3.2
Impact Factor
ISSN: 1449-1907
Int J Med Sci 2019; 16(2):221-230. doi:10.7150/ijms.29312 This issue Cite
Research Paper
A New Look at Causal Factors of Idiopathic Scoliosis: Altered Expression of Genes Controlling Chondroitin Sulfate Sulfation and Corresponding Changes in Protein Synthesis in Vertebral Body Growth Plates
1. Novosibirsk Research Institute of Traumatology and Orthopaedics n.a. Ya.L. Tsivyan, Novosibirsk, Russia
2. Meshalkin National Medical Research Center, Ministry of Health of the Russian Federation, Novosibirsk, Russia
3. Institute of Chemical Biology and Fundamental Medicine, Russian Academy of Science, Novosibirsk, Russia
4. Institute of Cytology and Genetics, Russian Academy of Science, Novosibirsk, Russia
5. University of Pittsburgh, Pittsburgh PA, USA
6. Arrowhead Pharmaceuticals, Madison WI, USA
Received 2018-8-17; Accepted 2018-12-7; Published 2019-1-1
Abstract
Background: In a previous report, we demonstrated the presence of cells with a neural/glial phenotype on the concave side of the vertebral body growth plate in Idiopathic Scoliosis (IS) and proposed this phenotype alteration as the main etiological factor of IS. In the present study, we utilized the same specimens of vertebral body growth plates removed during surgery for Grade III-IV IS to analyse gene expression. We suggested that phenotype changes observed on the concave side of the vertebral body growth plate can be associated with altered expression of particular genes, which in turn compromise mechanical properties of the concave side.
Methods: We used a Real-Time SYBR Green PCR assay to investigate gene expression in vertebral body growth plates removed during surgery for Grade III-IV IS; cartilage tissues from human fetal spine were used as a surrogate control. Special attention was given to genes responsible for growth regulation, chondrocyte differentiation, matrix synthesis, sulfation and transmembrane transport of sulfates. We performed morphological, histochemical, biochemical, and ultrastructural analysis of vertebral body growth plates.
Results: Expression of genes that control chondroitin sulfate sulfation and corresponding protein synthesis was significantly lower in scoliotic specimens compared to controls. Biochemical analysis showed 1) a decrease in diffused proteoglycans in the total pool of proteoglycans; 2) a reduced level of their sulfation; 3) a reduction in the amount of chondroitin sulfate coinciding with raising the amount of keratan sulfate; and 4) reduced levels of sulfation on the concave side of the scoliotic deformity.
Conclusion: The results suggested that altered expression of genes that control chondroitin sulfate sulfation and corresponding changes in protein synthesis on the concave side of vertebral body growth plates could be causal agents of the scoliotic deformity.
Keywords: idiopathic scoliosis, vertebral body growth plate, gene expression
Introduction
Scoliotic deformity is one of the most common spine pathologies affecting children and adolescents. Idiopathic scoliosis (IS) occurs in otherwise healthy children and adolescents, affecting 2-4 million people in the Russian Federation (extrapolated from [1]) and approximately 8 million in the United States, representing tremendous medical, social, and financial burden [2, 3]. While etiological factors of IS have not been identified [4], [5], which to some extent could be attributed to the absence of a proper animal model [6], several hypotheses have tried to delineate possible causative factors. The first hypotheses founded on a biomechanical model was offered by Somerville in 1952 [7] and further elaborated by Roaf [8]. In modern times, mechanical effects on vertebral growth have been investigated in detail by Ian Stokes (e.g. [9]).
While all agree that asymmetric growth of the concave and convex sides of vertebral body growth plates causes IS deformity (e.g. [9]) and implementation of the Hueter-Volkmann principle is intuitive[10], approaches based on biomechanical models were not able to offer radical cure or prevention. During the last few decades, the genetic nature of IS has been intensively investigated, but recent studies concluded that identification of genes determining the development of this disease is very difficult [11]. Some studies have even achieved rather contradictory data. Gorman et al., [12] analyzed 50 representative studies including 34 candidate gene studies and 16 full genome ones. The authors concluded that contemporary data on the genetics of IS do not explain its etiology and could not be used to determine the prognosis of the disease [12]. Different treatment strategies based on neurological models also were investigated, but general agreement is that additional research is needed (for a detailed account of IS hypotheses see [1, 13]). The analysis of Wang and co-authors on contemporary hypotheses and approaches to an IS cure concluded that “The current treatment at best is treating the morphologic and functional sequelae of AIS and not the cause of the disease” [14].
Driven by the fact that prevailing models cannot explain pathological features of IS [15, 16], Burwell and co-authors outlined a novel multifactorial Cascade Concept of IS pathogenesis [17], which together with previous ideas by the same group [18] put an emphasis on epigenetic factors affecting vertebral growth in infancy and early childhood.
We hypothesised that such epigenetic factors may affect vertebral structure development much earlier, during neural crest cell migration through somites, resulting in altered vertebral growth plate differentiation. In a previous report, we demonstrated the presence of cells with a neural/glial phenotype on the concave side of the vertebral body growth plate in IS and proposed this phenotype alteration as the main etiological factor of the IS [19]. In the present study we utilized selected specimens from the same study (vertebral body growth plates removed during surgery for Grade III-IV IS) to analyse gene expression. We suggested that phenotype changes observed on the concave side of the vertebral body growth plate can be associated with altered expression of particular genes, which in turn compromise mechanical properties of the concave side. This study included morphological and biochemical analyses of the vertebral growth plate of the deformity and investigation of the expression of genes whose products can influence IS development. The objective of the study was to conduct an expression analysis of the genes regulating differentiation and functioning of chondrocytes, as well as the synthesis of intracellular matrix components, with simultaneous morphological and biochemical analyses of the growth plate cartilage in IS.
Materials and Methods
Clinical specimens
Vertebral body growth plates from the curve apex and from above and below the curve apex were removed during the surgery of anterior release and interbody fusion in 12 patients aged 11-15 years with IS of Grade III-IV [19]. An ideal control for this study would be normal, non-hypoxic human growth plate specimens from non-scoliotic subjects of corresponding ages. However, such specimens are extremely rarely accessible; for example, these specimens may become available following urgent surgery for spinal trauma, when removal of vertebral body growth plates would be dictated by treatment requirements. In reality, however, such control specimens have never been achievable in our settings (or for other research groups, as far as we know). However, existing information allows for bridging gene expression patterns from vertebral body growth plates of different developmental stages and then using available specimens as a provisional control. Comparison of gene expression patterns of human vertebral fetal growth plate cartilage showed similarities between 8-12 and 12-20 week old fetal cartilage [20-22]. No obvious changes were observed in RAGE expression between fetal, juvenile, and young adolescent discs (until the age of 13 years) [23]. Therefore, as a provisional control, cartilage structural components of the human fetal spine at 10-12 weeks of development were used. Ten specimens were obtained from healthy women immediately after medical abortions performed in the clinics licensed by Ministry of Health of The Russian Federation, in accordance with the approved list of medical indications. All patients gave written informed consent to participate in the study. The study was performed in accordance with the ethical principles of the Helsinki Declaration and standards of the Institutional Bioethical Committee.
Morphology, histochemistry, biochemistry, ultrastructural analysis. Morphological, histochemical, biochemical, and ultrastructural studies of cells and matrix growth plates of the vertebral bodies of patients with IS and of the control samples were performed according to protocols described previously [24].
Isolation of cells from tissue specimens
Hyaline cartilage of the growth plates and fetal cartilage were washed in saline solution, milled to a size of 1-2 mm in a petri dish with a minimal volume of Roswell Park Memorial Institute (RPMI) medium, placed in a 1,5% solution of collagenase in siliconized dishes and incubated in a CO2 incubator at 37°C for 22-24 hours. The resulting cell suspension was passed through a nylon filter to remove the tissue pieces, and the cells were pelleted by centrifugation for 10 minutes at 2000 rpm. The pelleted cells were re-suspended in saline, and the total amount of cells was determined using a haemocytometer.
Isolation of RNA from cells and preparation of samples for PCR
Total cellular RNA was isolated from cells by the trizol method (TRI Reagent, Sigma, USA) according to the manufacturer's recommendations. The precipitated RNA was dissolved in 30-50 µl of RNAse-free water (Fermentas, Latvia).
To remove genomic DNA, the isolated RNA was treated with RNAse-free DNAse (Fermentas, Latvia) according to the manufacturer's recommendations. cDNA was obtained from reverse transcription of 2 μg of total RNA of each sample using the Oligo (dT)15 primer (BIOSSET, Russia), and the enzyme M-MLV Reverse Transcriptase (Promega, USA) according to the manufacturer's recommendations (200 u. M-MLV reaction, reaction volume 25 µl).
Determination of mRNA levels of the tested genes by quantitative PCR
All real-time PCR reactions were performed in a iCycler IQ5 thermocycler (Bio-Rad, USA) in the presence of the dye SYBR Green I. The volume of the reaction mixture was 30 µl: 8,6 µl of water, 0,2 µl of each forward and reverse primer (45 μM), 1 µl (5 units) of Taq polymerase (Fermentas, Latvia), and 5 µl of cDNA were added to 15 µl of 2x buffer (7 mM MgCl2, 130 mM Tris-HCl, pH 8,8, 32 mM (NH4)2SO4, 0,1% Tween-20, 0,5 mM of each dNTP). Primer sequences and PCR conditions are presented in Table 1.
List of genes, primers, and conditions of Real-Time SYBR Green I PCR
| № | Name of gene Genes, GenBank acc. N | Sequence of primers (5'->3'): | Size of fragment (nucleotides) | PCR conditions |
|---|---|---|---|---|
| 1 | GAPDH NM_002046.3 | F: TGAAGGTCGGAGTCAACGGATTTGGT R: CATCGCCCCACTTGATTTTGGAGGG | 258 | 1. 95º С - 3,5 min 2. 40 cycles 95ºС - 20 sec 66º С - 15 sec 72º С - 30 sec 84º С - 10 sec |
| 2 | ACAN NM_013227.3 | F: GGCGAGCACTGTAACATAGACCAGG R: CCGATCCACTGGTAGTCTTGGGCAT | 206 | 1. 95ºС - 3,5 min. 40 cycles 95ºС - 20 sec. 66ºС - 15 sec. 72ºС - 30 sec. 88ºС - 10 sec. |
| 3 | LUM NM_002345.3 | F:ACCTGGAGGTCAATCAACTTGAGAAGTTTG R: AGAGTGACTTCGTTAGCAACACGTAGACA | 172 | 1. 95º С - 3,5 min 2. 40 cycles 95º С - 20 sec. 64º С - 15 sec. 72º С - 30 sec 82º С - 10 sec |
| 4 | VCAN NM_004385.4 | F: CTGGCAAGTGATGCGGGTCTTTACC R: GGAGCCCGGATGGGATATCTGACAG | 278 | 1. 95º С - 3,5 min 2. 40 cycles 95º С - 20 sec 66º С - 15 sec 72º С - 30 sec 86º С - 10 sec |
| 5 | COL1A1 NM_000088.3 | F: GAAGACATCCCACCAATCACCTGCGTA R: GTGGTTTCTTGGTCGGTGGGTGACT | 227 | 1. 95º С - 3,5 min 2. 40 cycles 95º С - 20 sec 66º С - 15 sec 72º С - 30 sec 88º С - 10 sec |
| 6 | COL2A1 NM_001844.4 | F: AAGGAGACAGAGGAGAAGCTGGTGC R: AATGGGGCCAGGGATTCCATTAGCA | 299 | 1. 95º С - 3,5 min 2. 40 cycles 95º С - 15 sec 65º С - 10 sec 72º С - 20 sec 88º С - 10 sec |
| 7 | HAPLN1 NM_001884.3 | F: GGTAGCACTGGACTTACAAGGTGTGGT R: GGCTCTCTGGGCTTTGTGATGGGAT | 222 | 1. 95º С - 3 min 30 sec 2. 40 cycles 95º С - 20 sec 67º С - 15 sec 72º С - 20 sec 87º С - 10 sec |
| 8 | PAX1 NM_006192.3 | F: AACATCCTGGGCATCCGGACGTTTA R: AGGGTGGAGGCCGACTGAGTGTAT | 194 | 1. 95º С - 3,5 min 2. 40 cycles 95º С - 20 sec 68º С - 15 sec 72º С - 30 sec 89,5º С - 10 sec |
| 9 | PAX9 NM_006194.3 | F: CTCCATCACCGACCAAGTGAGCGA R: GAGCCATGCTGGATGCTGACACAAA | 212 | 1. 95º С - 3,5 min 2. 40 cycles 95º С - 20 sec 68º С - 15 sec 72º С - 30 sec 89,5º С - 10 sec |
| 10 | SOX9 NM_000346.3 | F: ACTACACCGACCACCAGAACTCCAG R: AGGTCGAGTGAGCTGTGTGTAGACG | 206 | 1. 95º С - 3,5 min 2. 40 cycles 95º С - 20 sec 68º С - 15 sec 72º С - 30 sec 88º С - 10 sec |
| 11 | IHH NM_002181.3 | F: GATGAACCAGTGGCCCGGTGTG R: CCGAGTGCTCGGACTTGACGGA | 233 | 1. 95º С - 3,5 min 2. 40 cycles 95º С - 12 sec 58º С - 08 sec 72º С - 20 sec 89º С - 10 sec |
| 12 | GHR NM_000163.2 | F: TGCCCCCAGTTCCAGTTCCAAAGAT R: AGGTTCACAACAGCTGGTACGTCCA | 284 | 1. 95º С - 3,5 min 2. 40 cycles 95º С - 20 sec 60º С - 15 sec 72º С - 30 sec 82º С - 10 sec |
| 13 | IGF1R NM_000875.3 | F: CGCACCAATGCTTCAGTTCCTTCCA R: CCACACACCTCAGTCTTGGGGTTCT | 266 | 1. 95º С - 3,5 min 2. 40 cycles 95º С - 20 sec 66º С - 15 sec 72º С - 30 sec 85º С - 10 sec |
| 14 | EGFR NM_005228.3 | F: ATAGACGACACCTTCCTCCCAGTGC R: GTTGAGATACTCGGGGTTGCCCACT | 177 | 1. 95º С - 3,5 min 2. 40 cycles 95º С - 20 sec 62º С - 15 sec 72º С - 30 sec 87º С - 10 sec |
| 15 | TGFBR1 NM_001130916.1 | F: GGGCGACGGCGTTACAGTGTT R: AGAGGGTGCACATACAAACGGCCTA | 179 | 1. 95º С - 3,5 min. 2. 40 cycles 95º С - 25 sec 59º С - 05 sec 72º С - 20 sec 83º С - 10 sec |
| 16 | SLC26A2 NM_000112.3 | F: CCTGTTTTGCAGTGGCTCCCAA R: CCACAGAGATGTGACGGGAGGT | 208 | 1. 95º С - 3,5 min. 2. 40 cycles 95º С - 25 sec 59º С - 05 sec 72º С - 20 sec 84º С - 10 sec |
| 17 | CHST1 NM_003654.5 | F: ATACGGCACCGTGCGAAACTCG R: AGGCTGACCGAGGGGTTCTTCA | 165 | 1. 95º С - 3,5 min. 2. 40 cycles 95º С - 15 sec 62º С - 10 sec 72º С - 20 sec 89º С - 10 sec |
| 18 | CHST3 NM_004273.4 | F: AGAAAGGACTCACTTTGCCCCAGGA R: TGAAGCTGGGAGAAGGCTGAATCGA | 268 | 1. 95º С - 3,5 min. 2. 40 cycles 95º С - 20 sec 68º С - 15 sec 72º С - 20 sec 84º С - 10 sec |
The PCR results were evaluated by the computer program iCycler IQ 5. The specificity of the reaction was determined by analyzing the melting curves of amplification products ranging from 65°C to 95°C in increments of 1°C. To control PCR cross-contamination, RNAse-free water was added to the RNA precipitate, which was then used as a negative control. The gene glyceraldehyde-3-phosphate dehydrogenase (GAPDH) was used as a reference housekeeping gene. PCR products obtained after amplification of cDNA with specific primers were used as standards.
To construct the calibration curves, serial dilutions were prepared from obtained standards, and the Real-Time SYBR Green I PCR reaction was conducted.
The GAPDH gene was chosen as a reference gene to evaluate the relative levels of mRNA expression of target genes. The average value of a target gene was divided by the average value of the GAPDH gene for normalization. To represent the data, the smallest value designated as a calibrator was taken from the obtained normalized data. To calculate the relative amount of a target gene, normalized values of this gene were divided by the value of the calibrator (Figures 3, 4).
Statistical analysis
Statistical analysis of the results was performed using the package Microsoft Office Excel 2007 and the standard software package STATISTICA 6,0. The arithmetic mean value (M) and standard error of the mean value (m) were determined. The nonparametric statistical Mann-Whitney U-test was used to identify the difference in the probability of compared averages. Differences were considered significant at the 5% significance level (p < 0.05). Factor analysis was performed using the software package STATISTICA 6,0.
Results and discussion
Justification of the choice of candidate genes determining the development of IS
One undeniable factor in the formation of scoliotic deformity is the asymmetry of growth, which rationalizes choosing the growth plate as a possible source of misbalanced genetic growth regulations. By the time of birth, the vertebral body undergoes enchondral osteogenesis, with the exception of the cartilaginous plate, which undergoes longitudinal spinal growth. The process of growth in the postnatal period is a step-morphogenesis, the essence of which is proliferative periodization and chondrocyte differentiation from minimal differentiation to terminally differentiated chondrocytes and subsequent osteogenesis. Because regulations of both embryonic and postnatal development follow the same pattern [25], embryonic growth plates (12 weeks of embryogenesis) were utilized as controls for the study of gene expression levels.
Morphological and biochemical criteria of growth asymmetry
Structural and functional organization of the growth plates on the convex and concave sides of the spinal deformity were studied to evaluate qualitative and quantitative differences.
Biochemical data
The levels of proteoglycans (PG) and of their constituent glycosaminoglycans (GAGs) on the convex and concave sides of the deformity apex quantified by biochemical methods are presented in Table 2. Decreases in the share of PG1 in the total pool of PG and in the level of sulfation, which reduces the amount of chondroitin sulfate (CS) and raises the amount of keratan sulfate (KS), were detected on the concave side of the deformity.
Characteristics of PG of the vertebral body growth plates from different sides of curvature in IS patients (PG output is calculated in µg per mg of tissue wet weight. The relative amount of PG2 in the pool is shown as a percentage in parentheses).
| Convex side of deformity | Concave side of deformity | |||
|---|---|---|---|---|
| n=18 | n=18 | |||
| PG1 | PG2 | PG1 | PG2 | |
| PG output | 18,4±2,25 | 38,2±3,89* (63,5±3,62 (%)) | 10,2±1,56*1 | 28,8±1,74*1.2 (74,1±6,85 (%)) |
| CS/KS | 1,28+0,098 | 0,75+0,058* | 0,81+0,065*1 | 0,59+0,041*1.2 |
| Degree of sulfation (%) | 30,2±2,78 | 5,7±0,65* | 18,4±0,15*1 | 7,7±0,84*2 |
PG1 - diffused PG; PG2 - PGs linked with collagen; * - significant difference р<0,05; *1 - significant difference from analogous pool of convex side *2 - Significant difference from PG of convex side
Electrophoretic separation of PG from vertebral body growth plates showed a reduction in the amount of CS and an increase in KS.
Light microscopy analysis of growth plates of the scoliotic deformity
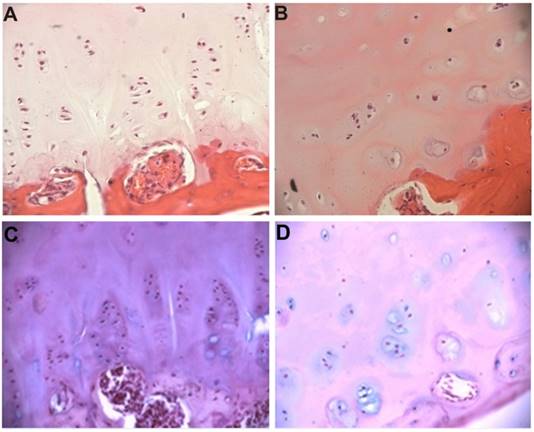
The growth plate on the convex side of the deformity showed preserved structural organization (Figure 1A). Columns of chondrocytes are arranged horizontally with respect to the axis of the spine and consist of 4-5 cells with large nuclei and narrow rims of cytoplasm. Groups of chondrocytes are embedded in homogeneous matrix. There are 1-2 nucleoli and dispersed chromatin in each nucleus. The ultrastructure of these cells corresponds to the differentiated stage. High polymeric CS are defined in chondrocytes and extracellular matrix (Figure 1C).
The concave side of the growth plate is devoid of zonal structuring. The poorly differentiated chondroblasts are scattered in the matrix. Rare cell groups, consisting of few cells, are localized in the lower layers (Figure 1B). Highly polymerized structures of CS are present in much low density in the matrix and cells (Figure 1D).
An acellular matrix containing high-polymerized PGs is located between the convex and concave sides of the growth plate deformity. Vessels penetrating vertebral growth plate are accompanied by osteogenesis (data not shown).
The vertebral body growth plate from the convex (A) and concave (B) sides of deformity in an IS patient. Hematoxylin & eosin staining, x200. Intensive staining for high polymeric CS on the convex side (C) and diminishing staining for high polymeric CS in the concave side, (Hale's reaction), x200.
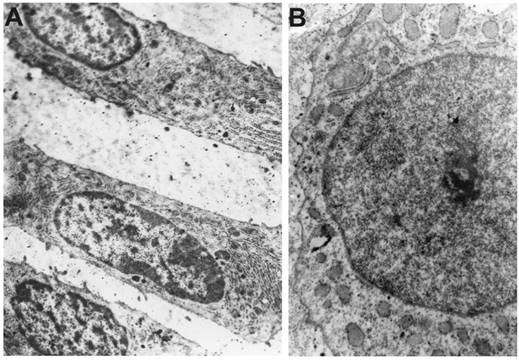
Ultrastructural organization of chondroblasts of the vertebral body growth plate from the convex (A) and concave (B) sides of a deformity in IS, x5000.
Ultrastructural analysis
Chondrocytes on the convex side (columnar arrangement) have off-centre nuclei, with both dispersed and condensed chromatin. The Golgi apparatus with numerous vacuoles is dispersed throughout the cytoplasm (Figure 2A). Ultrastructural arrangement of chondrocytes on the concave side is strongly modified: scarce Golgi apparatus, mainly located near nuclei, connected to inflated cisternae of the endoplasmic reticulum. Nuclei contain electron-dense chromatin assembly (Figure 2B).
A layer of hypertrophic cells consists of two types of chondrocytes: actively synthesizing and terminally differentiated cells, some of which undergo apoptosis. Cytoplasmic granules of CDH and NADH-diaphorase are found in the cells of the column layer and in the active hypertrophic cells. The cytoplasm of the lower hypertrophic cell layers is filled with granules of alkaline phosphatase (data not shown).
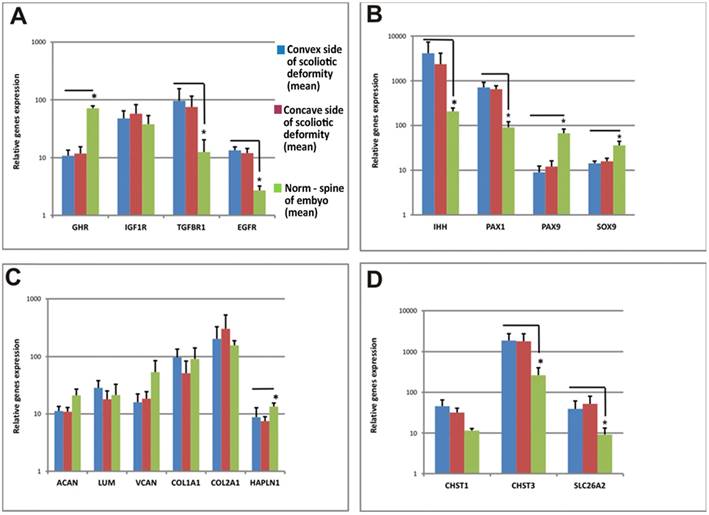
Growth factors and chondroblast gene expression levels in vertebral body growth plates and fetal vertebra are shown using SYBR-Green real time RT-PCR. Relative gene expression is calculated with respect to the GAPDH mRNA concentration as an internal control. Error bars represent standard deviation in each point. * - significant difference (р < 0,05). А. Genes encoding growth factors, B. Genes encoding transcription factors, C. Genes encoding PGs, D. Genes encoding sulfate group metabolism-related proteins.
Study of the expression of candidate genes conceivably determining IS
Alterations in the structural organization of cells and matrix on the concave side of the spinal deformity are the obvious cause of growth asymmetry. The presented data suggested the following basis for the selection of possible candidate genes determining IS. The expression levels of genes regulating the differentiation and metabolism of growth plate cartilage cells localized on the concave and convex sides of the deformity were investigated to identify genes whose hypo- or hyper-expression may cause the development of IS. The following genes were selected: genes involved in chondrocyte growth regulation: growth factors (GHR, EGFR, IGF1R, and TGFBR1), in differentiation signaling (IHH, PAX1, PAX9, and SOX9), in the regulation of essential protein synthesis - structural components of matrix PGs (ACAN, LUM, VCAN, COL1A1, СOL2A1, and HAPLN1), and in the sulfation and transmembrane transport of sulfates (DTDST, CHST1, and CHST3).
The expression levels of genes of interest measured relative to the expression level of the housekeeping gene GAPDH are presented in Figure 3. The studied genes can be divided into three groups according to their expression levels in cells of patients with IS relative to control cells: expression level does not differ from the norm (ACAN, LUM, VCAN, COL1A1, COL2A1, IGF1R, and GHST1), is below the norm (PAX9, SOX9, HAPLN1, and GHR), and is significantly higher than the norm (IHH, PAX1, TGFBR1, EGFR, SLC26A2, and CHST3).
The genes of the first group (ACAN, LUM, VCAN, COL1A1, COL2A1, IGF1R, and GHST1) are mainly represented by genes encoding proteins or PG core - peptide components of the matrix. The main structural components of the matrix are collagen and PGs [26, 27]. Collagen I is the major collagen type of bone tissue. It is present in fetal cartilage and initiates the differentiation of osteoblasts in the endochondral osteogenesis zone [28-30]. Collagen II is the major collagen of the mature cartilage matrix. It constitutes the structural basis of the chondron and forms a chondrometabolic barrier together with PGs [24]. Cartilage PGs perform metabolic, barrier, receptor, and other functions [31, 32]. Aggrecan is the most representative cartilage PG. It contains up to 100 CS chains covalently bound to a protein core [33, 34]. Versican is a component of the extracellular matrix containing long CS chains. It is involved in chondron formation, matrix stabilization, cell proliferation, adhesion and migration in early embryogenesis [35]. The functions of lumican are to organize and "bind" collagen fibers. Another gene in this group is CHST1 (carbohydrate (KS Gal-6) sulfotransferase 1), which transfers sulfate groups [36].
Unchanging levels of expression of these genes in patients with IS indicate that the protein matrix components in this group are synthesized normally and, apparently, cannot cause the development of IS.
A group of genes with low levels of expression in IS consists of genes with different functions. Key genes encoding the transcription factors PAX9, SOX9, and GHR and the link-protein gene HAPLN1 are found in this group. Low expression of HAPLN1 may reduce the contact between the structural components of the matrix, thus adversely affecting mechanical properties of the cartilage [37]. Growth hormone is one of the key hormones that regulate cartilage cell metabolism [38]. Reduced GH receptor expression in growth plate chondrocytes of the vertebral bodies, as compared with the control, notably diminishes binding of growth hormone by these cells and its efficiency. The genes PAX9 and SOX9, encoding transcription factors, are involved in the differentiation of chondrogenic cells in both somites and in growth plates during the postnatal period [39]. A high level of expression of PAX9 is typical for minimally differentiated cells [40], and the expression of SOX9 is necessary to induce the differentiation of chondrocytes and endochondral osteogenesis [41].
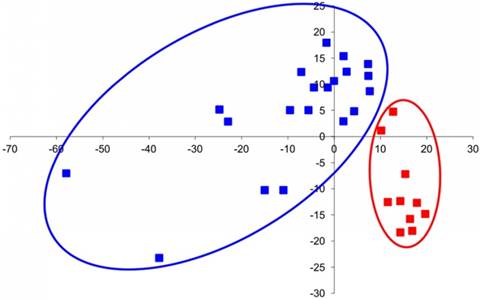
The group of genes that are hyper-expressed in IS includes IHH, PAX1, TGFBR1, EGFR, SLC26A2, and CHST3. The PAX1 gene determines the pattern of sclerotome segmentation and the development of the intervertebral disc during formation of the axial skeleton [41]. High expression of PAX1, which regulates chondrogenic differentiation, was observed in growth plate chondrocytes of patients with IS. It likely indicative of a low level of differentiation of chondrocytes because PAX1 is expressed at the early stages of sclerotome chondrogenic differentiation [42]. The IHH gene is the principle transcription factor that is involved in the recognition of activating signals from both Bmp and IGF and is normally expressed in prehypertrophic chondrocytes of the growth plate [43]. We discovered single hypertrophic cells on the concave sides of growth plates of IS patients, and a high level of expression of this gene suggests a trend towards chondrocyte hypertrophy in patients with IS. The EGFR and TGFBR1 genes facilitate the expression of the corresponding growth factor receptors, and their expression is characteristic of actively proliferating cells [41]. It is also known that both of these factors are essential for the chondrocyte differentiation of the vertebral column growth [42, 43]. Although data on the expression levels of these genes are ambiguous, certain patterns could be assumed. For example, biochemical data show a decrease in PG sulfation and in CS relative to KS on the concave side of the deformity, although the SHST3 gene (responsible for sulfation of CS) is overexpressed and CHST1 expression remains stable. These data may only suggest a different degree of sulfation of these molecules. In turn, unbalanced expression of genes can result in the disruption of differentiation and physiological activity of cells. Because normal expression patterns of the corresponding genes define normal morphogenesis, alteration of cycling and gene interactions lead to the formation of anomalous structures [43]. Indeed, factor analysis showed a fundamental difference between the groups of IS patient samples and control samples. Each of the IS patient samples had a combination of features distinguishing it from the control sample (Figure 4).
Factor analysis of chondroblast gene expression in IS vertebra and normal fetal vertebra. Control samples (isolated from normal fetal vertebra) are marked in red, and samples isolated from the concave and convex sides of damaged IS vertebra are marked in blue.
Conclusion
In a previous repot, we presented new data on the deposition of the neural crest cells into growth plates of vertebral bodies [19]. It is known that during migration, neural crest cells change their expression profiles [44]. We hypothesize that changes in expression of adhesion molecules [45] promoted neural crest cell settlement in the sclerotome mesenchymal environment [46]. As a putative signaling pathway we suggest upregulation of Pax1. Within the mesenchymal sclerotome, Pax1 is subsequently downregulated in cells that undergo chondrogenesis and only maintained in the mesenchymal anlagen of the intervertebral discs and the perichondrium of the vertebral bodies [47, 48]. Thus, even though Pax1 is required to initially trigger chondrogenesis in the early sclerotome [39], Pax1 overexpression prevents chondrocyte maturation in the differentiating sclerotome and inhibits Nkx3.2 expression and accumulation of proteoglycans [49], explaining why after the establishment of the chondrogenic lineage, Pax1 expression is supressed in chondrocytes.
In this study we were able to demonstrate altered expression of genes regulating CS sulfation and corresponding protein synthesis in scoliotic specimens compared with control tissues. Biochemical analysis revealed 1) a decrease in diffused PG in the total pool of PG; (2) reduced level of their sulfation; 3) a reduction in the amount of CS coinciding with an increased amount of KS; and 4) reduced levels of sulfation on the concave side of the scoliotic deformity. It was suggested that growth asymmetry in IS patients is associated with complex functional impairment of the vertebral cartilage cells. Such impairment may be indicative of the existence of cells with different phenotypes, which may not respond to normal signals of differentiation in the growth plate. Elevated expression levels of growth factor receptors suggest a lack of growth factors or intermediary molecules. Further study should be directed toward the analysis of cartilage cell subpopulations and their gene expression.
We strongly believe that identification of alterations in gene expression causing IS is the first step to break a theoretical barrier in finding the cure for IS. Of course, a logical solution for the correction of the altered gene expression would be local gene modulation or protein compensation (DNA transfection, RNA silencing, etc.), which is an extremely difficult task. However, no matter how difficult the suggested mission is, it will become a practical challenge rather than a theoretical barrier if we can identify the therapeutic targets.
Abbreviations
IS: idiopathic scoliosis; PG: proteoglycans; GAGs: glycosaminoglycans; CS: chondroitin sulfate; KS: keratan sulfate.
Acknowledgements
We thank Maria Afrazi and Lucas Trilling (Arrowhead Pharmaceuticals) for technical assistance. We also thank anonymous reviewers for the comments that allowed us to improve the manuscript.
Competing Interests
The authors have declared that no competing interest exists.
References
1. Fadzan M, Bettany-Saltikov J. Etiological Theories of Adolescent Idiopathic Scoliosis: Past and Present. The open orthopaedics journal. 2017;11:1466
2. Ogilvie J. Adolescent idiopathic scoliosis and genetic testing. Current opinion in pediatrics. 2010;22:67-70
3. Martin CT, Pugely AJ, Gao Y, Mendoza-Lattes SA, Ilgenfritz RM, Callaghan JJ. et al. Increasing hospital charges for adolescent idiopathic scoliosis in the United States. Spine (Phila Pa 1976). 2014;39:1676-82
4. Cheng JC, Castelein RM, Chu WC, Danielsson AJ, Dobbs MB, Grivas TB. et al. Adolescent idiopathic scoliosis. Nat Rev Dis Primers. 2015:1
5. Shinozaki K, Ebert O, Kournioti C, Tai YS, Woo SLC. Oncolysis of multifocal hepatocellular carcinoma in the rat liver by hepatic artery infusion of vesicular stomatitis virus. Molecular Therapy. 2004;9:368-76
6. Sharma S, Gao X, Londono D, Devroy SE, Mauldin KN, Frankel JT. et al. Genome-wide association studies of adolescent idiopathic scoliosis suggest candidate susceptibility genes. Human molecular genetics. 2011;20:1456-66
7. Somerville E. Rotational lordosis: the development of the single curve. The Journal of bone and joint surgery British volume. 1952;34:421-7
8. Roaf R. The basic anatomy of scoliosis. The Journal of bone and joint surgery British volume. 1966;48:786-92
9. Stokes I. Mechanical effects on skeletal growth. Journal of Musculoskeletal and Neuronal Interactions. 2002;2:277-80
10. Stokes IA, Burwell RG, Dangerfield PH. Biomechanical spinal growth modulation and progressive adolescent scoliosis-a test of the'vicious cycle'pathogenetic hypothesis: Summary of an electronic focus group debate of the IBSE. Scoliosis. 2006;1:16
11. Terhune EA, Baschal EE, Miller NH. Genetics and Functional Pathology of Idiopathic Scoliosis. In: (ed.) Kusumi K, Dunwoodie SL. The Genetics and Development of Scoliosis. Cham: Springer International Publishing. 2018 p. 159-78
12. Gorman KF, Julien C, Moreau A. The genetic epidemiology of idiopathic scoliosis. European spine journal: official publication of the European Spine Society, the European Spinal Deformity Society, and the European Section of the Cervical Spine Research Society. 2012;21:1905-19
13. Wajchenberg M, Astur N, Kanas M, Martins DE. Adolescent idiopathic scoliosis: current concepts on neurological and muscular etiologies. Scoliosis and spinal disorders. 2016;11:4
14. Wang WJ, Yeung HY, Chu WC, Tang NL, Lee KM, Qiu Y. et al. Top theories for the etiopathogenesis of adolescent idiopathic scoliosis. Journal of pediatric orthopedics. 2011;31:S14-27
15. Burwell RG, Aujla RK, Kirby AS, Dangerfield PH, Moulton A, Cole AA. et al. Body mass index of girls in health influences menarche and skeletal maturation: a leptin-sympathetic nervous system focus on the trunk with hypothalamic asymmetric dysfunction in the pathogenesis of adolescent idiopathic scoliosis? Studies in health technology and informatics. 2008;140:9-21
16. Burwell RG, Aujla RK, Grevitt MP, Dangerfield PH, Moulton A, Randell TL. et al. Pathogenesis of adolescent idiopathic scoliosis in girls - a double neuro-osseous theory involving disharmony between two nervous systems, somatic and autonomic expressed in the spine and trunk: possible dependency on sympathetic nervous system and hormones with implications for medical therapy. Scoliosis. 2009;4:24
17. Burwell RG, Clark EM, Dangerfield PH, Moulton A. Adolescent idiopathic scoliosis (AIS): a multifactorial cascade concept for pathogenesis and embryonic origin. Scoliosis and spinal disorders. 2016:11
18. Burwell RG, Dangerfield PH, Moulton A, Grivas TB. Adolescent idiopathic scoliosis (AIS), environment, exposome and epigenetics: a molecular perspective of postnatal normal spinal growth and the etiopathogenesis of AIS with consideration of a network approach and possible implications for medical therapy. Scoliosis. 2011;6:26
19. Zaydman AM, Strokova EL, Kiseleva EV, Suldina LA, Strunov AA, Shevchenko AI. et al. A New Look at Etiological Factors of Idiopathic Scoliosis: Neural Crest Cells. International journal of medical sciences. 2018;15:436-46
20. Zhang H, Marshall KW, Tang H, Hwang DM, Lee M, Liew CC. Profiling genes expressed in human fetal cartilage using 13,155 expressed sequence tags. Osteoarthritis and Cartilage. 2003;11:309-19
21. Tagariello A, Schlaubitz S, Hankeln T, Mohrmann G, Stelzer C, Schweizer A. et al. Expression profiling of human fetal growth plate cartilage by EST sequencing. Matrix biology: journal of the International Society for Matrix Biology. 2005;24:530-8
22. Pogue R, Sebald E, King L, Kronstadt E, Krakow D, Cohn DH. A transcriptional profile of human fetal cartilage. Matrix Biology. 2004;23:299-307
23. Nerlich AG, Bachmeier BE, Schleicher E, Rohrbach H, Paesold G, Boos N. Immunomorphological analysis of RAGE receptor expression and NF-kappaB activation in tissue samples from normal and degenerated intervertebral discs of various ages. Annals of the New York Academy of Sciences. 2007;1096:239-48
24. Zaidman A, Zaidman M, Strokova E, Korel A, Kalashnikova E, Rusova T. et al. The mode of inheritance of Scheuermann's disease. BioMed research international. 2013;2013:1-9
25. Hojo H, Ohba S, Yano F, Chung UI. Coordination of chondrogenesis and osteogenesis by hypertrophic chondrocytes in endochondral bone development. Journal of bone and mineral metabolism. 2010;28:489-502
26. James CG, Stanton LA, Agoston H, Ulici V, Underhill TM, Beier F. Genome-wide analyses of gene expression during mouse endochondral ossification. PloS one. 2010;5:e8693
27. Eames BF, de la Fuente L, Helms JA. Molecular ontogeny of the skeleton. Birth defects research Part C, Embryo today: reviews. 2003;69:93-101
28. Shepard JB, Gliga DA, Morrow AP, Hoffman S, Capehart AA. Versican knock-down compromises chondrogenesis in the embryonic chick limb. Anat Rec (Hoboken). 2008;291:19-27
29. Roughley PJ. The structure and function of cartilage proteoglycans. European cells & materials. 2006;12:92-101
30. Luo W, Guo C, Zheng J, Chen TL, Wang PY, Vertel BM. et al. Aggrecan from start to finish. Journal of bone and mineral metabolism. 2000;18:51-6
31. Carney S, Muir H. The structure and function of cartilage proteoglycans. Physiological reviews. 1988;68:858-910
32. Roughley P. The structure and function of cartilage proteoglycans. European cells & materials. 2006:12
33. Seyfried NT, McVey GF, Almond A, Mahoney DJ, Dudhia J, Day AJ. Expression and purification of functionally active hyaluronan-binding domains from human cartilage link protein, aggrecan and versican: formation of ternary complexes with defined hyaluronan oligosaccharides. The Journal of biological chemistry. 2005;280:5435-48
34. Rossi A, Superti-Furga A. Mutations in the diastrophic dysplasia sulfate transporter (DTDST) gene (SLC26A2): 22 novel mutations, mutation review, associated skeletal phenotypes, and diagnostic relevance. Human mutation. 2001;17:159-71
35. Nilsson O, Marino R, De Luca F, Phillip M, Baron J. Endocrine regulation of the growth plate. Hormone research. 2005;64:157-65
36. Kronenberg HM. Developmental regulation of the growth plate. Nature. 2003;423:332-6
37. Akiyama H, Kim JE, Nakashima K, Balmes G, Iwai N, Deng JM. et al. Osteo-chondroprogenitor cells are derived from Sox9 expressing precursors. Proc Natl Acad Sci U S A. 2005;102:14665-70
38. Hata K, Takashima R, Amano K, Ono K, Nakanishi M, Yoshida M. et al. Arid5b facilitates chondrogenesis by recruiting the histone demethylase Phf2 to Sox9-regulated genes. Nat Commun. 2013;4:2850
39. Rodrigo I, Hill RE, Balling R, Munsterberg A, Imai K. Pax1 and Pax9 activate Bapx1 to induce chondrogenic differentiation in the sclerotome. Development. 2003;130:473-82
40. Wang Q, Fang WH, Krupinski J, Kumar S, Slevin M, Kumar P. Pax genes in embryogenesis and oncogenesis. J Cell Mol Med. 2008;12:2281-94
41. Miraoui H, Oudina K, Petite H, Tanimoto Y, Moriyama K, Marie PJ. Fibroblast growth factor receptor 2 promotes osteogenic differentiation in mesenchymal cells via ERK1/2 and protein kinase C signaling. The Journal of biological chemistry. 2009;284:4897-904
42. Hall KC, Hill D, Otero M, Plumb DA, Froemel D, Dragomir CL. et al. ADAM17 controls endochondral ossification by regulating terminal differentiation of chondrocytes. Molecular and cellular biology. 2013;33:3077-90
43. Park M, Ohana E, Choi SY, Lee MS, Park JH, Muallem S. Multiple roles of the SO4(2-)/Cl-/OH- exchanger protein Slc26a2 in chondrocyte functions. The Journal of biological chemistry. 2014;289:1993-2001
44. Gammill LS, Bronner-Fraser M. Neural crest specification: migrating into genomics. Nature Reviews Neuroscience. 2003;4:795
45. Akitaya T, Bronner-Fraser M. Expression of cell adhesion molecules during initiation and cessation of neural crest cell migration. Developmental Dynamics. 1992;194:12-20
46. Scaal M. Early development of the vertebral column. Seminars in Cell & Developmental Biology: Elsevier. 2016 p. 83-91
47. Wallin J, Wilting J, Koseki H, Fritsch R, Christ B, Balling R. The role of Pax-1 in axial skeleton development. Development. 1994;120:1109-21
48. Takimoto A, Mohri H, Kokubu C, Hiraki Y, Shukunami C. Pax1 acts as a negative regulator of chondrocyte maturation. Experimental cell research. 2013;319:3128-39
49. Rainbow RS, Kwon H, Zeng L. The role of Nkx3. 2 in chondrogenesis. Frontiers in biology. 2014;9:376-81
Author contact
![]() Corresponding authors: Alla M. Zaydman, AZaydmanru. Vladimir M. Subbotin, vsubbotincom; vsbbtinedu. Office: 1-608-316-3924; Fax: 1-608-441-0741
Corresponding authors: Alla M. Zaydman, AZaydmanru. Vladimir M. Subbotin, vsubbotincom; vsbbtinedu. Office: 1-608-316-3924; Fax: 1-608-441-0741

 Global reach, higher impact
Global reach, higher impact