Impact Factor
ISSN: 1449-1907
Int J Med Sci 2018; 15(13):1443-1448. doi:10.7150/ijms.27341 This issue Cite
Review
Biology of MiR-17-92 Cluster and Its Progress in Lung Cancer
Lab for Noncoding RNA & Cancer, School of Life Sciences Shanghai University, Shanghai 200444
Received 2018-5-18; Accepted 2018-7-29; Published 2018-9-7
Abstract
MicroRNAs, a class of short endogenous RNAs, acting as post-transcriptional regulators of gene expression, mostly silence gene expression via binding imperfectly matched sequences in the 3'UTR of target mRNA. MiR-17-92, a highly conserved gene cluster, has 6 members including miR-17, miR-18a, miR-19a, miR-20a, miR-19b-1 and miR-92a. The miR-17-92 cluster, regarded as oncogene, is overexpressed in human cancers. Lung cancer is the leading cause of death all over the world. The molecular mechanism of lung cancer has been partly known at the levels of genes and proteins in last decade. However, new prognosis biomarkers and more target drugs should be developed in future. Therefore, noncoding RNAs, especially miRNAs, make them as new potentially clinical biomarkers for diagnosis and prognosis. In this review, we focus the current progress of miR-17-92 cluster in lung cancer.
Keywords: microRNAs, miR-17-92 cluster, oncogene, lung cancer, tumorigenesis
Introduction
MicroRNAs (miRNAs), highly conserved small non-coding RNAs, cause degradation of target mRNAs or inhibition of translation by binding with 3'-untranslated regions (3'-UTR), thus regulate the expression of genes at the post-transcriptional level [1]. It has been found that there are a large number of miRNAs, often located in a polycistronic gene called miRNA clusters, which affect cell growth, cell cycle and cell differentiation through co-expression in different tissues. Additionally, they can participate in the occurrence and development of tumors as new oncogenes or tumor suppressor genes, then affect the recurrence and metastasis of tumors. A growing body of research shows that miRNAs participate in the development of lung cancer [2-4].
Lung cancer is the most common cancer in the world, whose incidence and mortality are among the top three. Lung cancer is divided into two main histological groups including non-small cell lung cancer (NSCLC, 85%) and small cell lung cancer (SCLC, 15%) [5]. NSCLC can be further subclassified into adenocarcinomas, squamous cell, large cell and bronchoalveolar carcinomas (BAC). NSCLC has slower cell proliferation and relatively late diffusion and metastasis comparing with SCLC [6]. The 5-year survival rate is only 17.4% after the diagnosis of lung cancer patients in the United States, while the average results in developing countries are even worse [7]. Till now, miR-17-92 cluster, is significantly up-regulated in various solid tumors and affects the development of tumors [8], especially in lung cancer. In this review, we focus the roles of miR-17-92 and advance in lung carcinogenesis.
1. miR-17-92 cluster
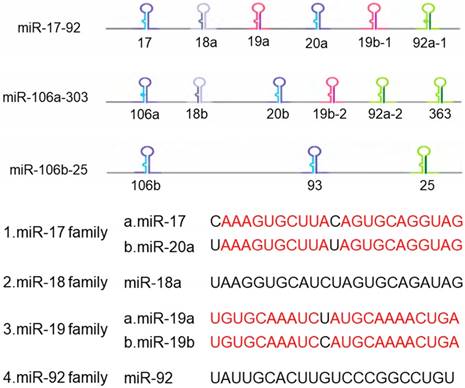
Now more than 300 miRNA clusters are found in the human genome, including miR-183-96-182 cluster, miR-35-41 cluster, miR-17-92 cluster and so on. MiR-17-92 cluster is involved in the development of multiple organs in mammals and closely related to the development and occurrence of tumors, thus it receives widespread attention in the world [9]. The miR-17-92 cluster is a highly conserved gene cluster containing a polycistronic promoter, located on human chromosome 13q31.3 and the third intron of the C13orf25 gene [10], which is the first miRNA oncogene [11]. The precursor transcripts contain a tandem stem-loop hairpin structure, which ultimately produce mature miRNAs: miR-17, miR-18a, miR-19a, miR-19b-1, miR-20a and miR-92a [12]. The six miRNAs are divided into four groups based on their sequences compositions: miR-17 and miR-20, miR-18, miR-19, and miR-92 (Figure 1). Among them, miR-18 shares homology with the sequences of miR-17 and miR-20. The miR-17-92 cluster has two paralogous clusters: miR-106b-25 and miR-106a-363, which can be used to increase the complexity and diversity of miR-17-92 cluster in mammalian cells by gene duplication [13]. These two homologous clusters encode 15 different miRNAs, in which the miR-106b-25 cluster is located in the 13th intron region on human chromosome 7, encoding miR-106b, miR-93 and miR-25. The miR-106a-363 cluster is located on the X chromosome and encodes miR-106a, miR-18b, miR-20b, miR-19b-2, miR-92-2, miR-363. These miRNAs share similar highly conserved gene sequences with the miR-17-92 cluster. At the same time, the miR-17-92 cluster is overexpressed in various solid tumors, so it is regarded as 'Oncogene'.
The structure of miR-17-92 cluster
2. miR-17-92 cluster and its targets in lung cancer
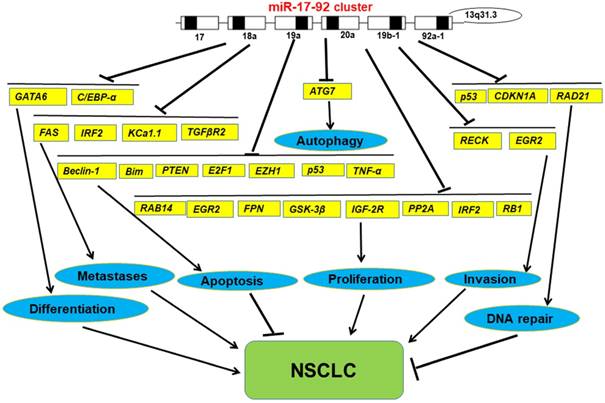
More than 30 targets of miR-17-92 cluster were validated, such as phosphatase and tensin homolog (PTEN), amyloid precursor protein (APP), cyclin D1 (CCND1), transcription factor E2F1, mitogen activation protein kinase 9 (MAPK9), lat gene 2 (LATS2) [2, 9, 14, 15]. We subsequently summarize confirmed targets of miR-17-92 in lung cancer by luciferase reporter assay in Table 1.
3. miR-17-92 cluster and lung tumorigenesis
3.1 miR-17
MiR-17-5p, the most prominent member in miR-17-92 cluster, is an essential regulator of fundamental cellular processes like proliferation, cell cycle, apoptosis, and autophagy, whose deficiency is lethal in the mouse. Large-scale miRNome analysis identifies miR-17-5p as an oncogene in tumors [40]. Moreover, serum miR-17-5p levels are inversely related to the survival of patients with lung cancer, that is, high levels correlated with shorter survival times [41]. In addition, inhibition of miR-17-5p by antisense oligonucleotides (ONs) can induce apoptosis selectively overexpressing miR-17-92 in lung cancer cells, suggesting the possibility of targeting an 'oncomiR addiction' to expression of these miRNAs in a subset of lung cancers. MiR-17-5p might also serve as a therapeutic target in lung cancer [42].
Validated targets of miR-17-92 cluster in lung cancer
| Member of miR-17-92 | Pathways affected | Targets of miR-17-92 | References |
|---|---|---|---|
| miR-17 | Akt/FoxO3a/Bim signaling | Bim | [16] |
| PI3K pathway | PTEN | [8] | |
| p38-HSP27 signaling | E2F1,E2F3 | [17] | |
| Initiation of autophagy | BECN1 | [18] | |
| TGFß/BMP signaling pathway | TGFβR2 | [19] | |
| Autophagy | ATG7 | [20] | |
| PI3K/AKT signaling pathway | EZH1 | [21] | |
| Cell cycle | CDKN1A | [22] | |
| Cell senescence | RAD21 | [22] | |
| Notch signaling pathway | RB1 | [23] | |
| BCL-2 signaling pathway | BCL-2 | [24] | |
| p53 signaling pathway | p53 | [25] | |
| Ca2+ signaling pathway | KCa1.1 | [26, 27] | |
| PI3K-Akt signaling pathway | PTEN | [28] | |
| miR-18a | Jak/Stat signaling pathway | IRF2 | [3] |
| EGFR signaling pathway | PP2A | [29] | |
| miR-19a/b | EGFR signaling pathway | Bim | [29] |
| TNF-α signaling pathway | TNF-α | [30] | |
| IGF1 signaling pathway | IGF-2R | [31] | |
| C/EBP-α,GATA6 | [32] | ||
| GSK-3β signaling pathway | GSK-3β | [33] | |
| Fas signaling pathway | FAS | [34] | |
| miR-20a | Cell membrane transport protein | FPN | [35] |
| EGFR signaling pathway | EGR2 | [36] | |
| BMP signaling pathway | TGFβR2 | [37] | |
| p53 signaling pathway | p53 | [27] | |
| miR-92a | Cell membrane transport protein | RAB14 | [38] |
| p53 signaling pathway | p53 | [27] | |
| RECK signaling pathway | RECK | [39] |
On the contrary, miR-17-5p is downregulated in NSCLC [43]. Moreover, downregulation of miR-17-5p upregulated BECN1, which resulted in apoptosis resistance of cancer cells via the treatment of paclitaxel [44]. In order to justify miR-17-5p can act as a tumor suppressor, a study proved that downregulation of miR-17-5p resulted in high expression of cyclin-dependent kinase inhibitor 1A (CDKN1A) and RAD21 cohesin complex component (RAD21) in cisplatin resistance of NSCLC [45]. In this case, miR-17-5p acts as a tumor suppressor.
Above all, miR-17-5p not only can act as an oncomiR, but also might also act as a tumor suppressor in lung cancer. The final effect of miR-17-5p seems to be highly context-dependent.
3.2 miR-18a
MiR-18a, as an oncogene, promotes the tumorigenesis and tumor angiogenesis [46, 47]. Besides, miR-18a promotes pathological angiogenesis by increasing VEGFA expression in hepatocellular carcinoma (HCC) [48]. In addition, some evidences have demonstrated that miR-18a promotes cell proliferation via stimulating CCND1 in oesophageal squamous cell carcinoma cells [49].
Previous study has already shown that upregulated miR-18a has a shorter survival in NSCLC patients [28]. In recent clinical studies, overexpression miR-18a is negatively associated with the clinical response of NSCLC via activating the serine/threonine-protein kinase 4 (STK4) pathway. Besides, miR-18a is also relevant to clinical tumor node metastasis (TNM) stage, tumor differentiation and regional lymph node metastasis (P < 0.005) [50]. Last but not least, our experimental data reveal miR-18a-5p can promote NSCLC by directly targeting IRF2 [3]. Further experiments indicated that IRF2 can promote cell apoptosis, inhibit cell proliferation and migration. Moreover, miR-18a-5p can active autophagy in NSCLC. Collectively, these results indicate that miR-18a-5p cannot only promote NSCLC via suppressing IRF2, but dysregulated miR-18a expression mediate oncogenes to affect tumorigenesis. It may be a convincing biomarker for the early detection of lung cancer [3].
3.3 miR-19a/b
The miR-19 family (miR-19a and miR-19b) are key oncogenic components of the miR-17-92 cluster. MiR-19 family are associated with poor prognosis of NSCLC patients and other carcinoma [51-54]. Subsequently, a finding suggests that miR-19 triggered epithelial-mesenchymal transition (EMT) in lung cancer [55]. What's more, the overexpression of serum miR-19 family are considered as independent prognostic factors for NSCLC patients [53]. MiR-19b enhances the proliferation of NSCLC cells by the epidermal growth factor receptor (EGFR) signaling pathway [29]. In addition, the study reveals novel mechanisms by grape seed procyanidin (GSE) in lung cancer [31].
3.4 miR-20a
MiR-20a can enhance cell proliferation and promote cell migration in various cancers, such as lung cancer, gastric cancer and hepatocellular carcinoma [56-58]. In addition, the upregulation of miR-20a promotes radio-resistance for NSCLC patients, which can be conductive to poor prognosis, as showed in other cancers [59, 60].
Likewise, previous reports have observed miR-17-5p and miR-20a target the transcription factor E2F1, a target of transcriptional regulator Myc-like (C-MYC) [61]. It is reported that all 3'UTR of E2Fs mRNA have miR-20a binding sites. Two functional E2F transcription factor binding sites are contained in the core promoter region of miR-17-92 cluster. E2Fs can directly activate transcription of miR-17-92 via binding the promoters and their effects similarly. However, miR-20a can reduce E2F1 expression, such a balance shift may be also contributed by MYC, which can trans activate E2Fs. E2Fs, MYC and miR-17-92 cluster together provide a complex regulatory for NSCLC cell proliferation and apoptosis [62-64].
3.5 miR-92a
It has been shown that miR-92a is also involved in the biology of tumors through a complex mechanism. Moderate overexpression of miR-92a causes a reduction of F-box and WD repeat domain containing 7 (FBXW7), resulting in cervical cancer proliferation and invasion [65]. In addition, according to the recent research, miR-92a plays an essential role in the development of colorectal cancer and may serve as a diagnostic and prognostic biomarker [66, 67]. Moreover, MiR-92a promotes the proliferation of gastric cancer stem cells [68]. Recent study demonstrates high expression of miR-92a in HCC [69]. MiR-92a partly promotes the migration and invasion of esophageal squamous cell carcinoma cells (ESCC) by inhibiting cadherin 1 (CDH1) [70].
Although a growing number of evidences mention the key role of miR-92a in tumor biology, its role is rarely reported in the development of lung cancer. Moreover, little is known about the relationship between the miR-92a and survival in patients with NSCLC. The results of these show that the mechanism underlying this relationship may be exported in the future. Our lab results showed miR-92a promoted the development of NSCLC via targeting sprouty RTK signaling antagonist 4 (SPRY4, under submitting). If miRNA-based therapeutics indeed become a reality, the miR-17-92 cluster and related miRNAs will undoubtedly be among the first to be targeted. Importantly, miR-17-92 cluster will play an irreplaceable role in lung cancer.
4. Conclusions and perspective
As modulation of miRNAs represents a novel approach for enhancing the therapeutic efficacies of cancer therapy, research efforts have been put forth to identify agents that induce or inhibit the expression of miRNA. The carcinogenic role of miR-17-92 cluster in different tumors has been confirmed [8].
At the first time, overexpression miR-17 promotes the proliferation and metastasis of hepatoma cells [71]. What's more, miR-17 promotes the progression of colorectal cancer via activating the Wnt/β-catenin pathway [72]. At the second time, miR-18a promotes the development of gastric cancer cells via inhibiting p53 and promoting the expression of signal transducer and activator of transcription 3 (STAT3) [73]. At the third time, high expression of miR-19 associates with TNM stage of lung cancer, which boosts to metastasis of lymph node [33, 53]. Besides, miR-19 promotes proliferation of esophageal cancer cells and prevents apoptosis of cancer cells via down-regulating the expression of TNF-α [30]; What's more, miR-20 promotes prostate cancer cells invasion and metastasis by targeting non-receptor tyrosine kinase ABL proto-oncogene 2, non-receptor tyrosine kinase (ABL2) [74]. Last but not least, miR-92, inhibiting PTEN, activating PTEN/PI3K/AKT signaling pathway, promotes invasion and metastasis in rectal cancer cells [75]. In summary, each member of the miR-17-92 cluster has a direct or indirect relationship with cancer, it can promote the occurrence and development of tumors by regulating the expression of genes.
With the advancement of precise medical theories and advances in technology, the research of miR-17-92 cluster has continued to deepen in tumor cells, particularly, the roles of miR-17-92 cluster have been continuously explored in lung cancer. Furthermore, the new study has observed that Docosahexaenoic acid (DHA), as a novel therapeutic, modulates expression of miR-17-92 and inhibits cell migration and viability in lung cancer [76]. Intriguingly, accumulating studies show that the roles of miR-17-92 cluster are not clear in lung cancer and need to explore continually in the future.
In general, we have found that miR-17-92 cluster, as tumor promoter, has a measurable impact on the development of lung cancer upon most occasions. However, in any particular case, miR-17-92 cluster also can impress the development of lung cancer (Figure 2). Undoubtedly, this discovery opens up a new way for us to study the relationship between miRNA and tumorigenesis, it certainly highlights the import roles in cancer biology and there may be a more complex relationships at the same time. Moreover, it also supports the continued study promotes the further development of cancers in the clinical outcome. Further studies on it may provide new ideas for the study of tumors, find new biomarkers for the clinical treatment of cancers, and provide a better basis for the precise treatment of cancers.
The functions of miR-17-92 cluster in NSCLC
Abbreviations
miRNAs: microRNAs; NSCLC: non-small cell lung cancer; SCLC: small cell lung cancer; BAC: bronchoalveolar carcinomas; PTEN: tensin homolog; APP: amyloid precursor protein; CCND1: cyclin D1; E2F1: E2F transcription factor 1; MAPK9: mitogen activation Protein kinase 9; LATS2: lat gene 2; Bim: BCL2 like 11; BECN1: Beclin-1; TGFβR2: transforming growth factor beta receptor 2; ATG7: autophagy related 7; EZH1: enhancer of zeste 1 polycomb repressive complex 2 subunit; CDKN1A: cyclin dependent kinase inhibitor 1A; RAD21: RAD21 cohesin complex component; RB1: RB transcriptional corepressor 1; BCL-2: B cell leukemia/lymphoma 2; KCa1.1: potassium calcium-activated channel subfamily M alpha 1; IRF2: interferon regulatory factor 2; PP2A: serine/threonine protein phosphatase 2A; TNF-α: tumor necrosis factor alpha; IGF-2R: insulin like growth factor 2 receptor; C/EBP-α: CCAAT enhancer binding protein alpha; GATA6: GATA binding protein 6; GSK-3β: glycogen synthase kinase 3 beta; FAS: Fas cell surface death receptor; FPN: FerroPortiN; EGR2: early growth response 2; RAB14: RAB14, member RAS oncogene family; RECK: reversion inducing cysteine rich protein with kazal motifs; ONs: oligonucleotides; HCC: hepatocellular carcinoma; STK4: serine/threonine-protein kinase 4; TNM: tumor node metastasis; EMT: epithelial-mesenchymal transition; EGFR: epidermal growth factor receptor; GSE: grape seed procyanidin; C-MYC: transcriptional regulator Myc-like; CDH1: cadherin 1; SPRY4: sprouty RTK signaling antagonist 4; STAT3: signal transducer and activator of transcription 3; ABL2: ABL proto-oncogene 2, non-receptor tyrosine kinase; DHA: Docosahexaenoic acid.
Acknowledgements
This study was funded by the National Natural Science Foundation (Grant no. 81572122).
Competing Interests
The authors have declared that no competing interest exists.
References
1. Ma ZL, Hou PP, Li YL, Wang DT, Yuan TW, Wei JL. et al. MicroRNA-34a inhibits the proliferation and promotes the apoptosis of non-small cell lung cancer H1299 cell line by targeting TGFbetaR2. Tumour biology: the journal of the International Society for Oncodevelopmental Biology and Medicine. 2015;36:2481-90
2. Fang L, Du WW, Yang WN, Rutnam ZJ, Peng C, Li HR. et al. MiR-93 enhances angiogenesis and metastasis by targeting LATS2. Cell Cycle. 2012;11:4352-65
3. Liang C, Zhang X, Wang HM, Liu XM, Zhang XJ, Zheng B. et al. MicroRNA-18a-5p functions as an oncogene by directly targeting IRF2 in lung cancer. Cell Death Dis. 2017:8
4. Ma ZL, Zhang BJ, Wang DT, Li X, Wei JL, Zhao BT. et al. Tanshinones suppress AURKA through up-regulation of miR-32 expression in non-small cell lung cancer. Oncotarget. 2015;6:20111-20
5. Shao Y, Shen YQ, Li YL, Liang C, Zhang BJ, Lu SD. et al. Direct repression of the oncogene CDK4 by the tumor suppressor miR-486-5p in non-small cell lung cancer. Oncotarget. 2016;7:34011-21
6. Wang P, Liu X, Shao Y, Wang H, Liang C, Han B. et al. MicroRNA-107-5p suppresses non-small cell lung cancer by directly targeting oncogene epidermal growth factor receptor. Oncotarget. 2017;8:57012-23
7. Li YL, Liu XM, Zhang CY, Zhou JB, Shao Y, Liang C. et al. MicroRNA-34a/EGFR axis plays pivotal roles in lung tumorigenesis. Oncogenesis. 2017;6:e372
8. Osada H, Takahashi T. let-7 and miR-17-92: small-sized major players in lung cancer development. Cancer science. 2011;102:9-17
9. Grillari J, Hackl M, Grillari-Voglauer R. miR-17-92 cluster: ups and downs in cancer and aging. Biogerontology. 2010;11:501-6
10. Ota A, Tagawa H, Karnan S, Tsuzuki S, Karpas A, Kira S. et al. Identification and characterization of a novel gene, C13orf25, as a target for 13q31-q32 amplification in malignant lymphoma. Cancer Res. 2004;64:3087-95
11. He L, Thomson JM, Hemann MT, Hernando-Monge E, Mu D, Goodson S. et al. A microRNA polycistron as a potential human oncogene. Nature. 2005;435:828-33
12. Zhou M, Cai J, Tang Y, Zhao Q. MiR-17-92 cluster is a novel regulatory gene of cardiac ischemic/reperfusion injury. Med Hypotheses. 2013;81:108-10
13. Doebele C, Bonauer A, Fischer A, Scholz A, Reiss Y, Urbich C. et al. Members of the microRNA-17-92 cluster exhibit a cell-intrinsic antiangiogenic function in endothelial cells. Blood. 2010;115:4944-50
14. Serva A, Knapp B, Tsai YT, Claas C, Lisauskas T, Matula P. et al. miR-17-5p Regulates Endocytic Trafficking through Targeting TBC1D2/Armus. Plos One. 2012:7
15. Dews M, Fox JL, Hultine S, Sundaram P, Wang W, Liu YY. et al. The myc-miR-17~92 axis blunts TGF{beta} signaling and production of multiple TGF{beta}-dependent antiangiogenic factors. Cancer Res. 2010;70:8233-46
16. Dai BB, Meng JR, Peyton M, Girard L, Bornmann WG, Ji L. et al. STAT3 Mediates Resistance to MEK Inhibitor through MicroRNA miR-17. Cancer Res. 2011;71:3658-68
17. Trompeter HI, Abbad H, Iwaniuk KM, Hafner M, Renwick N, Tuschl T. et al. MicroRNAs MiR-17, MiR-20a, and MiR-106b Act in Concert to Modulate E2F Activity on Cell Cycle Arrest during Neuronal Lineage Differentiation of USSC. Plos One. 2011:6
18. Chatterjee A, Chattopadhyay D, Chakrabarti G. miR-16 targets Bcl-2 in paclitaxel-resistant lung cancer cells and overexpression of miR-16 along with miR-17 causes unprecedented sensitivity by simultaneously modulating autophagy and apoptosis. Cell Signal. 2015;27:189-203
19. Jiang ZY, Yin J, Fu WF, Mo YJ, Pan YG, Dai L. et al. miRNA 17 Family Regulates Cisplatin-Resistant and Metastasis by Targeting TGFbetaR2 in NSCLC. Plos One. 2014:9
20. Sun W, Zu Y, Fu X, Deng Y. Knockdown of lncRNA-XIST enhances the chemosensitivity of NSCLC cells via suppression of autophagy. Oncology reports. 2017;38:3347-54
21. Zhang W, Lin J, Wang P, Sun J. miR-17-5p down-regulation contributes to erlotinib resistance in non-small cell lung cancer cells. Journal of drug targeting. 2017;25:125-31
22. Zhao J, Fu WF, Liao HY, Dai L, Jiang ZY, Pan YG. et al. The regulatory and predictive functions of miR-17 and miR-92 families on cisplatin resistance of non-small cell lung cancer. Bmc Cancer. 2015:15
23. Li KN, Li ZH, Zhao N, Xu YQ, Liu YJ, Zhou YS. et al. Functional analysis of microRNA and transcription factor synergistic regulatory network based on identifying regulatory motifs in non-small cell lung cancer. Bmc Syst Biol. 2013:7
24. Wang Q, Li YC, Wang J, Kong J, Qi Y, Quigg RJ. et al. miR-17-92 cluster accelerates adipocyte differentiation by negatively regulating tumor-suppressor Rb2/p130. Proceedings of the National Academy of Sciences of the United States of America. 2008;105:2889-94
25. Borkowski R, Du L, Zhao Z, McMillan E, Kosti A, Yang CR. et al. Genetic mutation of p53 and suppression of the miR-17 approximately 92 cluster are synthetic lethal in non-small cell lung cancer due to upregulation of vitamin D Signaling. Cancer Res. 2015;75:666-75
26. Cheng YY, Wright CM, Kirschner MB, Williams M, Sarun KH, Sytnyk V. et al. KCa1.1, a calcium-activated potassium channel subunit alpha 1, is targeted by miR-17-5p and modulates cell migration in malignant pleural mesothelioma. Mol Cancer. 2016:15
27. Borkowski R, Du LQ, Zhao ZZ, McMillan E, Kosti A, Yang CR. et al. Genetic Mutation of p53 and Suppression of the miR-17 similar to 92 Cluster Are Synthetic Lethal in Non-Small Cell Lung Cancer due to Upregulation of Vitamin D Signaling. Cancer Res. 2015;75:666-75
28. Xu X, Zhu S, Tao Z, Ye S. High circulating miR-18a, miR-20a, and miR-92a expression correlates with poor prognosis in patients with non-small cell lung cancer. Cancer medicine. 2018;7:21-31
29. Baumgartner U, Berger F, Hashemi Gheinani A, Burgener SS, Monastyrskaya K, Vassella E. miR-19b enhances proliferation and apoptosis resistance via the EGFR signaling pathway by targeting PP2A and BIM in non-small cell lung cancer. Mol Cancer. 2018;17:44
30. Liu M, Wang Z, Yang S, Zhang W, He S, Hu C. et al. TNF-alpha is a novel target of miR-19a. International journal of oncology. 2011;38:1013-22
31. Mao JT, Xue B, Smoake J, Lu QY, Park H, Henning SM. et al. MicroRNA-19a/b mediates grape seed procyanidin extract-induced anti-neoplastic effects against lung cancer. The Journal of nutritional biochemistry. 2016;34:118-25
32. Oeztuerk-Winder F, Guinot A, Ochalek A, Ventura JJ. Regulation of human lung alveolar multipotent cells by a novel p38alpha MAPK/miR-17-92 axis. The EMBO journal. 2012;31:3431-41
33. Zhu J, Wang S, Chen Y, Li X, Jiang Y, Yang X. et al. miR-19 targeting of GSK3beta mediates sulforaphane suppression of lung cancer stem cells. The Journal of nutritional biochemistry. 2017;44:80-91
34. Huang G, Nishimoto K, Yang Y, Kleinerman ES. Participation of the Fas/FasL signaling pathway and the lung microenvironment in the development of osteosarcoma lung metastases. Advances in experimental medicine and biology. 2014;804:203-17
35. Babu KR, Muckenthaler MU. miR-20a regulates expression of the iron exporter ferroportin in lung cancer. Journal of molecular medicine. 2016;94:347-59
36. Wei L, Ran F. MicroRNA-20a promotes proliferation and invasion by directly targeting early growth response 2 in non-small cell lung carcinoma. Oncology letters. 2018;15:271-7
37. Yang S, Cho YJ, Jin L, Yuan G, Datta A, Buckhaults P. et al. An epigenetic auto-feedback loop regulates TGF-beta type II receptor expression and function in NSCLC. Oncotarget. 2015;6:33237-52
38. Kanzaki H, Ito S, Hanafusa H, Jitsumori Y, Tamaru S, Shimizu K. et al. Identification of direct targets for the miR-17-92 cluster by proteomic analysis. Proteomics. 2011;11:3531-9
39. Lin HY, Chiang CH, Hung WC. STAT3 upregulates miR-92a to inhibit RECK expression and to promote invasiveness of lung cancer cells. Br J Cancer. 2013;109:731-8
40. Yu J, Ohuchida K, Mizumoto K, Fujita H, Nakata K, Tanaka M. MicroRNAmiR-17-5pis overexpressed in pancreatic cancer, associated with a poor prognosis, and involved in cancer cell proliferation and invasion. Cancer Biology & Therapy. 2014;10:748-57
41. Chen Q, Si Q, Xiao S, Xie Q, Lin J, Wang C. et al. Prognostic significance of serum miR-17-5p in lung cancer. Medical oncology. 2013;30:353
42. Ebi H, Sato T, Sugito N, Hosono Y, Yatabe Y, Matsuyama Y. et al. Counterbalance between RB inactivation and miR-17-92 overexpression in reactive oxygen species and DNA damage induction in lung cancers. Oncogene. 2009;28:3371-9
43. Lin S, Sun JG, Wu JB, Long HX, Zhu CH, Xiang T. et al. Aberrant microRNAs expression in CD133(+)/CD326(+) human lung adenocarcinoma initiating cells from A549. Molecules and cells. 2012;33:277-83
44. Chatterjee A, Chattopadhyay D, Chakrabarti G. miR-17-5p downregulation contributes to paclitaxel resistance of lung cancer cells through altering beclin1 expression. Plos One. 2014;9:e95716
45. Zhao J, Fu W, Liao H, Dai L, Jiang Z, Pan Y. et al. The regulatory and predictive functions of miR-17 and miR-92 families on cisplatin resistance of non-small cell lung cancer. Bmc Cancer. 2015;15:731
46. Hsu TI, Hsu CH, Lee KH, Lin JT, Chen CS, Chang KC. et al. MicroRNA-18a is elevated in prostate cancer and promotes tumorigenesis through suppressing STK4 in vitro and in vivo. Oncogenesis. 2014:3
47. Brunet Vega A, Pericay C, Moya I, Ferrer A, Dotor E, Pisa A. et al. microRNA expression profile in stage III colorectal cancer: circulating miR-18a and miR-29a as promising biomarkers. Oncology reports. 2013;30:320-6
48. Morita K, Shirabe K, Taketomi A, Soejima Y, Yoshizumi T, Uchiyama H. et al. Relevance of microRNA-18a and microRNA-199a-5p to hepatocellular carcinoma recurrence after living donor liver transplantation. Liver transplantation: official publication of the American Association for the Study of Liver Diseases and the International Liver Transplantation Society. 2016;22:665-76
49. Zhang W, Lei C, Fan J, Wang J. miR-18a promotes cell proliferation of esophageal squamous cell carcinoma cells by increasing cylin D1 via regulating PTEN-PI3K-AKT-mTOR signaling axis. Biochemical and biophysical research communications. 2016;477:144-9
50. Shen Z, Wu X, Wang Z, Li B, Zhu X. Effect of miR-18a overexpression on the radiosensitivity of non-small cell lung cancer. International journal of clinical and experimental pathology. 2015;8:643-8
51. Navarro A, Marrades RM, Vinolas N, Quera A, Agusti C, Huerta A. et al. MicroRNAs expressed during lung cancer development are expressed in human pseudoglandular lung embryogenesis. Oncology. 2009;76:162-9
52. Wu C, Cao Y, He Z, He J, Hu C, Duan H. et al. Serum levels of miR-19b and miR-146a as prognostic biomarkers for non-small cell lung cancer. The Tohoku journal of experimental medicine. 2014;232:85-95
53. Lin Q, Chen T, Lin Q, Lin G, Lin J, Chen G. et al. Serum miR-19a expression correlates with worse prognosis of patients with non-small cell lung cancer. Journal of surgical oncology. 2013;107:767-71
54. Wu Q, Yang Z, An Y, Hu H, Yin J, Zhang P. et al. MiR-19a/b modulate the metastasis of gastric cancer cells by targeting the tumour suppressor MXD1. Cell Death Dis. 2014:5
55. Li J, Yang S, Yan W, Yang J, Qin YJ, Lin XL. et al. MicroRNA-19 triggers epithelial-mesenchymal transition of lung cancer cells accompanied by growth inhibition. Lab Invest. 2015;95:1056-70
56. Zhang Y, Zheng L, Ding Y, Li Q, Wang R, Liu T. et al. MiR-20a Induces Cell Radioresistance by Activating the PTEN/PI3K/Akt Signaling Pathway in Hepatocellular Carcinoma. International journal of radiation oncology, biology, physics. 2015;92:1132-40
57. Li X, Zhang Z, Yu M, Li L, Du G, Xiao W. et al. Involvement of miR-20a in promoting gastric cancer progression by targeting early growth response 2 (EGR2). International journal of molecular sciences. 2013;14:16226-39
58. Hayashita Y, Osada H, Tatematsu Y, Yamada H, Yanagisawa K, Tomida S. et al. A polycistronic microRNA cluster, miR-17-92, is overexpressed in human lung cancers and enhances cell proliferation. Cancer Res. 2005;65:9628-32
59. Huang G, Chen X, Cai Y, Wang X, Xing C. miR-20a-directed regulation of BID is associated with the TRAIL sensitivity in colorectal cancer. Oncology reports. 2017;37:571-8
60. Huang D, Bian G, Pan Y, Han X, Sun Y, Wang Y. et al. MiR-20a-5p promotes radio-resistance by targeting Rab27B in nasopharyngeal cancer cells. Cancer cell international. 2017;17:32
61. O'Donnell KA, Wentzel EA, Zeller KI, Dang CV, Mendell JT. c-Myc-regulated microRNAs modulate E2F1 expression. Nature. 2005;435:839-43
62. Novotny GW, Sonne SB, Nielsen JE, Jonstrup SP, Hansen MA, Skakkebaek NE. et al. Translational repression of E2F1 mRNA in carcinoma in situ and normal testis correlates with expression of the miR-17-92 cluster. Cell death and differentiation. 2007;14:879-82
63. Sylvestre Y, De Guire V, Querido E, Mukhopadhyay UK, Bourdeau V, Major F. et al. An E2F/miR-20a autoregulatory feedback loop. Journal of Biological Chemistry. 2007;282:2135-43
64. Woods K, Thomson JM, Hammond SM. Direct regulation of an oncogenic micro-RNA cluster by E2F transcription factors. Journal of Biological Chemistry. 2007;282:2130-4
65. Gong JH, Cui Z, Li L, Ma Q, Wang QF, Gao YH. et al. MicroRNA-25 promotes gastric cancer proliferation, invasion, and migration by directly targeting F-box and WD-40 Domain Protein 7, FBXW7. Tumor Biol. 2015;36:7831-40
66. Liu GH, Zhou ZG, Chen R, Wang MJ, Zhou B, Li Y. et al. Serum miR-21 and miR-92a as biomarkers in the diagnosis and prognosis of colorectal cancer. Tumor Biol. 2013;34:2175-81
67. Zhou T, Zhang G, Liu Z, Xia S, Tian H. Overexpression of miR-92a correlates with tumor metastasis and poor prognosis in patients with colorectal cancer. International journal of colorectal disease. 2013;28:19-24
68. Wu Q, Yang Z, Wang F, Hu S, Yang L, Shi Y. et al. MiR-19b/20a/92a regulates the self-renewal and proliferation of gastric cancer stem cells. J Cell Sci. 2013;126:4220-9
69. Shigoka M, Tsuchida A, Matsudo T, Nagakawa Y, Saito H, Suzuki Y. et al. Deregulation of miR-92a expression is implicated in hepatocellular carcinoma development. Pathology international. 2010;60:351-7
70. Chen ZL, Zhao XH, Wang JW, Li BZ, Wang Z, Sun J. et al. microRNA-92a promotes lymph node metastasis of human esophageal squamous cell carcinoma via E-cadherin. The Journal of biological chemistry. 2011;286:10725-34
71. Jung YJ, Kim JW, Park SJ, Min BY, Jang ES, Kim NY. et al. c-Myc-mediated overexpression of miR-17-92 suppresses replication of hepatitis B virus in human hepatoma cells. Journal of medical virology. 2013;85:969-78
72. Sun R, Liang Y, Yuan F, Nie X, Sun H, Wang Y. et al. Functional polymorphisms in the promoter region of miR-17-92 cluster are associated with a decreased risk of colorectal cancer. Oncotarget. 2017;8:82531-40
73. Mohammadian F, Pilehvar-Soltanahmadi Y, Mofarrah M, Dastani-Habashi M, Zarghami N. Down regulation of miR-18a, miR-21 and miR-221 genes in gastric cancer cell line by chrysin-loaded PLGA-PEG nanoparticles. Artif Cell Nanomed B. 2016;44:1972-8
74. Qiang XF, Zhang ZW, Liu Q, Sun N, Pan LL, Shen J. et al. miR-20a promotes prostate cancer invasion and migration through targeting ABL2. Journal of cellular biochemistry. 2014;115:1269-76
75. Ke TW, Wei PL, Yeh KT, Chen WT, Cheng YW. MiR-92a Promotes Cell Metastasis of Colorectal Cancer Through PTEN-Mediated PI3K/AKT Pathway. Annals of surgical oncology. 2015;22:2649-55
76. Ali M, Heyob K, Jacob NK, Rogers LK. Alterative Expression and Localization of Profilin 1/VASPpS157 and Cofilin 1/VASPpS239 Regulates Metastatic Growth and Is Modified by DHA Supplementation. Mol Cancer Ther. 2016;15:2220-31
Author contact
![]() Corresponding author: zlmaedu.cn; Tel: 86-21-66137015; Fax: 86-21-66132177
Corresponding author: zlmaedu.cn; Tel: 86-21-66137015; Fax: 86-21-66132177

 Global reach, higher impact
Global reach, higher impact