Medium (α-MEM) with 15% fetal bovine serum (Life Technologies, Grand Island, NY, USA). Human mammary cancer cell line Hs578T, MCF-7, and MDA-MB-231 were purchased from American Type Culture Collection (USA) and were respectively maintained in Dulbecco's Modified Eagle Medium (DMEM), Minimum Essential Medium (MEM), and Leibovitz's L-15 Medium with 10% fetal bovine serum respectively. All culture medium contained (Life, 100 units/mL penicillin G, 100 μg/mL streptomycin, and 0.25 μg/mL amphotericin B Technologies, Grand Island, NY, USA). H184B5F5/M10, Hs578T, and MCF-7 were cultured in 5% CO2 air atmosphere and MDA-MB-231 was cultured at CO2-free air atmosphere at 37°C.
Bioinformatic analysis
The expression in SLC27A6 in different types of normal and tumor samples and overall survival curve was evaluated by GEPIA database (http://gepia. cancer-pku.cn/) which was established using gene expression data via RNA sequencing from Cancer Genome Atlas (TCGA) and Genotype-Tissue Expression (GTEx) and patient survival [21]. The relapse-free survival (RFS) was evaluated by Kaplan‑Meier (KM) plotter (http://kmplot.com) which was established using gene expression data via Affymetrix microarray expression profiles and survival information from Gene Expression Omnibus (GEO) database [22] and high‑ and low‑expression groups were divided according to the “median” expression levels. Moreover, the expression of SLC27A6 in different stages and subtypes of breast cancer samples was evaluated by the UALCAN database (http://ualcan.path. uab.edu) [23]. The functional protein association network of SLC27A6 was drawn via in stringAPP (version 1.4.0) in Cytoscape software version 3.6.1 [24, 25]. The biological process annotation was determined by DAVID Bioinformatics Resources 6.7 (https://david. ncifcrf.gov) [26, 27].
Western blot assay
Total protein was collected 48 hours after subculture and protein concentration was determined by Pierce BCA Protein Assay Kit (Thermo Fisher Scientific). Protein was separated on 10-15% SDS-PAGE and then transferred to PVDF membranes (Millipore). The PVDF membrane was then blocked with 5% dried skimmed milk in tris-buffered saline with 0.05 % Tween-20 (TBST) buffer for 1 hour. The membrane was hybridized with the primary antibodies including anti-GAPDH (1:5000, Cat. No. #MAB374) which was purchased from Millipore (USA); anti-CDK2 (1:1000, Cat. No. #2546), anti- CDK4 (1:1000, Cat. No. #12790), anti-CDK6 (1:2000, Cat. No. #3136S), anti-cyclin D1 (1:1000, Cat. No. #2978), and anti-p21 (1:1,000; catalog no. 2946) which were purchased from Cell Signaling Technology (USA); anti-SLC27A6 (1:1000, Cat. No. #ab72654) which was purchased from Abcam (UK) at 4 °C overnight. After TBST washing 3 times, the membrane was then hybridized with anti-rabbit IgG or anti-mouse IgG HRP-linked antibody (1:3000, Cell Signaling Technology, USA). The images were acquired on Alpha Innotech FluorChem FC2 imaging system (ProteinSimple; Bio-Techne, Minneapolis, MN, USA).
Knockdown of SLC27A6
Lentivirus shRNAs were prepared by the RNAi Core Facility (Taipei, Taiwan). The lentivurus-shRNA clones include: Lenti-emptyT (clone ID, TRCN0000089107; vector control); Lenti-shSLC27A6 #19 (clone ID, TRCN0000043419, targeting sequence: 5'- GCTCATTATAATTCGGCTGAA-3', targeting on SLC27A6); Lenti-shSLC27A6 #20 (clone ID, TRCN0000043420, targeting sequence: 5'-CCCATGTCTTCCTGAACCATT-3', targeting on SLC27A6). To silencing the gene expression, the H184B5F5/M10 and Hs578T cells lines were complete culture media containing 8 μg/ml polybrene (EMD Millipore, Billerica, MA, USA) in 6 cm dish at 37˚C for 30 min. Lentiviruses for H184B5F5/M10 and Hs578T were added for infection at multiplicity of infection = 5. The culture medium was refreshed with fresh culture media with 2 μg/ml puromycin (Sigma‑Aldrich; Merck KGaA, Darmstadt, Germany) after 24 hours of incubation. The infected cells then were maintained in medium with 2 μg/ml puromycin for 3-6 generations and used in assays.
Fatty acid uptake assay
Before fatty acid uptake assay, 1✕104 H184B5F5/M10 and Hs578T were seeded on a 96‑well plate overnight. The fatty acid uptake was evaluated via the Free Fatty Acid Uptake Assay Kit (Fluorometric) (cat. no. ab176768; Abcam, UK). After phosphate‑ buffered saline (PBS) washing and 1-hour preincubated in serum‑free media, cells were then incubated in a fluorescent fatty acid mixture for 30 minutes. The results were evaluated by using a microplate fluorescence reader at 485/528 nm (FLx800; BioTek Instruments Inc., Winooski, VT, USA). The fluorescence signal from vector control group was set to 100% for relative quantification.
Reactive oxygen species (ROS) detection
ROS levels were evaluated using a DCFDA Cellular Detection Assay kit (Cat. No. #ab113851, Abcam, UK) according to manufacturer's instruction. In 96-well plate, 1✕104 adherent H184B5F5/M10 and Hs578T cells were stained with 100 μl of 20 μM DCFDA solution at 37°C for 45 minutes in the dark. After washing with PBS, the results were evaluated by using a microplate fluorescence reader (FLx800; BioTek Instruments Inc., Winooski, VT, USA) at 485/528 nm.
Triglyceride quantification
2-4✕104 H184B5F5/M10 and Hs578T cells were suspended in 100 μl of PBS containing 1% Triton X-100 (Sigma-Aldrich, St. Louis, MO, USA). Cell was mixed on the vortex mixer for 1 minute and then was placed on ice for 30 minutes. After centrifugation at 10,000 ✕ g at 4°C for 15 minutes, the supernatant was collected and then the concentration of triglyceride was analyzed by a Triglyceride Quantification Kit (Cat. No. #ab65336; Abcam, UK) according to manufacturer's instruction. The results were evaluated by using a microplate reader (PowerWaveTM 340; BioTek Instruments Inc., Winooski, VT, USA) at 570 nm.
Assessment of cell growth
The short-term cell proliferation of H184B5F5/ M10 andHs578T was evaluated by WST‑1 assay (4‑[3‑(4‑iodophenyl)‑2‑(4‑nitrophenyl)‑2H‑5‑tetrazolio]‑1,3‑benzene disulfonate) (Clontech, Mountain View, CA, USA) according to manufacturer's instruction. Before WST-1 assay, 3x103 cells were respectively seeded in 96‑well plates overnight. The culture media were then replaced with 100 μl mixture consisting of 95 μl fresh culture media and 5 μl WST-1 reagent. For 24 and 48 hours incubation, the absorbance at 450 nm was determined on a microplate spectrophotometer (PowerWaveTM 340; BioTek, Winooski, VT, USA). The long-term cell growth was evaluated by colony formation assay. 500 cells were seeded in a 6-well plate with 2.5 ml of fresh culture medium. Cell culture media were replaced every 3 day until 14 days after seeding. Colonies were stained with crystal violet (0.4 g/L; Sigma, St. Louis, MO, USA) and the colony number was counted.
Assessment of cell migration
3 ✕ 105 H184B5F5/M10 cells were seeded into 24-well plates. When cells reached a 100 percent confluent monolayer, a scratch was made by a 200 μL pipette tip. Cell debris was washed by phosphate- buffered saline (PBS) washing. Subsequently, the cells were cultured in culture media with 1% FBS for 12 h. The images were captured via a Leica inverted microscope. Migration area was quantitated by TScratch software (version 1.0. Available at http:// www.cse-lab.ethz.ch).
Cell cycle analysis
H184B5F5/M10 cells were maintained in culture medium and harvested at 48 hours incubation after subculture. Cells were fixed with 70% ethanol overnight at 4°C. After PBS washing, cells were incubated with 1 U/ml of DNase-free RNase A and 5 µg/ml of propidium iodide for 10 min at 4°C in the dark (Sigma-Aldrich, St. Louis, MO, USA). The cell cycle distribution was evaluated on BD Accuri C6 flow cytometer (BD Biosciences). The distribution of G0/G1, S and G2/M phase cells were determined as a percentage of the total number of cells.
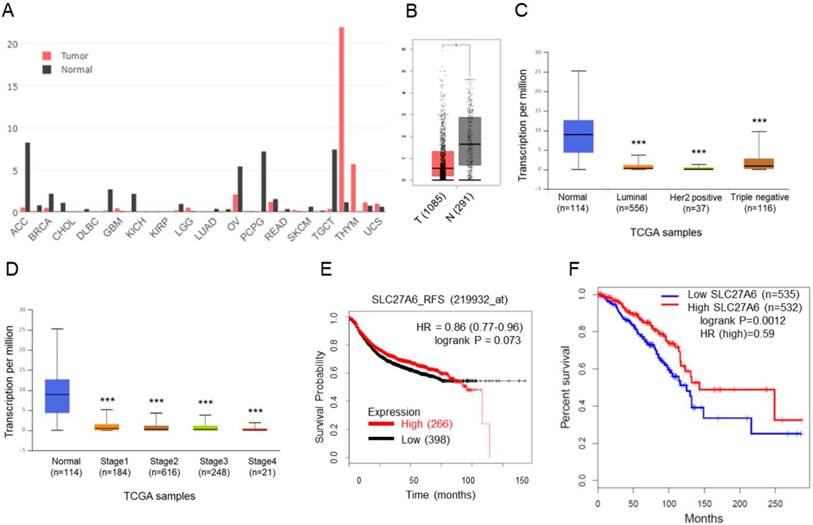
SLC27A6 expression in tumoral and non-tumoral breast tissues and the association between SLC27A6 expression and clinical outcomes. (A) The expression of SLC27A6 in different types of tumor and non-tumor tissues. Abbreviation of each cancer type: adrenocortical carcinoma (ACC), breast invasive carcinoma (BRCA), cholangiocarcinoma (CHOL), lymphoid neoplasm diffuse large B-cell lymphoma (DLBC), glioblastoma multiforme (GBM), kidney chromophobe (KICH), kidney renal papillary cell carcinoma (KIRP), brain lower grade glioma (LGG), lung adenocarcinoma (LUAD), ovarian serous cystadenocarcinoma (OV), pheochromocytoma and paraganglioma (PCPG), rectum adenocarcinoma (READ), skin cutaneous ,elanoma (SKCM), testicular germ cell tumors (TGCT), thymoma (THYM), uterine carcinosarcoma (UCS). (B) The expression of SLC27A6 in breast tumor and non-tumor tissues. The number in parentheses indicated sample size. Above results were obtained from GEPIA database. (C) The SLC27A4 expression in different subtypes and (D) different stages of breast tumor tissues via the UALCAN database. (E) The correlation between SLC27A6 expression (microarray) and relapse-free survival via KM Plotter. (F) The correlation between SLC27A6 expression (RNA sequencing) and overall survival via GEPIA database. * p < 0.05, *** p < 0.001 as compared with the normal.
Statistics
All graphs and statistics were made by the GraphPad Prism 7 software (GraphPad Software, Inc., La Jolla, CA, USA). To examine statistical difference among all groups, a one‑way analysis of variance (ANOVA) with bonferroni multiple comparison test was used. p<0.05 was considered to indicate a statistically significant difference.
Result
The SLC27A6 expression in non-tumor tissues was higher than that in tumor tissues
The SLC27A6 expression in tumoral and non-tumoral tissues in clinical samples was analyzed through GEPIA database. Higher SLC27A6 was detected in non-tumor tissues when compared with tumor tissue in breast cancer and some types of cancer (Figure 1A and 1B). Furthermore, the expression of SLC27A6 in non-tumor tissue is higher than that in different subtypes and different stages of breast cancer (Figure 1C and 1D) via analysis of UALCAN database. To evaluate whether SLC27A6 expression was associated with survival of breast cancer patients, it was evaluated via two different databases including the Kaplan‑Meier (KM) plotter and GEPIA. The gene expression of KM plotter and GEPIA was determined through Affymetrix microarray expression profiles and RNA sequencing, respectively. The trend toward better relapse-free survival (RFS) and overall survival in breast cancer patients with higher SLC27A6 expression (Figure 1E and 1F, p=0.073 and 0.012, respectively).
SLC27A6 expression was repressed in non-tumorigenic and tumorigenic breast cells
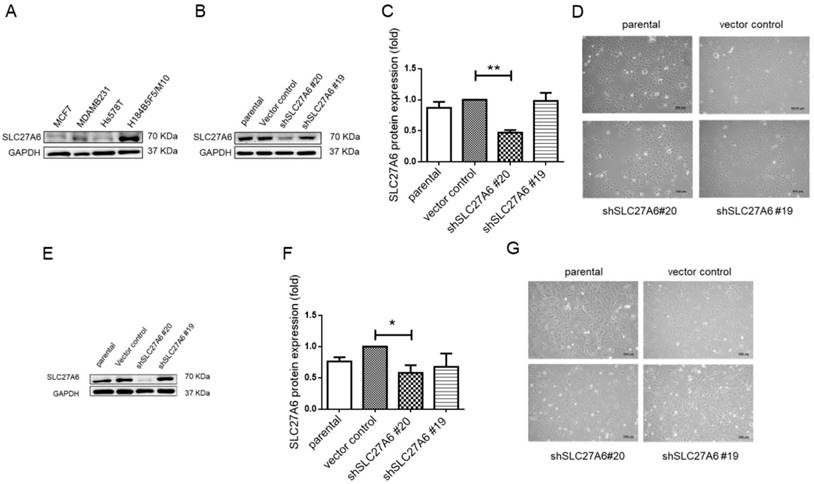
To further investigate the role of SLC27A6 in vitro, the SLC27A6 expression was evaluated by Western blot assay in an immortal and non-tumorigenic human mammary epithelial cell H184B5F5/ M10 and in different type of breast cancer cell lines, MCF-7, Hs578T, and MDA-MB-231. In Figure 2A, the highest SLC27A6 expression was observed in H184B5F5/M10 and relatively low SLC27A6 expression was observed in Hs578T cells. Thus, the H184B5F5/M10 and Hs578T were chosen for investigating the role of SLC27A6 in non-tumorigenic and tumorigenic breast cells. H184B5F5/M10 and Hs578T cells were transduced with lentivirus short hairpin RNA (shRNA) targeting two different sequence of SLC27A6 (shSLC27A6#19 and shSLC27A6#20). The results showed that the expression of SLC27A6 in H184B5F5/M10 and Hs578T was significantly repressed after transduction of lentivirus shSLC27A6#20 but not shSLC27A6#19. The cell morphology of both cells was not significantly changed after repressing SLC27A6 expression (Figure 2B to 2G).
Repressing SLC27A6 decreased capacity of fatty acid uptake in non-tumorigenic breast cells
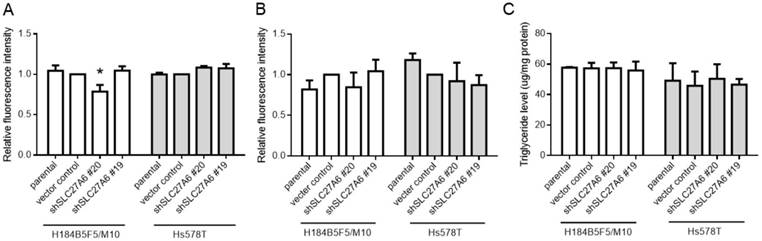
SLC27A6 is a bifunction enzyme with long-chain fatty acids transport and acyl-CoA synthetase (ACS) activity [15, 16]. ACS enzyme activity is associated with acyl-CoA metabolic pathways including β-oxidation and triglyceride synthesis [9]. Therefore, the fatty acid uptake capacity, reactive oxygen species (ROS) level, and intracellular triglyceride concentration were determined in both cell lines. Our results revealed that the fatty acid uptake capacity was inhibited in H184B5F5/M10 with lentivirus shSLC27A6#20 group. By contrast, there was no significant difference among all groups in Hs578T (Figure 3A). In addition, repressing SLC27A6 did not alter the ROS level and triglyceride concentration in H184B5F5/M10 and Hs578T (Figure 3B and 3C).
Repressing SLC27A6 inhibited cell growth in non-tumorigenic breast cells
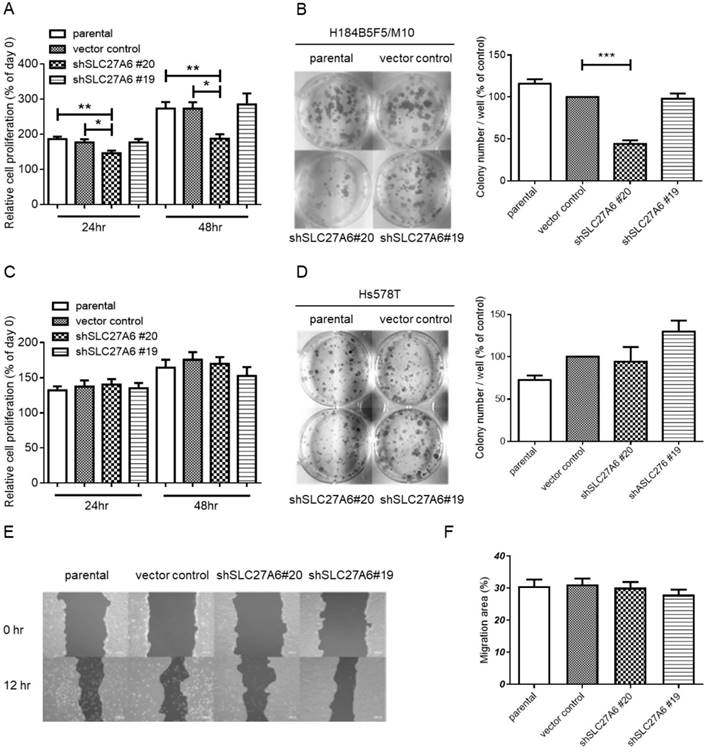
To investigate whether SLC27A6 expression level affects cell growth in non-tumorigenic and tumorigenic breast cells, the WST-1 assay and colony formation were performed. In H184B5F5/M10, slower cell growth was observed in the shSLC27A6#20 group when compared to vector control and parental groups (Figure 4A and 4B). However, the cell growth of Hs578T was not altered by repressing SLC27A6 expression (Figure 4C and 4D). Because long-chain fatty transport is associated with metastasis, the cell migration capacity was evaluated by wound-healing assay. The results showed that silencing SLC27A6 did not significantly affect cell migration of H184B5F5/ M10 (Figure 4E and 4F). Therefore, the effect of growth inhibition is associated with silencing efficiency of SLC27A6 in non-tumorigenic breast cell.
Knockdown of SLC27A6 in the tumorigenic and non-tumorigenic breast cell line. (A) Screening SLC27A6 expression in different cell lines. (B) Detection of protein expression, (C) quantification of protein expression, and (D) cell morphology in SLC27A6-silencing H184B5F5/M10. The shSLC27A6#20 and shSLC27A6#19 indicated two short hairpin RNA targeting two different sequences of human SLC27A6. (E) Detection of protein expression, (F) quantification of protein expression, and (G) cell morphology in SLC27A6-silencing Hs578T. * p < 0.05, ** p < 0.01 as compared with the vector control. Scare bar = 100 μm.
The effect of SLC27A6-silencing on fatty acid uptake capacity, ROS, and triglyceride levels. (A) Fatty acid uptake assay, (B) ROS levels, and (C) triglyceride concentration in H184B5F5/M10 and Hs578T. * p < 0.05 as compared with the vector control.
The effect of SLC27A6-silencing on cell proliferation and migration. (A) Short-term cell growth of H184B5F5/M10 was evaluated by WST-1 assay at 24 and 48 hours after cell seeding, and (B) long-term cell growth was evaluated by colony formation assay at 14 days after cell seeding in H184B5F5/M10. The quantification of colonies was showed at the right panel. The proliferation of Hs578T was evaluated by (C) WST-1 and (D) colony formation assay. (E) The migration capacity of H184B5F5/M10 was evaluated by wound-healing assay, and (F) quantification of wound-healing assay. * p < 0.05, ** p < 0.01, *** p < 0.001, as compared with the vector control.
The effect of SLC27A6-silencing on cell cycle regulators. (A) Cell cycle of SLC27A6-silencing H184B5F5/M10 was evaluated through propidium iodide (PI) staining on flow cytometry. (B) The quantitative result of PI staining assay. (C) The expression of cell cycle regulators cyclin D1, CDK2, CDK4, CDK6, and p21. (D) The quantitative result of cyclin D1, CDK4, and CDK6. * p < 0.05, *** p < 0.001 as compared with the vector control.
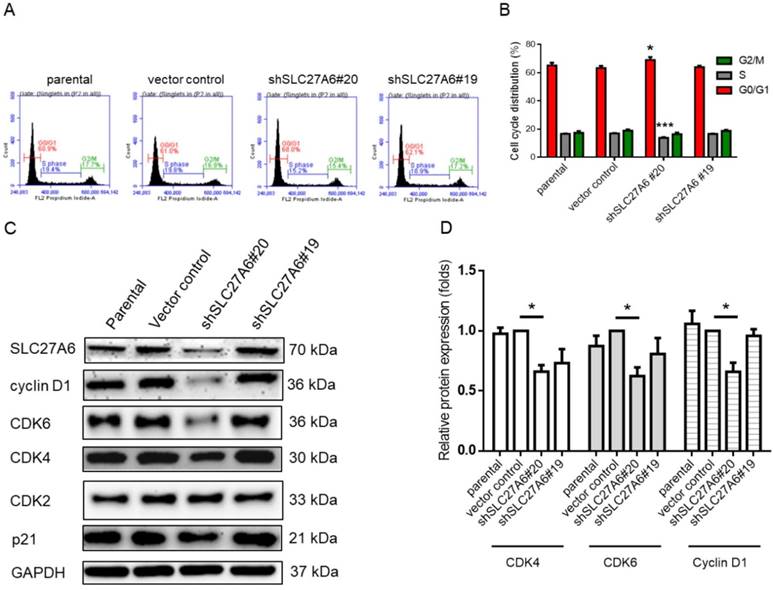
Repressing SLC27A6 inhibited cell growth in non-tumorigenic breast cells through mediating cell cycle regulators
Because cell growth of H184B5F5/M10 was affected by SLC27A6 repression, the cell cycle status was analyzed via the propidium iodide staining assay on flow cytometry. In Figure 5A and 5B, the results showed that increasing cell population in G0/G1 phase and decreasing cell population in S phase in the shSLC27A6#20 group. The protein expression of cell cycle regulator including cyclin D1, cell division protein kinase 4 (CDK4), and CDK6 is relatively low in the shSLC27A6#20 group when compared to the control group. The expression of CDK4 and p21 which was a cell cycle inhibitor was not significantly changed (Figure 5C and 5D). The result might imply the low expression of these cell cycle regulators is associated with low SLC27A6 expression.
Functional protein-associated networks of SLC27A6 in non-tumorigenic breast cells
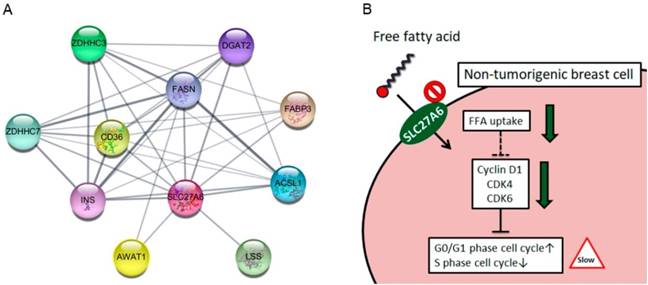
The SLC27A6 protein-associated network was analyzed via STRING database. In Figure 6A, the SLC27A6-associated proteins including ACSL1, AWAT1, CD36, DGAT2, FABP3, FASN, INS, LSS, ZDHHC3, and ZDHHC7 was shown. The full name of each protein was listed in Table 1. The biological process of these genes was performed through DAVID Bioinformatics Resources (Table 2). These genes involve in the process of lipid biosynthesis, fatty acid metabolic process, and fatty acid transport, etc. In addition, the function of ZDHHC3 and ZDHHC7 were related to palmitoyltransferase activity and protein-cysteine S-palmitoyltransferase activity which play important role in the process of fatty acid oxidation [28]. Thus, repressing SLC27A6 expression might significantly affect lipid metabolic pathways in non-tumoral breast cells. The summarized graph of the present study was shown in Figure 6B.
Discussion
Breast cancer is one of the most threatening disease [29]. Fatty acids are demonstrated to affect the behaviors of breast cancer cells. Activation of short-chain fatty acid receptors via short chain fatty acids indices mesenchymal to epithelial transition which drives cells toward non-invasive phenotypes in breast cancer cells [30]. In addition, recent studies suggest that the long-chain fatty transport is related to metastasis and proliferation of breast cancer cells [11-14]. Thus, SLC27 family proteins might also play a role in breast cancer progression. Interestingly, our bioinformatic analysis revealed that SLC27A6 expression in non-tumoral tissue was higher than that in tumoral tissue in clinical samples. We suppose that low SLC27A6 expression in tumoral tissue might be associated with the specific substrate preference of SLC27A6. Unsaturated fatty acid, oleic acid (C18:1), arachidonic acid (C20:4), and saturated fatty acid, lignoceric acid (C24:0) are known substrate of SLC27A6 [16]. The antitumor effect of oleic acid was reported in several types of cancer [31]. In breast cancer, oleic acid treatment results in induction of apoptosis and suppression of proliferation [31]. Therefore, attenuation of SLC27A6 expression might be beneficial for cancer cells although arachidonic acid metabolic pathways are linked to inflammation, angiogenesis, tumor proliferation and metastasis [32]. A further investigation for the regulatory mechanism of SLC27A6 expression between tumoral and non-tumoral breast cell is necessary.
(A) Functional interacting networks of SLC27A6 via the STRING database. (B) The summary scheme based on the results of H184B5F5/M10. Knockdown of SLC27A6 decreased the capacity of free fatty acid uptake and inhibited cell growth via several cell cycle regulators in the non-tumorigenic breast cells.
Full name of SLC27A6-associated proteins
| Gene symbol | Official full name |
|---|---|
| ACSL1 | Acyl-CoA Synthetase Long Chain Family Member 1 |
| AWAT1 | Acyl-CoA Wax Alcohol Acyltransferase 1 |
| CD36 | CD36 molecule |
| DGAT2 | Diacylglycerol O-acyltransferase 2 |
| FASN | Fatty acid synthase |
| FABP3 | Fatty acid-binding protein |
| INS | Insulin |
| LSS | Lanosterol synthase |
| ZDHHC3 | Zinc Finger DHHC-Type Containing 3 |
| ZDHHC7 | Zinc Finger DHHC-Type Containing 7 |
The gene list of biological processes analysis
| GOTERM_BP_FAT | P Value | Genes |
|---|---|---|
| lipid biosynthetic process | 2.07E-05 | AWAT1, DGAT2, FABP3, FASN, LSS |
| fatty acid metabolic process | 1.64E-04 | ACSL1, FABP3, FASN, SLC27A6 |
| lipid transport | 0.0031 | CD36, FABP3, SLC27A6 |
| lipid localization | 0.0036 | CD36, FABP3, SLC27A6 |
| long-chain fatty acid transport | 0.0123 | CD36, FABP3 |
| fatty acid transport | 0.0164 | CD36, FABP3 |
| regulation of fatty acid metabolic process | 0.0286 | ACSL1, INS |
| monocarboxylic acid transport | 0.0291 | CD36, FABP3 |
| regulation of cellular ketone metabolic process | 0.0332 | ACSL1, INS |
Note: The subset “GOTERM_BP_FAT” of biological process in gene ontology analysis was performed.
Similar expression pattern of SLC27A6 was observed in non-tumorigenic H184B5F5/M10 and breast cancer cell lines. Because H184B5F5/M10 is derived from primary mammary cells, the Hs578T which is derived primary tumor was chosen for following experiments [33, 34]. Repressing SLC27A6 leads to decrease fatty acid uptake capacity, inhibit cell proliferation, and delay cell cycle in H184B5F5/ M10. CDK4, CDK6, and cyclin D1 expression decreased in SLC27A6-silencing H184B5F5/M10. Except to lipid metabolism, long-chain fatty acids serve as ligands of peroxisome proliferator-activated receptors (PPAR), nuclear receptors including retinoid-X receptor (RXR), liver-X receptor (LXR), hepatocyte nuclear factor 4 (HNF4), and free fatty acid receptors (FFAR) which regulate downstream metabolic pathways such as β-oxidation, ketogenesis, and triglyceride synthesis [35]. Therefore, the ROS and triglyceride levels in H184B5F5/M10 was determined; however, the levels were not significantly changed after repressing SLC27A6. We speculate that the knockdown of SLC27A6 might alter uptake of some specific long-chain fatty acids which is essential for H184B5F5/M10 proliferation, and might not significantly alter cellular pool of long-chain fatty acids because the other transport proteins compensate the effect of SLC27A6 silencing. The specific fatty acids-mediated by SLC27A6 might affect the regulation of CDK4, CDK6, and cyclin D1. CDK4 can regulate cell cycle and metabolism. The CDK4- pocket protein retinoblastoma (pRB)-transcription factors E2F1 pathway participates in metabolism control such as glucose homeostasis and fatty synthesis through modulation PPARγ [36]. A recent study demonstrates that CDK4 inhibits fatty acid oxidation via modulation of AMP-activated protein kinase (AMPK) [37]. The detailed regulatory mechanism between cell cycle regulators and specific fatty acids is worthy of future investigation. By contrast, Hs578T expresses relatively low level of SLC27A6. Repression of SLC27A6 did not affect phenotypes of Hs578T. We suppose that the other members of SLC27 family protein and other fatty acid transporter proteins might compensate the effect of SLC27A6 repression. SLC27A4 is another member of SLC27 family and our recent study demonstrates that high expression of SLC27A4 is associated with breast cancer tissues [20]. When SLC27A4 was silenced, the proliferation, migration, and invasion of Hs578T were suppressed [20]. Thus, the result might suggest that SLC27A6 plays a minor role in progression of breast cancer.
Currently, the interaction between SLC27A6 and other proteins is not fully-understood. Therefore, the functional protein association networks were evaluated by STRING database. According to the analysis, repressing SLC27A6 might affect several lipid metabolic pathways including lipid biosynthesis, transport, and β-oxidation, etc. Thus, SLC27A6- silencing should affect the other cellular metabolic pathways. Blocking fatty acids, increasing fatty acids degradation, increasing fatty acids storage in neutral triglyceride, and decreasing fatty acids from triglyceride storage are potential strategies to reduce tumor cell proliferation [38]. Therefore, silencing SLC27A6 might disturb multiple lipid metabolic pathways and cell cycle regulation even though H184B5F5/M10 is a non-tumorigenic cell line.
Although H184B5F5/M10 and Hs578T could not fully reflect physiological non-tumoral and tumoral breast tissues, this study still reveals that inverse correlation between SLC27A6 expression and tumoral tissues and provides a new insight into SLC27A6- mediated cell growth and cell cycle regulation in non-tumorigenic breast cells.
Acknowledgements
This study was supported by grants from the Ministry of Science and Technology (MOST 104-2314- B-037-053-MY4; MOST 105-2314-B-037-037-MY3; MOST 106-2314-B-037-046; MOST 106-2320-B-037-029- MY3), the Kaohsiung Medical University Hospital (KMUHS10701; KMUHS10712; KMUH106-6R34; KMUH106-6R77), and the Kaohsiung Medical University (KMU-DK108008). The authors thank the Center for Research Resources and Development of Kaohsiung Medical University.
Competing Interests
The authors have declared that no competing interest exists.
References
1. Wang DQ. Regulation of intestinal cholesterol absorption. Annu Rev Physiol. 2007;69:221-48
2. Wang TY, Liu M, Portincasa P, Wang DQ. New insights into the molecular mechanism of intestinal fatty acid absorption. Eur J Clin Invest. 2013;43:1203-23
3. Reshef L, Olswang Y, Cassuto H, Blum B, Croniger CM, Kalhan SC. et al. Glyceroneogenesis and the triglyceride/fatty acid cycle. J Biol Chem. 2003;278:30413-6
4. Marten B, Pfeuffer M, Schrezenmeir J. Medium-chain triglycerides. Int Dairy J. 2006;16:1374-82
5. Schwenk RW, Holloway GP, Luiken JJ, Bonen A, Glatz JF. Fatty acid transport across the cell membrane: regulation by fatty acid transporters. Prostaglandins Leukot Essent Fatty Acids. 2010;82:149-54
6. Ehehalt R, Fullekrug J, Pohl J, Ring A, Herrmann T, Stremmel W. Translocation of long chain fatty acids across the plasma membrane-lipid rafts and fatty acid transport proteins. Mol Cell Biochem. 2006;284:135-40
7. Dutta-Roy AK. Cellular uptake of long-chain fatty acids: role of membrane-associated fatty-acid-binding/transport proteins. Cell Mol Life Sci. 2000;57:1360-72
8. Kihara A. Very long-chain fatty acids: elongation, physiology and related disorders. J Biochem. 2012;152:387-95
9. Faergeman NJ, Knudsen J. Role of long-chain fatty acyl-CoA esters in the regulation of metabolism and in cell signalling. Biochem J. 1997;323( Pt 1):1-12
10. Hirschey MD, DeBerardinis RJ, Diehl AME, Drew JE, Frezza C, Green MF. et al. Dysregulated metabolism contributes to oncogenesis. Semin Cancer Biol. 2015;35(Suppl):S129-S50
11. Liu RZ, Graham K, Glubrecht DD, Germain DR, Mackey JR, Godbout R. Association of FABP5 expression with poor survival in triple-negative breast cancer: implication for retinoic acid therapy. Am J Pathol. 2011;178:997-1008
12. Liu RZ, Graham K, Glubrecht DD, Lai R, Mackey JR, Godbout R. A fatty acid-binding protein 7/RXRbeta pathway enhances survival and proliferation in triple-negative breast cancer. J Pathol. 2012;228:310-21
13. Zhao J, Zhi Z, Wang C, Xing H, Song G, Yu X. et al. Exogenous lipids promote the growth of breast cancer cells via CD36. Oncol Rep. 2017;38:2105-15
14. Pascual G, Avgustinova A, Mejetta S, Martin M, Castellanos A, Attolini CS. et al. Targeting metastasis-initiating cells through the fatty acid receptor CD36. Nature. 2017;541:41-5
15. Anderson CM, Stahl A. SLC27 fatty acid transport proteins. Mol Aspects Med. 2013;34:516-28
16. Watkins PA. Very-long-chain acyl-CoA synthetases. J Biol Chem. 2008;283:1773-7
17. Gimeno RE, Ortegon AM, Patel S, Punreddy S, Ge P, Sun Y. et al. Characterization of a heart-specific fatty acid transport protein. J Biol Chem. 2003;278:16039-44
18. Auinger A, Helwig U, Pfeuffer M, Rubin D, Luedde M, Rausche T. et al. A variant in the heart-specific fatty acid transport protein 6 is associated with lower fasting and postprandial TAG, blood pressure and left ventricular hypertrophy. Br J Nutr. 2012;107:1422-8
19. Chassen SS, Ferchaud-Roucher V, Gupta MB, Jansson T, Powell TL. Alterations in placental long chain polyunsaturated fatty acid metabolism in human intrauterine growth restriction. Clin Sci (Lond). 2018;132:595-607
20. Yen MC, Chou SK, Kan JY, Kuo PL, Hou MF, Hsu YL. Solute Carrier Family 27 Member 4 (SLC27A4) Enhances Cell Growth, Migration, and Invasion in Breast Cancer Cells. Int J Mol Sci. 2018:19
21. Tang Z, Li C, Kang B, Gao G, Li C, Zhang Z. GEPIA: a web server for cancer and normal gene expression profiling and interactive analyses. Nucleic Acids Res. 2017;45:W98-W102
22. Gyorffy B, Lanczky A, Eklund AC, Denkert C, Budczies J, Li Q. et al. An online survival analysis tool to rapidly assess the effect of 22,277 genes on breast cancer prognosis using microarray data of 1,809 patients. Breast Cancer Res Treat. 2010;123:725-31
23. Chandrashekar DS, Bashel B, Balasubramanya SAH, Creighton CJ, Ponce-Rodriguez I, Chakravarthi B. et al. UALCAN: A Portal for Facilitating Tumor Subgroup Gene Expression and Survival Analyses. Neoplasia. 2017;19:649-58
24. Shannon P, Markiel A, Ozier O, Baliga NS, Wang JT, Ramage D. et al. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13:2498-504
25. Szklarczyk D, Morris JH, Cook H, Kuhn M, Wyder S, Simonovic M. et al. The STRING database in 2017: quality-controlled protein-protein association networks, made broadly accessible. Nucleic Acids Res. 2017;45:D362-D8
26. Huang da W, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 2009;4:44-57
27. Huang da W, Sherman BT, Lempicki RA. Bioinformatics enrichment tools: paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res. 2009;37:1-13
28. Qu Q, Zeng F, Liu X, Wang QJ, Deng F. Fatty acid oxidation and carnitine palmitoyltransferase I: emerging therapeutic targets in cancer. Cell Death Dis. 2016;7:e2226
29. Siegel RL, Miller KD, Jemal A. Cancer statistics, 2018. CA Cancer J Clin. 2018;68:7-30
30. Thirunavukkarasan M, Wang C, Rao A, Hind T, Teo YR, Siddiquee AA. et al. Short-chain fatty acid receptors inhibit invasive phenotypes in breast cancer cells. PloS one. 2017;12:e0186334
31. Carrillo C, Cavia Mdel M, Alonso-Torre SR. Antitumor effect of oleic acid; mechanisms of action: a review. Nutr Hosp. 2012;27:1860-5
32. Borin TF, Angara K, Rashid MH, Achyut BR, Arbab AS. Arachidonic Acid Metabolite as a Novel Therapeutic Target in Breast Cancer Metastasis. Int J Mol Sci. 2017:18
33. Chavez KJ, Garimella SV, Lipkowitz S. Triple negative breast cancer cell lines: one tool in the search for better treatment of triple negative breast cancer. Breast Dis. 2010;32:35-48
34. Yang TC, Stampfer MR, Tobias CA. Radiation studies on sensitivity and repair of human mammary epithelial cells. Int J Radiat Biol. 1989;56:605-9
35. Nakamura MT, Yudell BE, Loor JJ. Regulation of energy metabolism by long-chain fatty acids. Prog Lipid Res. 2014;53:124-44
36. Blanchet E, Annicotte JS, Fajas L. Cell cycle regulators in the control of metabolism. Cell cycle. 2009;8:4029-31
37. Lopez-Mejia IC, Lagarrigue S, Giralt A, Martinez-Carreres L, Zanou N, Denechaud PD. et al. CDK4 Phosphorylates AMPKalpha2 to Inhibit Its Activity and Repress Fatty Acid Oxidation. Mol Cell. 2017;68:336-49 e6
38. Currie E, Schulze A, Zechner R, Walther TC, Farese RV Jr. Cellular fatty acid metabolism and cancer. Cell Metab. 2013;18:153-61
Author contact
![]() Corresponding authors: Professor Ming-Feng Hou, Graduate Institute of Clinical Medicine, College of Medicine, Kaohsiung Medical University, No. 100, Shih-Chuan 1st Road, Kaohsiung 807, Taiwan, R.O.C. E-mail: mifehoedu.tw or Professor Ya-Ling Hsu, Graduate Institute of Medicine, College of Medicine, Kaohsiung Medical University, No. 100, Shih-Chuan 1st Road, Kaohsiung 807, Taiwan, R.O.C. E-mail: hsuyl326com
Corresponding authors: Professor Ming-Feng Hou, Graduate Institute of Clinical Medicine, College of Medicine, Kaohsiung Medical University, No. 100, Shih-Chuan 1st Road, Kaohsiung 807, Taiwan, R.O.C. E-mail: mifehoedu.tw or Professor Ya-Ling Hsu, Graduate Institute of Medicine, College of Medicine, Kaohsiung Medical University, No. 100, Shih-Chuan 1st Road, Kaohsiung 807, Taiwan, R.O.C. E-mail: hsuyl326com

 Global reach, higher impact
Global reach, higher impact