Impact Factor
ISSN: 1449-1907
Int J Med Sci 2018; 15(12):1312-1319. doi:10.7150/ijms.27505 This issue Cite
Research Paper
Significant association of long non-coding RNAs HOTAIR genetic polymorphisms with cancer recurrence and patient survival in patients with uterine cervical cancer
1. Department of Obstetrics and Gynaecology, Hsinchu Mackay Memorial Hospital, Hsinchu City, Taiwan
2. Department of Medicine, Mackay Medical College, New Taipei City, Taiwan
3. Mackay Junior College of Medicine, Nursing and Management College, Taipei, Taiwan
4. Institute of Medicine, Chung Shan Medical University, Taichung, Taiwan
5. Department of Medical Research, Chung Shan Medical University Hospital, Taichung, Taiwan
6. School of Medicine, Chung Shan Medical University, Taichung, Taiwan
7. Department of Obstetrics and Gynecology, Changhua Christian Hospital, Changhua, Taiwan
8. Department of Obstetrics and Gynecology, Chung Shan Medical University Hospital, Taichung, Taiwan
Received 2018-5-27; Accepted 2018-6-30; Published 2018-8-6
Abstract
Up to date, no study explores the relationship of single nucleotide polymorphisms (SNPs) of long non-coding RNAs HOTAIR (lncRNAs HOTAIR) with cancer recurrence and patient survival in uterine cervical cancer for Taiwanese women. We therefore designed this study to investigate the clinical roles of lncRNAs HOTAIR SNPs in cervical cancer. One hundred and sixteen patients with cervical invasive cancer and 96 patients with preinvasive lesions as well as 318 control women were consecutively recruited. LncRNAs HOTAIR SNPs rs920778, rs12427129, rs4759314 and rs1899663 were analyzed and their genotypic frequencies were examined by real-time polymerase chain reaction. The results indicated that there were no genotypic differences between patients with cervical neoplasia and normal controls as well as among patients with invasive and invasive cancer, and normal controls. However, genotype GG in lncRNAs HOTAIR SNP rs920778 was demonstrated to be a predictor for poorer cancer recurrence probability [p=0.001, hazard ratio (HR): 7.25, 95% CI: 2.19-23.96]. Furthermore, cervical cancer patients with genotype GG in lncRNAs HOTAIR rs920778 had worse overall survival (p=0.002, HR: 7.22, 95% CI: 2.09-24.92). No significant associations exhibited between lncRNAs HOTAIR SNP rs920778 and clinicopathological parameters. In conclusion, this studied lncRNAs HOTAIR SNPs are not associated with cervical carcinongensis. However, lncRNAs HOTAIR SNP rs920778 may be regarded as an independent predictor of cancer recurrence probability and overall survival in cervical cancer patients.
Keywords: long non-coding RNAs HOTAIR, single nucleotide polymorphism, uterine cervical cancer, recurrence-free survival, overall survival
Introduction
Uterine cervical cancer was the fifth common type of cancer and most common type of gynecological cancer if carcinoma in situ was included according to Taiwan 2013 cancer registry annual report. Its mortality was the tenth in Taiwanese cancers. Cytologic diagnoses of cervical lesions include low-grade squamous intraepithelial lesions (LSILs) which are compatible with cervical intraepithelial neoplasia 1 (CIN1, low-grade CIN) or mild dysplasia histologically, as well as high-grade squamous intraepithelial lesions (HSILs), which are histolocially diagnosed as CIN 2 and CIN3 (high-grade CIN) or moderate dysplasia and severe dysplasia, respectively[1]. Cervical carcinogenesis is currently considered as a continuum of neoplastic transition from CIN to invasive squamous cell carcinoma and approximate 20-30% of HSILs may develop to invasive cancer [2, 3].
Long non-coding RNAs (lncRNAs) are found to be a group of transcribed RNA molecules and they are longer than 200 nucleotides and unable to translate protein obviously [4]. They are involved in a variety of biological function, including cell proliferation and differentiation as well as tumorigenesis [5-7]. The HOX transcript antisense intergenic RNA (HOTAIR), firstly discovered by Rinn et al., is located on the antisense strand of the HOXC gene cluster on chromosome 12q13.13 and is identified as a 2158 nucleotides lncRNAs with 6 exons [8, 9]. These lncRNAs, with regulatory functions of transcription, was implicated in the proximal-distal axis during development. It was firstly reported by Gupta et al. that increased HOTAIR expression was implicated in breast cancer as well as tumor invasiveness and metastasis [10]. Moreover, its dysregulation was demonstrated to be related to colon and lung cancers [11, 12] .
Single nucleotide polymorphism (SNP) occurs if a single nucleotide in the shared sequence of a gene changes more than 1% in the individuals of a species or paired chromosomes in an individual. Genetic polymorphism are considered to have an influence on the promoter activity and gene expression, and is involved in disease development [13]. Genetic polymorphisms of the lncRNAs HOTAIR were known to be significantly related to the susceptibility of some human cancers such as breast cancer and gastric cancer [14, 15]. Because no study associates lncRNAs HOTAIR SNPs with patient prognosis in cervical cancer up to date, we conduct this study to investigate the association of lncRNAs HOTAIR SNPs with cervical carcinogenesis and explore its relationships with clinicopathological variables of cervical cancer, cancer recurrence and patient survival.
Materials and Methods
Subjects
We consecutively enrolled 116 patients with invasive cancer and 96 patients with high-grade CIN of uterine cervix as well as 318 normal controls at the Department of Obstetrics and Gynecology in Chung Shan Medical University in Taichung, Taiwan from February 1994 to October 2014. Cervical cancer patients received routine treatment protocols in this hospital. Patients with high-grade CIN were recruited as cervical preinvasive lesions and received abdominal total hysterectomy, vaginal total hysterectomy, large loop excision of transformation zone or simple trachelectomy. The diagnoses of all patients with cervical invasive cancer or preinvasive lesions were further verified based on the pathologic report of cervical punch biopsy under colposcopy before the treatment started. Normal controls received Papanicolaou smears at outpatient department for general examination in Chung Shan Medical University Hospital and the normal cytologic diagnosis was further confirmed under colposcopy. All studied individuals were Taiwanese women who resided in central Taiwan. This study was approved by the institutional review board of Chung Shan Medical University Hospital (CSMUH No: CS14014). Our staffs received informed consents from all subjects.
Selection of lncRNAs HOTAIR single nucleotide polymorphisms and DNA extraction from all subjects
In this study, four lncRNAs HOTAIR SNPs were selected according to International HapMap Project data and the study of Guo et al. [16]. Genetic polymorphisms rs920778, rs12427129, rs4759314 and rs1899663 were included. Blood specimens were drawn by the staffs from all participatants with a standard venipuncture technique and deposed into Vacutainer tubes containing ethylenediaminetetraacetic acid. The materials were stored at 4℃ immediately after obtainment. DNA was extracted from peripheral vein blood leukocytes according to manufacturer's protocol as described in detail previously [17]. Moreover, we dissolved DNA in pH 7.8 TE buffer and then quantified it by a measurement of OD260. The ratio OD260 /OD280 was also calculated and DNA sample within the range of 1.8-2.0 was considered as pure to avoid the cross reactivity with the homologous RNA present in the sample. The final preparation was stored at -20 °C and was used to act as templates for the PCR.
Single nucleotide polymorphisms (SNPs) by real time-PCR and genotyping
Genotypes of lncRNAs HOTAIR SNPs rs920778, rs12427129, rs4759314 and rs1899663 were determined by ABI StepOne Real-Time PCR System (Applied Biosystems, Foster City, CA, USA), and analyzed with SDS vers. 3.0 software, as described previously [18].
Statistical analysis
ANOVA was used to compare the age difference among patients with invasive cancer and patients with preinvasive lesions of uterine cervix as well as control women and then Bonferroni test was used for post hoc analysis. Hardy-Weinberg equilibrium was applied to check the genotypic frequencies of lncRNAs HOTAIR SNPs rs920778, rs12427129, rs4759314 and rs1899663 in the normal controls (degree of freedom = 2). Chi-square or Fisher's exact tests were used to associate genotypic distributions of lncRNAs HOTAIR SNPs with the incidence of cervical neoplasias (including preinvasive lesions and invasive cancer). The adjusted odds ratios (AORs) with their 95% confidence intervals (CIs) were applied to evaluate the relationships among genotypic frequencies of lncRNAs HOTAIR SNPs and the incidence of cervical neoplasias (including preinvasive lesions and invasive cancer) using the logistic and multinomial logistic regression models after age control. Chi-square or Fisher's exact tests were applied to assess cancer recurrence event and patient death event by lncRNAs HOTAIR SNPs. The risks of cancer recurrence event and patient death event for significant lncRNAs HOTAIR SNP were adjusted by various clinicopathological parameters, such as clinical stage (I or ≥ II), histopathologic types inluding squamous cell carcinoma or adenocarcinoma, cell grading (well, or moderate and poor differentiation), invasion depth of cervical stroma (≤10 mm or >10 mm of stromal invasion depth), tumor diameter (≤4 or >4 cm), parametrium and vaginal invasion and pelvic lymph node metastasis, using logistic regression forward stepwise model. In addition, chi-square or Fisher's exact tests were also used to evaluate the relationship of significant lncRNAs HOTAIR SNP with clinicopathological variables. Kaplan-Meier model was used to relate significant lncRNAs HOTAIR SNPs to cancer recurrence free-survival and overall survival of patients with cervical cancer relative to recurrence and survival time or until closing date of the study, December 4, 2017. A Cox proportional hazard model was used to evaluate the effects of significant lncRNAs HOTAIR SNP on the recurrence probability or overall survival after adjusting for various clinicopathological variables in multivariate analysis relative to recurrence or survival time. The SPSS, version 12.0 and WinPepi Software, version 10.0 were used for statistical analysis. P <0.05 was considered as statistically significant difference.
Results
There was significant difference for age distribution between patients with cervical neoplasia and normal control women (50.2 ± 13.8 vs. 43.9 ± 10.1, p<0.001). The age distribution was significantly different between patients with cervical invasive cancer and those with preinvasive lesion (55.4 ± 12.5 vs. 44.0 ± 12.6, p<0.001) as well as between patients with cervical cancer and normal control women (55.4 ± 12.5 vs. 43.9 ± 10.1, p<0.001) but not significantly different between patients with preinvasive lesions and control women (44.0 ± 12.6 vs. 43.9 ± 10.1, p=1.000).
The genotypic distribution of lncRNAs HOTAIR SNP rs920778 satisfied Hardy- Weinberg equilibrium in the normal controls [χ2 value: 1.66 < 5.99, p>0.05, degree of freedom (d.f.)=2]. Genotypic frequencies of other lncRNAs HOTAIR SNPs rs12427129, rs4759314 and rs1899663 all conformed to the equilibrium (χ2 value: 0.21, p>0.05, d.f.=2; χ2 value: 1.27, p>0.05, d.f.=2; χ2 value: 1.13, p>0.05, d.f.=2; respectively).
Relationship of lncRNAs genetic polymorphisms distributions with uterine cervical carcinogenesis
The genotypic frequencies of lncRNAs SNPs in the Taiwanese women with cervical neoplasias and normal control women are listed in Table 1. There were no significantly different distributions of lncRNAs SNPs rs920778, rs12427129, rs4759314 and rs1899663 between patients with cervical neoplasias and normal control women. No significant differences were found for these SNPs between patients with cervical neoplasias and normal controls even after controlling for age. Although the cervical neoplasia group was subdivided into subgroups of invasive cancer and preinvasive lesions, we could not demonstrate genotypic differences of these 4 lncRNAs SNPs among patients with cervical invasive cancer and patient with preinvasive lesions as well as normal controls (Table 2). After controlling for age, these conditions still presented.
The prediction of lncRNAs HOTAIR genetic polymorphisms for cancer prognosis events
Genotypic frequency of AA, AG and GG of lncRNAs HOTAIR SNP rs920778 was significantly related to recurrence event (p=0.021) and death event (p=0.03) of patients with uterine cervical cancer. Further analysis revealed that cervical cancer patients with genotype GG had more risk to have recurrence event (OR: 6.37, 95% CI: 0.96-46.20; p=0.028) and death event (OR: 8.67, 95% CI: 1.28-63.57; p=0.012), as compared to those with AA/AG (Table 3). However, there were no associations of lncRNAs HOTAIR SNPs rs12427129, rs4759314 and rs1899663 with cancer recurrence event and patient death event.
Genotypic distribution of single nucleotide polymorphisms of long non-coding RNAs HOTAIR in patients with uterine cervical neoplasia and normal control women.
| Variables | Normal controls (n = 318) | Cervical neoplasiaa (n = 212) | p value | OR (95% CI) | AOR (95% CI)b |
|---|---|---|---|---|---|
| rs920778 | |||||
| Co-dominant | |||||
| AAc | 165 | 107 | 0.950 | 1.00 | 1.00 |
| AG | 134 | 92 | 1.06 (0.74-1.52) | 1.02 (0.70-1.47) | |
| GG | 19 | 13 | 1.06 (0.50-2.23) | 0.89 (0.41-1.92) | |
| Dominant | |||||
| AAc | 165 | 107 | 0.749 | 1.00 | 1.00 |
| AG/GG | 153 | 105 | 1.06 (0.75-1.50) | 1.00 (0.70-1.43) | |
| Recessive | |||||
| AA/AGc | 299 | 199 | 0.941 | 1.00 | 1.00 |
| GG | 19 | 13 | 1.03 (0.50-2.13) | 0.88 (0.42-1.87) | |
| rs12427129 | |||||
| Co-dominant | |||||
| CCc | 275 | 181 | 0.629 | 1.00 | |
| CT | 42 | 29 | 1.05 (0.63-1.75) | 1.12 (0.66-1.89) | |
| TT | 1 | 2 | 3.04 (0.27-33.76) | 3.11 (0.27-36.03) | |
| Dominant | |||||
| CCc | 275 | 181 | 0.720 | 1.00 | 1.00 |
| CT/TT | 43 | 13 | 1.10 (0.67-1.80) | 1.17 (0.70-1.95) | |
| Recessive | |||||
| CC/CTc | 317 | 210 | 0.567 | 1.00 | 1.00 |
| TT | 1 | 2 | 3.02 (0.27-33.51) | 3.06 (0.26-35.44) | |
| rs4759314 | |||||
| Co-dominant | |||||
| AAc | 266 | 171 | 0.124 | 1.00 | 1.00 |
| AG | 48 | 41 | 1.33 (0.84-2.10) | 1.34 (0.83-2.15) | |
| GG | 4 | 0 | u.a. | u.a. | |
| Dominant | |||||
| AAc | 266 | 171 | 0.376 | 1.00 | 1.00 |
| AG/GG | 52 | 41 | 1.23 (0.78-1.93) | 1.20 (0.75-1.92) | |
| Recessive | |||||
| AA/AGc | 314 | 212 | 0.154 | 1.00 | 1.00 |
| GG | 4 | 0 | u.a. | u.a. | |
| rs1899663 | |||||
| Co-dominant | |||||
| GGc | 209 | 140 | 0.992 | 1.00 | 1.00 |
| GT | 101 | 67 | 0.99 (0.68-1.44) | 0.97 (0.66-1.44) | |
| TT | 8 | 5 | 0.93 (0.30-2.91) | 0.79 (0.25-2.53) | |
| Dominant | |||||
| GGc | 209 | 140 | 0.940 | 1.00 | 1.00 |
| GT/TT | 109 | 72 | 0.99 (0.68-1.42) | 0.96 (0.66-1.40) | |
| Recessive | |||||
| GG/GTc | 310 | 207 | 0.909 | 1.00 | 1.00 |
| TT | 8 | 5 | 0.94 (0.30-2.90) | 0.80 (0.25-2.53) |
Statistical analysis: logistic regression model, chi-square or Fisher's exact tests.
aCervical neoplasia included preinvasive lesions and invasive cancer of uterine cervix.
bThe adjusted odds ratios with their 95% confident intervals were calculated by logistic regression after controlling for age.
cRegarded as a reference for comparison to evaluate the odds ratio of other genotypes.
AOR, adjusted odds ratio; 95% CI, 95% confidence interval; u.a., unavailable.
Genotypic distribution of single nucleotide polymorphisms of long non-coding RNAs HOTAIR in patients with invasive cancer or patients with preinvasive lesions of uterine cervix and normal control women
| Variables | Normal controls (n = 318 ) | Preinvasive lesions (n = 96 ) | Invasive cancer (n = 116 ) | p value | AOR (95% CI)a | AOR (95% CI)b |
|---|---|---|---|---|---|---|
| rs920778 | ||||||
| Co-dominant | ||||||
| AAc | 165 | 50 | 57 | 0.858 | 1.00 | 1.00 |
| AG | 134 | 42 | 50 | 1.03 (0.65-1.65) | 1.02 (0.63-1.65) | |
| GG | 19 | 4 | 9 | 0.69 (0.23-2.14) | 1.07 (0.43-2.68) | |
| Dominant | ||||||
| AAc | 165 | 50 | 57 | 0.867 | 1.00 | 1.00 |
| AG/GG | 153 | 46 | 59 | 0.99 (0.63-1.57) | 1.03 (0.65-1.64) | |
| Recessive | ||||||
| AA/AGc | 299 | 92 | 107 | 0.549 | 1.00 | 1.00 |
| GG | 19 | 4 | 9 | 0.68 (0.23-2.06) | 1.06 (0.43-2.58) | |
| rs12427129 | ||||||
| Co-dominant | ||||||
| CCc | 275 | 85 | 96 | 0.635 | 1.00 | 1.00 |
| CT | 42 | 10 | 19 | 0.77 (0.37-1.60) | 1.57 (0.83-3.00) | |
| TT | 1 | 1 | 1 | 3.24 (0.20-52.29) | 3.22 (0.17-60.28) | |
| Dominant | ||||||
| CCc | 275 | 85 | 96 | 0.451 | 1.00 | 1.00 |
| CT/TT | 43 | 11 | 20 | 0.83 (0.41-1.68) | 1.61 (0.86-3.04) | |
| Recessive | ||||||
| CC/CTc | 317 | 95 | 115 | 0.352 | 1.00 | 1.00 |
| TT | 1 | 1 | 1 | 3.34 (0.21-53.85) | 3.01 (0.16-56.03) | |
| rs4759314 | ||||||
| Co-dominant | ||||||
| AAc | 266 | 76 | 95 | 0.467 | 1.00 | 1.00 |
| AG | 48 | 20 | 21 | 1.46 (0.82-2.61) | 1.21 (0.66-2.24) | |
| GG | 4 | 0 | 0 | u.a. | u.a. | |
| Dominant | ||||||
| AAc | 266 | 76 | 95 | 0.590 | 1.00 | 1.00 |
| AG/GG | 52 | 20 | 21 | 1.35 (0.76-2.39) | 1.06 (0.58-1.94) | |
| Recessive | ||||||
| AA/AGc | 314 | 96 | 116 | 0.481 | 1.00 | 1.00 |
| GG | 4 | 0 | 0 | u.a. | u.a. | |
| rs1899663 | ||||||
| Co-dominant | ||||||
| GGc | 209 | 66 | 74 | 0.848 | 1.00 | 1.00 |
| GT | 101 | 29 | 38 | 0.91 (0.55-1.45) | 1.06 (0.65-1.75) | |
| TT | 8 | 1 | 4 | 0.40 (0.48-3.22) | 1.18 (0.32-4.37) | |
| Dominant | ||||||
| GGc | 209 | 66 | 74 | 0.748 | 1.00 | 1.00 |
| GT/TT | 109 | 30 | 42 | 0.87 (0.53-1.42) | 1.08 (0.66-1.74) | |
| Recessive | ||||||
| GG/GTc | 310 | 95 | 112 | 0.620 | 1.00 | 1.00 |
| TT | 8 | 1 | 4 | 0.41 (0.05-3.30) | 1.15 (0.31-4.23) |
Statistical analysis: multinomial logistic regression or chi-square or Fisher's exact tests.
aThe adjusted odds ratio with its 95% CI was calculated by multinomial logistic regression model after controlling for age between patients with cervical preinvasive lesions and control women.
bThe adjusted odds ratio with its 95% CI was calculated by multinomial logistic regression models after controlling for age between patients with cervical invasive cancer and control women.
cRegarded as a reference for comparison to evaluate the odds ratios of other genotypes.
AOR, adjusted odds ratio; 95% CI, 95% confidence interval; u.a., unavailable.
Impacts of long non-coding RNAs HOTAIR (lncRNAs HOTAIR) genetic polymorphism rs920778 on cancer recurrence event and death event of the patients with uterine cervical cancer
| Variablesa | Recurrence event | p value | OR & 95% CIc | |
|---|---|---|---|---|
| no recurrence | recurrence | |||
| lncRNAs HOTAIR rs920778 | 0.028 | |||
| AA/AGb | 86 | 18 | 1.00 | |
| GG | 3 | 4 | 6.37 (0.96-46.20) | |
| Death event | p value | OR & 95% CIb | ||
| survival | death | |||
| lncRNAs HOTAIR rs920778 | 0.012 | |||
| AA/AGb | 91 | 14 | 1.00 | |
| GG | 3 | 4 | 8.67 (1.28-63.57) | |
Statistical analysis: chi-square or Fisher's exact tests
aSome data could not be obtained from the patients with cervical cancer due to incomplete medical charts or records.
bAs a comparison reference
cOR and 95% CI, odds ratio and 95% confidence interval for lncRNAs HOTAIR genetic polymorphism rs920778, compared to its respective control.
Multivariate analysis of long non-coding RNAs HOTAIR (lncRNAs HOTAIR) genetic polymorphism rs920778 on cancer recurrence event and death event of the patients with uterine cervical cancer
| Variablesa | Recurrence event | p value | OR & 95%CIc | |
|---|---|---|---|---|
| no recurrence | recurrence | |||
| lncRNAs HOTAIR rs920778 | 0.049 | |||
| AA/AGb | 86 | 18 | 1.00 | |
| GG | 3 | 4 | 1.13 (1.01-37.04) | |
| Stromal invasion depth | 0.006 | |||
| ≤10 mmb | 59 | 5 | 1.00 | |
| >10 mm | 30 | 17 | 5.13 (1.60-16.39) | |
| Pelvic lymph node metastasis | 0.021 | |||
| negativeb | 73 | 10 | 1.00 | |
| positive | 16 | 12 | 3.62 (1.22-10.75) | |
| Death event | p value | OR & 95%CIb | ||
| survival | death | |||
| lncRNAs HOTAIR rs920778 | 0.014 | |||
| AA/AGb | 91 | 14 | 1.00 | |
| GG | 3 | 4 | 9.09 (1.57-52.63) | |
| Pelvic lymph node metastasis | 0.002 | |||
| negativeb | 76 | 7 | 1.00 | |
| positive | 18 | 11 | 5.95 (1.88-18.87) | |
Statistical analysis: multivariate logistic regression model
aSome data could not be obtained from the patients with cervical cancer due to incomplete medical charts or records.
bAs a comparison reference
cOR and 95% CI, odds ratio and 95% confidence interval for lncRNAs HOTAIR genetic polymorphism rs920778 and clinicopathological variables, compared to its respective control.
Multivariate analysis for the association of significant lncRNAs HOTAIR SNP and clinicopathological parameters with cancer recurrence and patient survival
Cervical cancer patients with genotype GG in lncRNAs HOTAIR rs920778 displayed more of the risk of cancer recurrence event (p=0.049; OR: 1.13, 95% CI: 1.01-37.04; Table 4). Additionally, deep stromal and positive pelvic lymph node metastasis also increased the risk of recurrence event (p=0.006; OR: 4.61, 95% CI: 1.60-16.39 and p=0.021; OR: 3.62, 95% CI: 1.22-10.75, respectively). Considering patient death event, cervical cancer patients with genotype GG in lncRNAs HOTAIR SNP rs920778 displayed more risk to have poorer survival event (logistic regression model; p=0.014; OR: 9.09, 95% CI: 1.57-52.63; Table 4). Moreover, positive pelvic lymph node metastasis (p=0.002; OR: 5.95, 95% CI: 1.88-18.87) was also a predictive factor for patient death event.
The association of significant lncRNAs HOTAIR SNP with clinicopathological variables
No significant associations existed between lncRNAs HOTAIR SNP rs920778 and clinicopathological parameters. However, cervical patients with genotype GG tended to display more risk to have parametrium invasion than those with AA/AG (p=0.064; OR: 4.50, 95% CI: 0.69-33.30; Table 5).
Association of genotypic distribution of long non-coding RNAs HOTAIR genetic polymorphism rs920778 with clinicopathological variables of the patients with invasive cancer of uterine cervix
| Variablesa | rs920778 recessive | p value | OR (95% CI) | |
|---|---|---|---|---|
| GG | AA/AGb | |||
| Clinical stage | 0.257 | |||
| stage Ib | 3 | 68 | 1.00 | |
| ≥ stage II | 4 | 37 | 2.45 (0.39-17.48) | |
| Pathologic type | 1.000 | |||
| squamous cell carcinomab | 6 | 89 | 1.00 | |
| adenocarcinoma | 1 | 16 | 1.88 (0.02-8.49) | |
| Cell grading | 1.000 | |||
| well (grade 1)b | 1 | 25 | 1.00 | |
| moderate & poor (grades 2/3) | 6 | 80 | 0.93 (0.21-89.65) | |
| Invasion depth of cervical stroma | 0.459 | |||
| ≤10 mmb | 3 | 61 | 1.00 | |
| >10 mm | 4 | 44 | 1.85 (0.29-13.18) | |
| Tumor diameterb | 0.235 | |||
| ≤ 4cm | 3 | 70 | 1.00 | |
| >4cm | 4 | 35 | 2.67 (0.42-19.03) | |
| Parametrium | 0.064 | |||
| no invasionb | 3 | 81 | 1.00 | |
| invasion | 4 | 24 | 4.50 (0.69-33.30) | |
| Vagina | 0.188 | |||
| no invasionb | 4 | 82 | 1.00 | |
| invasion | 3 | 22 | 2.80 (0.38-17.69) | |
| Pelvic lymph node | 0.372 | |||
| no metastasisb | 4 | 79 | 1.00 | |
| metastasis | 3 | 26 | 2.28 (0.31-14.33) | |
Statistical analyses: chi-square or Fisher's exact tests,
aSome clinicopathological data could not be obtained from the patients with cervical cancer due to incomplete medical charts or records.
bAs a reference. OR, odds ratio; 95% CI, 95% confidence interval.
Influence of lncRNAs HOTAIR genetic polymorphism and clinicopathological characteristics on cancer recurrence probability and overall survival of the patients with uterine cervical cancer
When time interval was included for analysis, cervical cancer patients with genotype GG in lncRNAs HOTAIR SNP rs920778 were found to have worse recurrence-free survival, as compared to those with AA/AG based on Kaplan-Meier curve, using univariate analysis [p=0.005, hazard ratio (HR)= 4.34, 95% CI: 1.43-13.11; Figure 1A]. Moreover, patients with GG had worse overall survival than those with AA/AG (5-year survival rate: 42.9 vs. 87.9%, p=0.003, HR= 4.98, 95% CI: 1.57-15.87; Figure 1B).
Kaplan-Meier curves for recurrence-free survival rate according to long non-coding RNAs HOTAIR genetic polymorphism rs920778 (A, genotype GG vs. AA/AG; p=0.005), as well as for overall survival rate according to rs920778 (B, genotype GG vs. AA/AG; p=0.003). Log-rank test was applied for statistical significance.
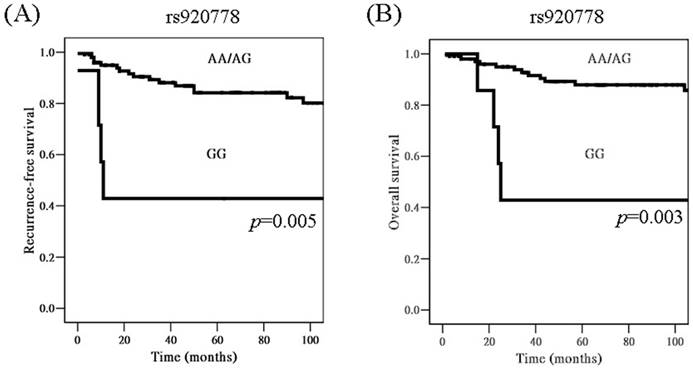
Impacts of long non-coding RNAs HOTAIR (lncRNAs HOTAIR) genetic polymorphism rs920778 and clinicopathological characteristics on cancer recurrence probability and overall survival of the patients with uterine cervical cancer
| Variablesa | Recurrence probability | Overall survival | ||
|---|---|---|---|---|
| p value | OR & 95%CIc | p value | OR & 95%CIc | |
| lncRNAs HOTAIR rs920778 | 0.001 | 0.002 | ||
| AA/AGb | 1.00 | 1.00 | ||
| GG | 7.25 (2.19-23.96) | 7.22 (2.09-24.92) | ||
| Stromal invasion depth | <0.001 | 0.026 | ||
| ≤10 mmb | 1.00 | 1.00 | ||
| >10 mm | 8.91 (2.80-28.33) | 4.09 (1.18-14.13) | ||
| Pelvic lymph node metastasis | >0.05 | 0.027 | ||
| negativeb | 1.00 | |||
| positive | 3.28 (1.15-9.39) | |||
Statistical analysis: multivariate Cox regression model
aSome data could not be obtained from the patients with cervical cancer due to incomplete medical charts or records.
bAs a comparison reference
cOR and 95% CI, odds ratio and 95% confidence interval for lncRNAs HOTAIR genetic polymorphism rs920778 and clinicopathological variables, compared to its respective control.
Genotype GG in lncRNAs HOTAIR SNP rs920778 and deep stromal invasion were demonstrated to be predictors for poorer recurrence probability in multivariate analysis [p=0.001, HR: 7.25, 95% CI: 2.19-23.96 and p<0.001, HR: 8.91, 95% CI: 2.80-28.33, respectively; Table 6]. Furthermore, cervical cancer patients with genotype GG in lncRNAs HOTAIR rs920778 had worse overall survival (p=0.002, HR: 7.22, 95% CI: 2.09-24.92). In addition, deep stromal invasion and positive lymph node metastasis exhibited significant associations with worse overall survival (p=0.026, HR: 4.09, 95% CI: 1.18-14.13 and p=0.027, HR: 3.28, 95% CI: 1.15-9.39, respectively; Table 6).
Discussion
In this study we could not demonstrate different genotypic distributions of 4 lncRNAs HOTAIR SNPs between patients with cervical neoplasia and normal control women in Taiwanese women. Even after subdivision into invasive and preinvasive subgroups from the cervical neoplasia patients and controlling for age, there were no genotypic differences among patients with invasive and patients with preinvasive cancer and normal controls. However, a meta-analysis indicated a significant association between lncRNAs HOTAIR SNP rs920778 and cancer risk [19]. Chu et al. reported that lncRNAs HOTAIR SNP rs920778 increases the cancer risk in recessive model [20]. Furthermore, it has been revealed that individuals with TT (AA) in HOTAIR rs920778 polymorphism were associated with an increased risk of gastric cancer and esophageal squamous cell carcinoma in China [14, 21]. In contrast, genotype CC (GG) in HOTAIR rs920778 polymorphism was demonstrated to increase the risk of breast cancer significantly in a Turkish population [15]. Moreover, a similarly elevated risk for SNP rs4759314 was showed in the gastric cancer subjects [19]. By contrast, no significant relationship was found between SNP rs1899663 and cancer susceptibility [19]. It has been reported a significant correlation of HOTAIR SNP rs920778 with susceptibility to cervical cancer in Chinese people [16]. Women carrying allele T (A) have increased susceptibility to cervical cancer than those carrying allele C (G).
Although we could not find the involvement of lncRNAs HOTAIR SNPs in cervical carcinogens, a different genotypic distribution of SNP rs920778 was found for the prediction of cervical cancer recurrence and patient death events. It was attributable to the recessive effect of homozygote GG that significantly different genotypic frequency of AA, AG and GG of lncRNAs HOTAIR SNP rs920778 resulted in different recurrence and survival events. No other lncRNAs HOTAIR SNPs could be found to be correlated with patient prognosis in this study.
Our study revealed that genotype GG of lncRNAs HOTAIR SNP rs920778 was associated with poorer cancer recurrence and worse patient survival events for uterine cervical cancer in Taiwanese women using multivariate analyses. Because there were no associations of lncRNAs HOTAIR SNP rs920778 with clinicopathological parameters, lncRNAs HOTAIR SNP rs920778 could be regarded as an independent predictor of patient prognosis. Guo et al. reported an association between the SNP rs920778, being in the intronic enhancer of the HOTAIR, and cervical cancer [16]. It has been described by Hosseini et al. about the dysregulated lncRNA expression that is classified into intronic based on the nearest protein-coding transcripts [22]. Guo et al. further delineated that women carrying allele T (A) have increased expression levels of HOTAIR than those carrying allele C (G). However, they also found that cervical cancer patients with homozygote TT were related to tumor-node-metastasis (TNM) stage significantly.
To our knowledge, this study is the first to investigate the relationship of lncRNAs HOTAIR SNP with cancer recurrence and patient survival in cervical cancer in Taiwanese women. When time interval was considered, we found that patients with genotype GG of lncRNAs HOTAIR SNP rs920778 had worse recurrence-free survival and worse overall survival in univariate analysis based on unviariate Kaplain-Meier model. Moreover, patients with genotype GG of SNP rs920778 had poorer recurrence probability and worse overall survival based on multiviariate Cox proportional hazard model. Our findings are reasonable based on previous report that SNP rs920778 was in the intronic enhancer of the lncRNAs HOTAIR and could affect the expression of HOTAIR [16]. Lee et al. reported that elevated expression of HOTAIR is associated with reduced disease-free survival and overall survival in cervical cancer patients [23]. Their results indicated that elevated expression of HOTAIR was significantly related to tumor size and lymph node metastasis. Huang et al. showed that higher HOTAIR expression was not only significantly associated with FIGO stage and lymph node metastasis, but also significantly associated with decreased overall survival and disease-free survival [24]. A meta-analysis showed that higher HOTAIR expression is associated with large tumor size and positive lymph node metastasis in uterine cervical cancer [25]. HOTAIR overexpression was a risk factor for worse patient overall survival. Vascular endothelial growth factor and matrix metalloproteinase-9, which have an important role in cancer progression by increasing invasion, were up-regulated by HOTAIR in cervical cancer [26]. Therefore, HOTAIR was reported to be associated with recurrence of cervical cancer. In addition, Gupta et al. reported that increased lncRNA HOTAIR expression is noted in primary breast tumor and is a powerful predictor of breast cancer patient death as well [10].
In conclusion, our study reveals that lncRNAs HOTAIR SNP rs920778, rs12427129, rs4759314 and rs1899663 are not associated with cervical carcinongensis. However, lncRNAs HOTAIR SNP rs920778 is related to recurrence-free survival and overall survival in cervical cancer patients in univariate analysis. Genotype GG in lncRNAs HOTAIR SNP rs920778 is associated with poorer recurrence probability and worse overall survival in multivariate analysis. We infer that the influence of rs920778 on patient prognosis is mediated by the impact of rs920778 on the increased expression of HOTAIR. Because lncRNAs HOTAIR SNP rs920778 is not associated with clinicopathological variables of cervical cancer, it may be regarded as an independent predictor of patient prognosis in cervical cancer patients.
Acknowledgements
This study was supported by research grants from Ministry of Science and Technology (MOST 105-2314-B-040-016-MY2) and Chung Shan Medical University Hospital (CSH-2017-D-002). This study was also supported by Chung Shan Medical University and Hsinchu MacKay Memorial Hospital (CSMU-HCMMH-106-01).
Competing Interests
The authors have declared that no competing interest exists.
References
1. Khan MJ, Castle PE, Lorincz AT, Wacholder S, Sherman M, Scott DR, Rush BB, Glass AG, Schiffman M. The elevated 10-year risk of cervical precancer and cancer in women with human papillomavirus (HPV) type 16 or 18 and the possible utility of type-specific HPV testing in clinical practice. J Natl Cancer Inst. 2005;97:1072-1079
2. Bharti AC, Shukla S, Mahata S, Hedau S, Das BC. Anti-human papillomavirus therapeutics: facts & future. Indian J Med Res. 2009;130:296-310
3. Baak JP, Kruse AJ, Robboy SJ, Janssen EA, van Diermen B, Skaland I. Dynamic behavioural interpretation of cervical intraepithelial neoplasia with molecular biomarkers. J Clin Pathol. 2006;59:1017-1028
4. Derrien T, Johnson R, Bussotti G, Tanzer A, Djebali S, Tilgner H, Guernec G, Martin D, Merkel A, Knowles DG, Lagarde J, Veeravalli L, Ruan X, Ruan Y, Lassmann T, Carninci P, Brown JB, Lipovich L, Gonzalez JM, Thomas M, Davis CA, Shiekhattar R, Gingeras TR, Hubbard TJ, Notredame C, Harrow J, Guigo R. The GENCODE v7 catalog of human long noncoding RNAs: analysis of their gene structure, evolution, and expression. Genome Res. 2012;22:1775-1789
5. Mercer TR, Dinger ME, Mattick JS. Long non-coding RNAs: insights into functions. Nat Rev Genet. 2009;10:155-159
6. Gibb EA, Brown CJ, Lam WL. The functional role of long non-coding RNA in human carcinomas. Mol Cancer. 2011;10:38
7. Su SC, Reiter RJ, Hsiao HY, Chung WH, Yang SF. Functional Interaction between Melatonin Signaling and Noncoding RNAs. Trends Endocrinol Metab. 2018;29:435-445
8. Rinn JL, Kertesz M, Wang JK, Squazzo SL, Xu X, Brugmann SA, Goodnough LH, Helms JA, Farnham PJ, Segal E, Chang HY. Functional demarcation of active and silent chromatin domains in human HOX loci by noncoding RNAs. Cell. 2007;129:1311-1323
9. Hung T, Chang HY. Long noncoding RNA in genome regulation: prospects and mechanisms. RNA Biol. 2010;7:582-585
10. Gupta RA, Shah N, Wang KC, Kim J, Horlings HM, Wong DJ, Tsai MC, Hung T, Argani P, Rinn JL, Wang Y, Brzoska P, Kong B, Li R, West RB, van de Vijver MJ, Sukumar S, Chang HY. Long non-coding RNA HOTAIR reprograms chromatin state to promote cancer metastasis. Nature. 2010;464:1071-1076
11. Wu ZH, Wang XL, Tang HM, Jiang T, Chen J, Lu S, Qiu GQ, Peng ZH, Yan DW. Long non-coding RNA HOTAIR is a powerful predictor of metastasis and poor prognosis and is associated with epithelial-mesenchymal transition in colon cancer. Oncol Rep. 2014;32:395-402
12. Zhao W, An Y, Liang Y, Xie XW. Role of HOTAIR long noncoding RNA in metastatic progression of lung cancer. Eur Rev Med Pharmacol Sci. 2014;18:1930-1936
13. Shastry BS. SNPs: impact on gene function and phenotype. Methods Mol Biol. 2009;578:3-22
14. Pan W, Liu L, Wei J, Ge Y, Zhang J, Chen H, Zhou L, Yuan Q, Zhou C, Yang M. A functional lncRNA HOTAIR genetic variant contributes to gastric cancer susceptibility. Mol Carcinog. 2016;55:90-96
15. Bayram S, Sumbul AT, Batmaci CY, Genc A. Effect of HOTAIR rs920778 polymorphism on breast cancer susceptibility and clinicopathologic features in a Turkish population. Tumour Biol. 2015;36:3863-3870
16. Guo L, Lu X, Zheng L, Liu X, Hu M. Association of Long Non-Coding RNA HOTAIR Polymorphisms with Cervical Cancer Risk in a Chinese Population. PLoS One. 2016;11:e0160039
17. Wu HH, Liu YF, Yang SF, Lin WL, Chen SC, Han CP, Wang HL, Lin LY, Wang PH. Association of single-nucleotide polymorphisms of high-mobility group box 1 with susceptibility and clinicopathological characteristics of uterine cervical neoplasia in Taiwanese women. Tumour Biol. 2016
18. Su SC, Hsieh MJ, Lin CW, Chuang CY, Liu YF, Yeh CM, Yang SF. Impact of HOTAIR Gene Polymorphism and Environmental Risk on Oral Cancer. J Dent Res. 2018:22034517749451
19. Zhang J, Liu X, You LH, Zhou RZ. Significant association between long non-coding RNA HOTAIR polymorphisms and cancer susceptibility: a meta-analysis. Onco Targets Ther. 2016;9:3335-3343
20. Chu H, Chen Y, Yuan Q, Hua Q, Zhang X, Wang M, Tong N, Zhang W, Chen J, Zhang Z. The HOTAIR, PRNCR1 and POLR2E polymorphisms are associated with cancer risk: a meta-analysis. Oncotarget. 2017;8:43271-43283
21. Zhang X, Zhou L, Fu G, Sun F, Shi J, Wei J, Lu C, Zhou C, Yuan Q, Yang M. The identification of an ESCC susceptibility SNP rs920778 that regulates the expression of lncRNA HOTAIR via a novel intronic enhancer. Carcinogenesis. 2014;35:2062-2067
22. Hosseini ES, Meryet-Figuiere M, Sabzalipoor H, Kashani HH, Nikzad H, Asemi Z. Dysregulated expression of long noncoding RNAs in gynecologic cancers. Mol Cancer. 2017;16:107
23. Lee M, Kim HJ, Kim SW, Park SA, Chun KH, Cho NH, Song YS, Kim YT. The long non-coding RNA HOTAIR increases tumour growth and invasion in cervical cancer by targeting the Notch pathway. Oncotarget. 2016;7:44558-44571
24. Huang L, Liao LM, Liu AW, Wu JB, Cheng XL, Lin JX, Zheng M. Overexpression of long noncoding RNA HOTAIR predicts a poor prognosis in patients with cervical cancer. Arch Gynecol Obstet. 2014;290:717-723
25. Liu S, Zhang M, Qu P. Expression level and clinical significance of HOX transcript antisense intergenic RNA in cervical cancer: a meta-analysis. Sci Rep. 2016;6:38047
26. Kim HJ, Lee DW, Yim GW, Nam EJ, Kim S, Kim SW, Kim YT. Long non-coding RNA HOTAIR is associated with human cervical cancer progression. Int J Oncol. 2015;46:521-530
Author contact
![]() Corresponding author: Po-Hui Wang, MD, PhD, Institute of Medicine, Chung Shan Medical University, 110, Section 1, Chien-Kuo North Road, Taichung, 40201, Taiwan. Tel.: 886-4-24739595 ext. 21721; Fax: 884-4-24738493; E-mail: ginhow84921344com.tw
Corresponding author: Po-Hui Wang, MD, PhD, Institute of Medicine, Chung Shan Medical University, 110, Section 1, Chien-Kuo North Road, Taichung, 40201, Taiwan. Tel.: 886-4-24739595 ext. 21721; Fax: 884-4-24738493; E-mail: ginhow84921344com.tw

 Global reach, higher impact
Global reach, higher impact