Impact Factor
ISSN: 1449-1907
Int J Med Sci 2018; 15(9):840-848. doi:10.7150/ijms.25004 This issue Cite
Research Paper
A human origin strain Lactobacillus acidophilus DDS-1 exhibits superior in vitro probiotic efficacy in comparison to plant or dairy origin probiotics
1. School of Health Sciences, College of Health and Medicine, University of Tasmania, Launceston, Tasmania 7250 Australia
2. School of Land and Food, University of Tasmania, Launceston, Tasmania 7250 Australia.
3. UAS Laboratories, Madison, Wisconsin, USA.
4. School of Health and Biomedical Sciences, RMIT University, Bundoora, Victoria, 3082 Australia.
Received 2018-1-18; Accepted 2018-4-13; Published 2018-5-26
Abstract
Background: The health benefits of probiotics are well established and known to be strain-specific. However, the role of probiotics obtained from different origins and their efficacy largely remains unexplored. The aim of this study is to investigate the in vitro efficacy of probiotics from different origins.
Methods: Probiotic strains utilized in this study include Lactobacillus acidophilus DDS-1 (human origin), Bifidobacterium animalis ssp. lactis UABla-12 (human origin), L. plantarum UALp-05 (plant origin) and Streptococcus thermophilus UASt-09 (dairy origin). Screening assays such as in vitro digestion simulation, adhesion, cell viability and cytokine release were used to evaluate the probiotic potential.
Results: All strains showed good resistance in the digestion simulation process, especially DDS-1 and UALp-05, which survived up to a range of 107 to 108 CFU/mL from an initial concentration of 109 CFU/mL. Two human colonic mucus-secreting cells, HT-29 and LS174T, were used to assess the adhesion capacity, cytotoxicity/viability, and cytokine quantification. All strains exhibited good adhesion capacity. No significant cellular cytotoxicity or loss in cell viability was observed. DDS-1 and UALp-05 significantly upregulated anti-inflammatory IL-10 and downregulated pro-inflammatory TNF-α cytokine production. All the strains were able to downregulate IL-8 cytokine levels.
Conclusion: Of the 4 strains tested, DDS-1 demonstrated superior survival rates, good adhesion capacity and strong immunomodulatory effect under different experimental conditions.
Keywords: Probiotics, adhesion, gastrointestinal survival, immunomodulation
Introduction
Probiotic microorganisms are living, natural and safe modulators of gut microbiota. The term probiotic has been defined as “Live microorganisms that, when administered in adequate amounts, confer a health benefit on the host” [1]. Probiotics restore the beneficial composition of the gut microbiota by adhering to the intestinal epithelium, establishing microbial-host crosstalk and modulating immune responses [2]. Moreover, many strains produce regulatory metabolites, such as short chain fatty acids, which strengthen the intestinal epithelium in order to maintain a healthy state. Lactic acid bacteria (LAB) such as Lactobacillus, Bifidobacterium, Streptococcus, and Enterococcus species are widely used as probiotics, as they were noted to have a clinical effect in reducing the symptoms of diarrhea, inflammatory bowel disease and irritable bowel syndrome [3,4,5]. To confer any health benefits, probiotics need to survive through the hostile environment of the digestion process in sufficient numbers (at least 106 colony forming units (CFU)), tolerating acids, bile, and pancreatic digestive enzymes, and finally adhere to the intestinal epithelium in the colon. Interestingly, most of the LAB probiotics have demonstrated high survival rates under simulated conditions of gastric juice and high bile salts concentrations [6, 7].
Adherence of probiotics to the intestinal mucosal surfaces is also considered an essential probiotic trait as adhesion to colonic mucosa is the first step for gut colonization [6, 7]. The possible mechanism of adherence is via close interaction between surface adhesion proteins of bacteria and host cells [2, 6, 8]. Probiotics as generally considered safe as functional foods. A number of studies show that LAB strains are not toxic to intestinal epithelial cells [9, 10]. The safety of probiotics for human use has been the subject of a number of reviews by experts in food safety [11, 12, 13]. These reviews support the safety and suitability of LAB for use as oral probiotics, a conclusion that is largely based on their long history of safe use in food and as dietary supplements.
Experimental and human studies suggest that increased levels of pro-inflammatory cytokines TNF-α and IL-8 play a significant role in inducing intestinal inflammatory response [14]. LAB strains have been reported to reduce inflammation by downregulating IL-8 and TNF-α and by upregulating IL-10 levels [7, 14, 15]. This immuno-modulatory effect of probiotic bacteria offers additional potential benefits for the prevention and management of gastrointestinal inflammation.
Probiotic strain selection is often debated and linked to their origin. To date, many in vitro studies have reported the efficacies/functional properties of various probiotics obtained from different origins such as human, plant, dairy, and animal. However, most of these studies evaluated the efficacy of probiotic strain from a single origin strain, such as human [16, 17], dairy [15, 18], plant [19] or animal [6] to study their probiotic potential. To our knowledge, the current study is the first to compare probiotics of different origins. In our study, we investigated the efficacies of probiotics isolated from human, plant and dairy sources. To do this, the resistance and survival of these strains during the digestion process was assessed throughout buccal, stomach and intestinal conditions in a simulated in vitro setting. Subsequently, adhesion capacity and cytotoxicity studies were conducted on HT-29 and LS174T cells, and finally, the immunomodulation effect of these strains, particularly on IL-8, TNF-α and IL-10, was investigated on HT-29 cell supernatants. These findings would be of great importance to identify right probiotic strain for gut health.
Materials and Methods
Bacterial strains and origins
Bacterial strains utilized in the study include Lactobacillus acidophilus DDS-1(human origin), Bifidobacterium animalis ssp. lactis UABla-12 (human origin), L. plantarum UALp-05 (plant origin) and Streptococcus thermophilus UASt-09 (dairy origin), and were obtained in freeze-dried, free-flowing lyophilized form from UAS labs, Madison, WI, USA. All the strains were routinely grown on De Man Rogosa agar supplemented with 0.05% (w/v) of L-cysteine (MRS-C) except UASt-09, which was grown on M17 agar supplemented with 10% (w/v) lactose, under anaerobic conditions at 37 0C for 48 h.
Survival of strains in static in vitro digestion model
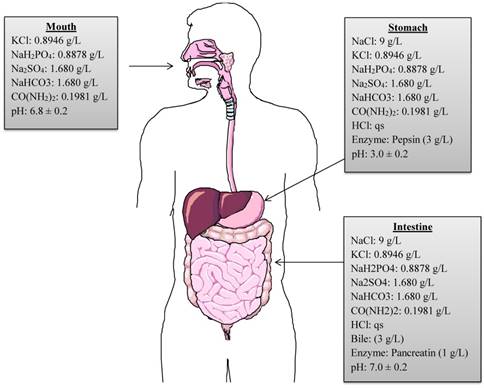
A continuous, static in vitro digestion model was employed to assess the survival of selected strains as described by Versantvoort et al. [20]. The model includes a step-wise human digestive process, simulating mouth, stomach, and intestinal compartments. However, it does not represent the continuous process of digestion. In order to mimic the continuous in vitro process of the human gastrointestinal tract, the present model was modified and adapted from Chavarri et al. [21] and Belguesmia et al. [17]. The chemical compositions of digestive fluids, enzymes, pH, temperature and residence time period in each compartment are reproduced in the modified model (Figure 1). Firstly, to reproduce the buccal conditions, 1 mL of each bacterial strain comprising approximately 109 CFU were individually added to 9 mL of simulated saliva juice (pH adjusted to 6.8 ± 0.2) and incubated anaerobically at 37 0C for 5 min. After incubation, the suspension was centrifuged at 3000 rpm for 5 min at 4 0C to recover the pellet containing each strain and to maintain the continuous process. The pellet was then re-suspended in 9 mL of simulated gastric juice comprised of 9 g/L of NaCl containing 0.3% (w/v) of porcine pepsin (Sigma Aldrich, St. Louis, MO, USA), and the pH adjusted to 3.0 ± 0.2 and incubated anaerobically at 37 0C for 120 min to mimic gastric conditions. After incubation, the gastric fluid was neutralized to pH 7.0 with phosphate buffered saline to stop the pepsin digestion. The supernatants were discarded after centrifugation and intestinal digestion process was initiated. The intestinal compartment included 0.3% (w/v) bovine bile (Sigma Aldrich, St. Louis, MO, USA) and 0.1% (w/v) pancreatic enzymes (MP Biomedical, CA, USA), pH was adjusted to 7.0 ± 0.2 before incubating anaerobically at 37 0C for 120 min. Overall, the bacterial strains were exposed to the various digestive conditions for a total of 240 min. After each step of the simulated digestion process, bacterial samples were collected and diluted in saline buffer. Lastly, 100 µL of each sample was plated on MRS-C/M17 agar plates and incubated at 37 0C for 24-72 h anaerobically to allow sufficient growth. The number of CFU/mL of each probiotic strain was calculated.
Cell culture
Human colonic epithelial cells (HT-29 and LS174T) were purchased from American Type Cell Culture (ATCC, Virginia, USA). For initial growth, HT-29 and LS174T were cultured in 75 cm2 tissue culture flasks and grown to confluence in complete medium [McCoy and RPMI 1640 medium respectively supplemented with 10% fetal bovine serum, 2 mM L-glutamine and 100 U/mL of antibiotics (penicillin and streptomycin)]. At confluence, the adhered cells were washed with PBS, dissociated using 0.1 w/v TrypLE® Express (Gibco, Victoria, Australia) and re-suspended in 24-well cell culture plates at a density of approximately 5 x 104 cells/mL. Cells were incubated for 24 h and grown to confluency before exposing them to different treatments for quantitative measurement of bacterial cell adhesion, cytotoxicity, and cytokine quantification.
Graphical representation of chemical compositions, concentrations, digestive enzymes and pH used in in vitro simulated digestion process. (qs = quantity sufficient).
Adhesion assay
Each wells containing cells was washed thrice with serum-free culture medium before adding the 1 mL of the bacterial suspension. Approximately 109 CFU/mL of each bacterial strain was first suspended in the respective cell culture medium without serum and antibiotics. The cells containing bacterial suspensions were incubated for 4 h with 5% CO2. After 4 h incubation, cell monolayers were washed thrice with Hank's balanced salt solution to remove the non-adherent bacteria in the wells. The cells were aspirated, lysed using 0.1% w/v TrypLE® Express for 10-15 min and collected in saline solution. All cell lysates were serially diluted, plated on MRS-C/M17 agar and incubated anaerobically for 48-72 h. The percentage of adhesion was calculated by the log of the number of adherent bacteria (log CFU) divided by the log of the total number of bacteria inoculated, multiplied by 100. Each determination was performed in triplicate.
Cell cytotoxicity and viability
Each bacterial suspension at approximately 109 CFU/mL in respective serum-free cell culture medium was added to the cell monolayer in a 24-well plate and incubated for 8 h. After incubation, the supernatants were collected for determination of cytotoxicity using the lactate dehydrogenase (LDH) assay [22] and cell viability was assessed by standard trypan blue exclusion assay.
The cellular cytotoxicity was assessed by the LDH in-vitro cytotoxicity assay (TOX7, Sigma-Aldrich, St. Louis, MO, USA). Briefly, the culture supernatants were centrifuged at 250×g for 4 min. An aliquot containing 50 µL of either blank (complete medium) or control (cells only) and cells treated with 100 µL of each bacterial supernatants obtained after 8 h incubation, was mixed with 100 µL of a solution containing LDH assay mixture (LDH substrate, LDH dye, and LDH cofactor). The mixture was then incubated at room temperature for 20-30 min and the reaction was quenched by the addition of 1N hydrochloric acid (15 μL). The absorbance was measured spectrophotometrically using a plate reader (Spectra Max M2 microplate reader, Sunnyvale, CA, USA) at a wavelength of 490 nm. The cellular viability was examined by Trypan Blue exclusion staining assay using a Countess Automated Cell Counter (Thermo-Fisher, Waltham, MA, USA).
Quantification of cytokines
HT-29 cells were used for quantification of IL-8, TNF-α, and IL-10 cytokine levels. The cells were subjected to probiotic/lipopolysaccharide (LPS) treatment under three different conditions, i.e., no-, co- and post- treatments, and LPS alone (control) in individual 24-well plates [23]. For no-treatment (probiotic only + HT-29 cells) conditions, 1 mL of each bacterial suspension (109 CFU/mL) was first added to each well containing cells and incubated at 37 0C, 5 % CO2 for 8 h. For co-treatment, each bacterial suspension was added to the cells and simultaneously challenged with LPS (100 ng/mL) (Sigma-Aldrich St. Louis, MO, USA) followed by incubation for 8 h. Lastly, in post-treatment, the cells were first challenged with LPS (100 ng/mL) and incubated for 4 h, after which, each bacterial suspension (1 mL of 109 CFU/mL) was added and re-incubated for 4 h. The supernatants from each well were collected and used for quantification of cytokines using Bio-Plex® Pro human cytokine assay kit (Bio-Rad®), according to the manufacturer's protocol. Briefly, 50 µL of cytokine beads were added to the 96-well plate and incubated for 30 min before washing twice with wash buffer. Then 50 µL of each standard, blank and samples were added to the respective wells and incubated at room temperature on a shaker at 850 rpm for 30 min. After incubation, the wells were washed thrice and 25 µL of detection antibody was added to each well and incubated at room temperature on a shaker at 850 rpm for 30 min. 50 µL of streptavidin-PE was then added to each well and incubated at room temperature in a shaker at 850 rpm for 10 min. After three washes, 125 μL assay buffer was added to each well and incubated at room temperature for 30 sec. After incubation, the plates were read on the Bio-Plex® 200 system and data was analyzed in Bio-Plex Data ProTM Software. All the experiments were performed in triplicates.
Statistical analysis
All the data are expressed as mean ± SEM calculated over three independent experiments with triplicates within each experiment. All the statistical analyses were performed using Graph Pad Prism software (version 6.0). The statistical differences between groups were measured using One-way ANOVA and Tukey's multiple comparison tests for adhesion and cytotoxicity assays. Two-way ANOVA was used for digestion simulation and cytokine quantification.
Results
In vitro simulated digestion process
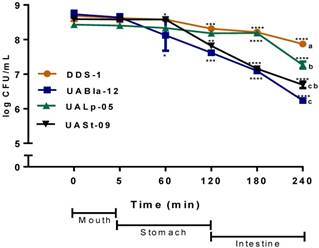
All tested bacterial strains survived passage through the mouth compartment followed by the stomach and intestinal compartments during the simulated digestion process. The strains retained over 75% of their initial concentration of 109 CFU/mL (Figure 2). DDS-1 exhibited the highest survival rates. No significant change in bacterial numbers of the strains was observed in the mouth compartment. During the transit from mouth to stomach, only UABla-12 (p < 0.001) and UASt-09 (p = 0.03) strains displayed a decrease of one logarithmic unit, which was significant when compared to DDS-1. In the intestinal compartment, DDS-1 and UALp-05 showed a decrease of one logarithmic unit, while UABla-12 and UASt-09 further reduced by two logarithmic units. Overall, DDS-1 (p = 0.034), showed the highest resistance, followed by UALp-05 (p = 0.008), UASt-09 (p = 0.002) and UABla-12 (p < 0.001) compared to initial concentration. The differences were statistically significant for UABla-12 (p < 0.001), UALp-05 (p = 0.01) and UASt-09 (p < 0.001) when compared to DDS-1.
Adhesion capacity
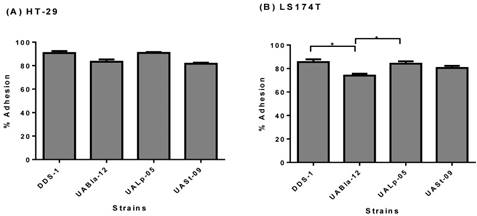
The adhesion assay demonstrated the adherence capability of each probiotic strain to HT-29 and LS174T human colonic cells. All the strains were able to adhere efficiently to both colonic cell types. Although, DDS-1 and UALp-05 showed the highest adhesion capacity (87-93% and 89-92%, respectively) followed by UABla-12 and UASt-09 (79-86% and 80-83%, respectively) for HT-29 cells (Figure 3A) the difference was not statistically significant. Comparatively, DDS-1 and UALp-05 showed relatively more adherence (81-88% and 80-87% respectively, Figure 3B) to LS174T cells. The adhesion capacities for the strain UABla-12 (p = 0.02) was significantly lower than DDS-1 and UALp-05 (p = 0.03).
Survival of each bacterial strain in the mouth, stomach and intestinal simulated in vitro digestion process. The samples were taken at each step and the number of CFU/ml was evaluated on agar medium. The date represents mean ± SEM of three replicates. Statistical analysis was by Two-way ANOVA. The values of groups designated with different letters are significantly different in the mouth and intestinal compartments (p < 0.05). For overall survival of individual strain *p < 0.01, **p < 0.005, ***p < 0.001 compared to DDS-1.
Cell cytotoxicity and viability
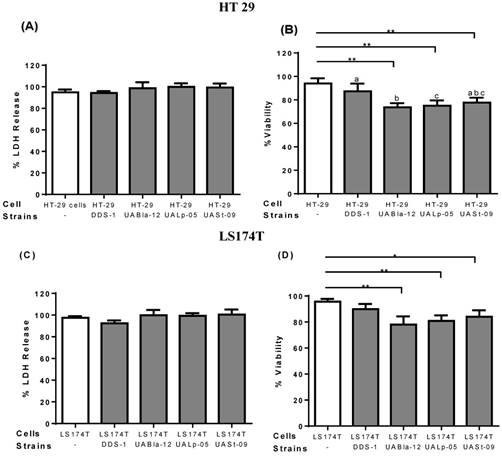
To assess the safety of the tested strains and any cytotoxicity, LDH and trypan blue assays were performed on epithelial cell culture supernatants. After 8 h incubation, all the other strains except DDS-1 showed a slightly increased LDH level, but, no significant cytotoxicity was observed. Similarly, after 8 h incubation, DDS-1 showed no significant loss of cell viability but a slight loss of cell viability was observed with UABla-12, UASt-09, and UALp-05 when compared to untreated cells.
Adhesion capacity of each bacterial strain to HT-29 (A) & LS 174T cells (B) after 4 h incubation period. Data represents mean ± SEM from three independent measurements. Statistical analysis was performed by One-way ANOVA with Tukey's multiple comparison (pairwise) tests. *p < 0.01.
Immuno-modulatory effects by probiotic strains
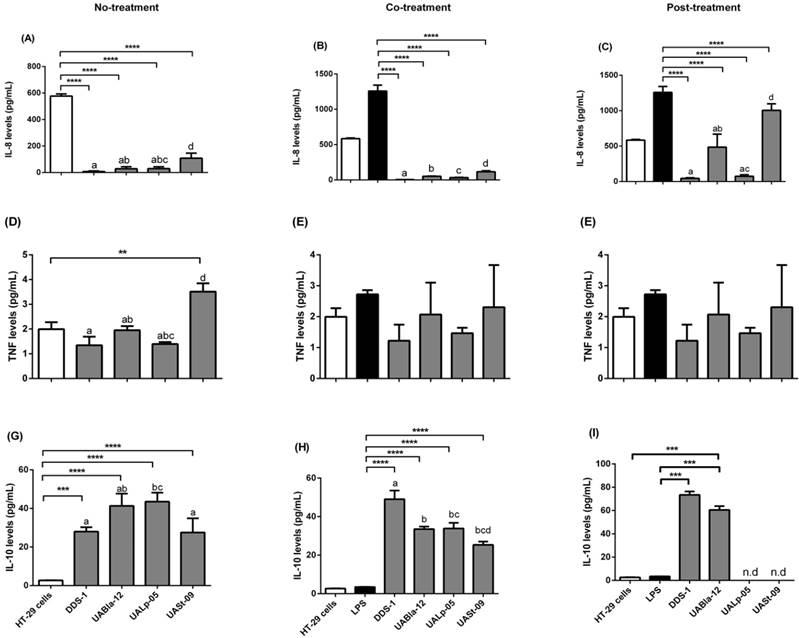
All strains modulated specific pro and anti-inflammatory cytokines (Figure 5). Each strain had a significant effect on the release of pro-inflammatory IL-8, and TNF-α as well as anti-inflammatory IL-10. When HT-29 cells were challenged with LPS for 8 h, this resulted in upregulation of IL-8 and TNF-α levels (1182.19 ± 165.78 and 2.56 ± 0.24 pg/mL, respectively). IL-8 secretion was significantly downregulated during no-treatment (Figure 5A) when compared to HT-29 cells alone as a control (574.24 ± 24.25 pg/mL). During co- (Figure 5B) and post-treatment (Figure 5C), all the strains significantly downregulated IL-8 levels (p < 0.0001) when compared to LPS control with the greatest effect shown by DDS-1 followed by UALp-05, UABla-12, and UASt-09. Overall, there was a statistical by significant difference observed between DDS-1 and UASt-09 (p = 0.002) under no- and post-treatment, while there was no change in the IL-8 secretion between strains under co-treatment.
Similar to the observed inhibition of release of IL-8, DDS-1 marked reduced TNF-α levels under the three tested conditions (Figures 5D-F). For no-treatment condition, TNF-α levels were downregulated by 1.22 ± 0.51 pg/mL by DDS-1 and 0.98 ± 0.49 pg/mL by UALp-05 compared to the cells alone control (2.0 ± 0.32 pg/mL). Interestingly, UASt-09 significantly upregulated the TNF-α levels (3.50 ± 0.50 pg/mL, p = 0.002). Upon co-treatment, TNF-α levels were found to be downregulated to 1.0 ± 0.51 pg/mL by DDS-1, and (1.0 ± 0.26 pg/mL) by UALp-05, while no major change in TNF-α levels was observed with UABla-12 compared to LPS treated control (2.32 ± 0.51 pg/mL). Relative to LPS treated control during the post-treatment, DDS-1 downregulated the TNF-α levels from 2.32 ± 0.51 pg/mL to 0.68 ± 0.10 pg/mL followed by UALp-05 which reduced levels to 1.19 ± 0.51 pg/mL. UABla-12 and UASt-09 did not affect the TNF-α levels. The differences were statistically significant between DDS-1 and UASt-09 (p = 0.002) under no-treatments and between DDS-1, UABl-12 (p = 0.01) and UASt-09 (p = 0.01) under post-treatment.
All the strains, to some extent, upregulated the levels of anti-inflammatory cytokine IL-10 under no-treatment condition (as shown in Figure 5G). The basal levels of IL-10 on cells alone (control) were 2.72 ± 0.51 pg/mL and the strains upregulated by 25.25 ± 1.99 pg/mL (DDS-1, p = 0.0003), 38.65 ± 6.40 pg/mL (UABla-12, p < 0.0001), 40.65 ± 3.75 pg/mL (UALp-05, p < 0.001) and 24.85 ± 8.12 pg/mL (UASt-09, p < 0.001). Upon co-treatment, DDS-1 significantly upregulated the levels of IL-10 by 45.53 ± 6.71, followed by UABla-12 (30.02 ± 4.26 pg/mL), UALp-05 (30.32 ± 6.21 pg/mL) and UASt-09 (21.82 ± 5.6 pg/mL) (Figure 5H) compared to LPS treated control (3.45 ± 0.5 pg/mL). During post-treatment condition, only DDS-1 and UABla-12 were able to upregulate IL-10 levels by 76 pg/mL and 62.84 pg/mL (p < 0.0001) respectively (Figure 5I). The statistically significant differences in IL-10 secretions were found between DDS-1 and UALp-05 (p < 0.001) under no-treatment and between DDS-1, UABla-12 (p < 0.001), UALp-05 (p < 0.001) and UASt-09 (p < 0.001) under co-treatment. There was no difference between strains under post-treatments.
Discussion
This study provides insightful information on the differences in the probiotic potential of individual LAB strains obtained from different origins.
The role of the origin of probiotic strain selection is often debated. Theoretically, human gut colonization is facilitated by selecting probiotics of human origin. However, numerous studies have shown that maximum probiotic effect after human consumption is strain-specific but not restricted to human origin strains. For example, B. animalis ssp. lactis of animal origin proved to be safe and modulated the gut microbiota in human subjects [24]. Similarly, B. lactis CNCM I‐2494 of dairy origin have alleviated minor digestive symptoms in healthy women [25]. Although there are numerous in vitro studies showcasing the functional properties of probiotics, there are no studies comparing probiotic efficacies with origins. To our knowledge, this the first study to elucidate the link between origin and efficacy of the probiotics in vitro with an assessment of the survival of each strain under simulated digestion process, their adhesion capacities to human colonic cells and their immunomodulatory effects.
Effect of probiotics on cellular cytotoxicity/viability of intestinal epithelial cells. Cell cytotoxicity and viability on HT 29 (A-B) and LS174T cells (C-D) was determined after 8 h incubation with each probiotic strain. Cell cytotoxicity was measured using LDH assay and data was represented as percentage LDH release. Viability was measured by trypan blue exclusion assay and data was represented as percentage viability. All the data is presented as mean ±SEM. Statistical analysis was performed by Two-way ANOVA test. * p < 0.05.
Resistance to gastric and intestinal environments after oral administration is considered as one of the most important traits of probiotics. Many LAB strains have been reported to exhibit good resistance to pH around 2.5 ± 3.0 and 6.5 ± 0.5 in gastric and intestinal conditions respectively [7, 17, 26, 27]. Generally, the L. acidophilus species is considered well adapted to survive harsh conditions of digestion. In fact, due to this ability, L. acidophilus has been considered an ideal vehicle for mucosal-targeted delivery of vaccines and biotherapeutics [28, 29]. In the present study, probiotic strains obtained from different origins were tested for their survivability in an in vitro simulated digestion static model and all of them displayed good survivability (106-108 CFU/mL) compared to their initial concentrations of 109 CFU/mL. However, the concentrations of UABla-12 and UASt-09 decreased around one logarithmic unit compared to their initial concentrations after exposure the strains to low pH and digestive enzyme in the gastric compartment. The viability loss of UABla-12 may be linked to its obligate anaerobic nature, and for UASt-09 its acid tolerance capacity is strain-specific [6, 7, 8, 15, 17, 36]. The acid/enzyme tolerance by lactobacilli (and most of the gram-positive strains) can be attributed, in part, to the presence of a constant gradient between extracellular and cytoplasmic pH [30, 31]. However, the strains in our study showed significantly better resistance in the gastric compartment compared to other studies [15, 17].
In our study, the strains were not only exposed to different pH ranges, salivary, gastric and intestinal juices, but also to bile and digestive enzymes such as pepsin, and pancreatic enzymes in a continuous process. Overall, all the strains were decreased by only one logarithmic unit in the intestinal compartment. Specifically, DDS-1 maintained higher concentrations followed by UALp-05, UABla-12, and UASt-09. This phenomenon may be linked to the ability of these strains to produce bile salt hydrolase (BSH) and previous studies have demonstrated BSH genes in lactobacilli strains which are believed to be responsible for their resistance [32, 33, 34, 35].
Adhesion capacity to colonic cells also plays a role in the selection of probiotic bacteria [17]. The human colonic cells such as HT-29, LS174T, and Caco2 are commonly used to test probiotic bacterial adhesion capacity [7, 15, 36]. High adhesion of LAB strains to colonic cells, as seen in our study, is reported in most studies [7, 17, 15, 36]. Toscano and colleagues [7] tested 3 different strains of Bifidobacterium on HT-29 cells and found moderate to low adhesion capacity. The relatively low adhesion capacity of Bifidobacteria [7] was quite similar to that observed with UABla-12 strain in our study. The probiotic adhesion ability involves various biophysical and biochemical properties of probiotics and epithelial cells, includes electrostatic forces, hydrophobicity, steric & passive forces and specific cellular structures [15].
Detection of cytokines released in the supernatants of HT-29 cells (control) after three conditions using Bio-Plex® 200 system. Detection of IL-8 levels (A, B, C), TNF levels (D, E, F) and IL-10 levels (G, H, I) after no (probiotic only), co (LPS and probiotics together) and post-treatment (challenged with LPS first for 4 h and later probiotics for another 4 h). The values of groups designated with different letters are significantly different (p < 0.05). Statistical analysis was done by Two-way ANOVA. * indicates the significant differences. *p <0.05, **p <0.005, ***p < 0.001, ****p < 0.0001. nd = not detected.
Bu and colleagues [37] compared the differences in biochemical and cellular properties of LS174T and HT-29 cells, is a study extensively based on mucin expressions among the cells involving their role in adhesion. Mucus is mostly composed of mucins protein blocks which are encoded with MUC genes. Out of 20 identified human mucins, MUC 2 and MUC 5 are implicated in various ligand-receptor interactions. Especially, MUC 2, a major gastrointestinal mucin secreted by goblet cells, plays a particularly important role in conferring the mucin barrier function of the gut. Morphologically, LS174T cells have many surface microvilli and more intercellular spaces than HT-29 cells. Surface microvilli play a role in the adhesion capacity of microbes to epithelial surfaces, and Bennett and colleagues [38] demonstrated that more surface microvilli establishes an electrostatic barrier which may repel the microbes from adhering to epithelial cells. High adhesion capacity of the strains used in the present study might be linked to the presence of MUC 2 gene and/or comparatively less surface microvilli.
Lactobacillus and Bifidobacterium species strains are generally considered to be safe [10, 17]. In the present study, none of the tested strains showed cytotoxic effects on HT-29 and LS174T cells. This is in agreement with the safety profile observed clinically with these strains [39, 40]. Furthermore, many studies have revealed the ability of various lactobacilli and bifidobacteria strains to limit the cytotoxicity caused by many entero-pathogens [41, 42]. In particular, the human origin strains, DDS-1 and UABla-12, have previously been shown to be safe and efficacious in multiple randomized controlled clinical studies. In a randomized controlled trial enrolling 96 preschool children with moderate-to-severe atopic dermatitis, subjects receiving DDS-1 and UABla-12 experienced a more rapid improvement in atopic dermatitis questionnaire scores, as well as improved immune markers, when compared to placebo over an 8 week intervention period [39]. Similarly, in another randomized controlled trial involving 225 children, short-term use of DDS-1 and UABla-12 was found to significantly shorten the duration of acute respiratory tract infection, when compared to placebo [40].
Immunomodulatory capacity is considered as an additional factor for the selection of probiotics. A number of in vitro probiotic studies have investigated the anti-inflammatory properties of probiotics to modulate the release of pro- and anti-inflammatory cytokines including IL-6, IL-8, TNF-α and IL-10 [15, 17, 43, 44]. In our study, most of the tested strains downregulated the pro-inflammatory IL-8, and TNF-α and upregulated IL-10 levels when compared to controls, but the rate and extent of immunomodulatory effect varied between the strains and under different conditions. The upregulation of pro-inflammatory cytokines upon LPS treatment could be attributed to T cell proliferation and activation, and the upregulation of anti-inflammatory cytokines in presence of these strains could be associated with counteracting molecular mechanisms leading to T cell activation [23]. As shown in previous studies, probiotics could influence the activation of dendritic cells (DCs) and macrophages of the innate immune system [3, 45]. Upon activation, DCs release anti-inflammatory TGF-β and IL-10, which allows the proliferation of adaptive T cells.
All the tested strains significantly suppressed IL-8 levels under no-, co- and post-treatment conditions. NF-ĸB and IκB pathways are crucial targets in the initiation of inflammatory responses evoked by cytokines including IL-6, IL-8, and pathogens. Ohkusa and colleagues [44] showed that VSL#3 probiotic suppressed IL-8 levels by inhibiting nuclear translocation of NF-ĸB. Reduction of IL-8 levels in our study could be attributed to inhibition of NF-ĸB. Only DDS-1 and UALp-05 were able to downregulate TNF-α levels in all conditions, which is in agreement with previous probiotic studies [15, 23]. Both UABla-12 and UASt-09 having no stimulatory effect on TNF levels under co- and post-treatment was consistent with the previous studies which actually showed an increase in TNF-α levels with probiotic treatment [15, 43]. The mechanism of downregulation of TNF-α levels by LAB strains has been linked to activation of certain pathways like extracellular-signal-regulated kinase (ERK), p38 and phosphoinositide 3-kinase (PI3K) pathways [46]. The beneficial effects of probiotics can be immuno-stimulatory rather than immuno-suppressive, suggesting that probiotics can restore the breach of the innate immune system and prevent the onset of inflammation by stimulation of TNF-α [2, 3]. All the tested strains induced the release of anti-inflammatory cytokine IL-10 during no- and co-treatment itself. However, only DDS-1 and UABla-12 were able to induce the secretion of IL-10 levels during post-treatment, which is consistent with previous studies conducted in colonic cells [23, 46]. In this particular case, a few studies reported that release of IL-10 by probiotics is a strain, origin, and dose-dependent [15, 43, 47]. Overall, all the strains demonstrated strong immunomodulatory effects; but specifically, the strain DDS-1, which strengthens its probiotic potential.
Taken together, all the strains showed good survival in the simulated digestion process, strongly adhered to intestinal epithelium and demonstrated an adequate immunomodulatory effect. However, the comparative analysis of the data suggests that origin may affect their probiotic potential. This was particularly in the case of human origin DDS-1 strain, which showed superior characteristics compared to other strains and could be an interesting candidate for more intensive research to elucidate and harness the bioactive effects.
Authors Contribution
RCV, RE, and TS designed the research, RCV, TS, RG, AP, ST, and MS, conducted the research, KDK and MB performed statistical analysis and paper editing; RCV and CM wrote the paper, and RCV and RE had primary responsibility for final content. All authors critically revised the manuscript for important intellectual content and read and approved the final manuscript.
Competing Interests
The authors have declared that no competing interest exists.
References
1. Hill C, Guarner F, Reid G, Gibson GR, Merenstein DJ, Pot B. et al. Expert consensus document: The International Scientific Association for Probiotics and Prebiotics consensus statement on the scope and appropriate use of the term probiotic. Nat Rev Gastroenterol Hepatol. 2014;11:506-14
2. Vemuri R, Gundamaraju R, Eri R. Role of lactic acid probiotic bacteria in IBD. Curr Pharm Design. 2017;23:2352-55
3. Vemuri RC, Gundamaraju R, Shinde T, Eri R. Therapeutic interventions for gut dysbiosis and related disorders in the elderly: antibiotics, probiotics or faecal microbiota transplantation? Benef Microbes. 2017;8:179-92
4. Akoglu B, Loytvedb A, Nuidingb H, Zeuzemc S, Faustb D. Probiotic Lactobacillus casei Shirota improves kidney function, inflammation and bowel movements in hospitalized patients with acute gastroenteritis - A prospective study. J Funct Foods. 2015;17:305-13
5. Orel R, Kamhi-Trop T. Intestinal microbiota, probiotics and prebiotics in inflammatory bowel disease. World J Gastroenterol. 2014;20:11505-24
6. Valeriano VD, Parungao-Balolong MM, Kang DK. In vitro evaluation of the mucin-adhesion ability and probiotic potential of Lactobacillus mucosae LM1. J Appl Microbiol. 2014;117:485-97
7. Toscano M, De Vecchi E, Gabrieli A, Zuccotti GV, Drago L. Probiotic characteristics and in vitro compatibility of a combination of Bifidobacterium breve M-16 V, Bifidobacterium longum subsp. infantis M-63 and Bifidobacterium longum subsp. longum BB536. Ann Microbiol. 2015;65:1079-86
8. Arai T, Obuchi S, Eguchi K, Seto Y. In vitro investigation of molecules involved in Lactobacillus gasseri SBT2055 adhesion to host intestinal tract components. J Appl Microbiol. 2016;120:1658-67
9. Messaoudi S, Madi A, Prévost H, Feuilloley M, Manai M, Dousset X. et al. In vitro evaluation of the probiotic potential of Lactobacillus salivarius SMXD51. Anaerobe. 2012;18:584-9
10. Sevda ER, Koparal AT, Kivanç M. Cytotoxic effects of various lactic acid bacteria on Caco-2 cells. Turkish J Biol. 2015;39:23-30
11. Bernardeau M, Guguen M, Vernoux JP. Beneficial lactobacilli in food and feed: long-term use, biodiversity and proposals for specific and realistic safety assessments. FEMS Microbiol Rev. 2006;30:487-513
12. Adams M. Safety of industrial lactic acid bacteria. J Biotechnol. 1999;68:171-8
13. Salminen S, von Wright A, Morelli L, Marteau P, Brassart D, de Vos WM. et al. Demonstration of safety of probiotics - a review. Int J Food Microbiol. 1998;44:93-106
14. Park JS, Joe I, Rhee PD, Jeong CS, Jeong GA. Lactic acid bacterium isolated from kimchi ameliorates intestinal inflammation in DSS-induced colitis. J Microbiol. 2017;55:304-10
15. Ranadheera CS, Evans CA, Adams MC, Baines SK. Effect of dairy probiotic combinations on in vitro gastrointestinal tolerance, intestinal epithelial cell adhesion and cytokine secretion. J Funct Foods. 2014;8:18-25
16. Bahrami B, Macfarlane S, Macfarlane GT. Induction of cytokine formation by human intestinal bacteria in gut epithelial cell lines. J Appl Microbiol. 2011;110:353-63
17. Belguesmia Y, Domenger D, Caron J, Dhulster P, Ravallec R, Drider D. et al. Novel probiotic evidence of lactobacilli on immunomodulation and regulation of satiety hormones release in intestinal cells. J Funct Foods. 2016;24:276-86
18. Thakur K, Tomar SK. Invitro study of riboflavin producing lactobacilli as potential probiotic. LWT- Food Sci Technol. 2016;68:570-78
19. Nomura M, Kobayashi M, Narita T, Kimoto-Nira H, Okamoto T. Phenotypic and molecular characterization of Lactococcus lactis from milk and plants. J Appl Microbiol. 2006;101:396-405
20. Versantvoort CH, Oomen AG, Van de Kamp E, Rompelberg CJ, Sips AJ. Applicability of an in vitro digestion model in assessing the bioaccessibility of mycotoxins from food. Food Chem Toxicol. 2005;43:31-40
21. Chávarri M, Marañón I, Ares R, Ibáñez FC, Marzo F, del Carmen Villarán M. Microencapsulation of a probiotic and prebiotic in alginate-chitosan capsules improves survival in simulated gastro-intestinal conditions. Int J Food Microbiol. 2010;142:185-89
22. Weidmann E, Brieger J, Jahn B, Hoelzer D, Bergmann L, Mitrou PS. Lactate dehydrogenase-release assay: a reliable, nonradioactive technique for analysis of cytotoxic lymphocyte-mediated lytic activity against blasts from acute myelocytic leukemia. Ann Hematol. 1995;70:153-58
23. Duary RK, Batish VK, Grover S. Immunomodulatory activity of two potential probiotic strains in LPS-stimulated HT-29 cells. Genes Nutr. 2014;9:1-16
24. Holmes E, Kinross J, Gibson GR, Burcelin R, Jia W, Pettersson S. et al. Therapeutic modulation of microbiota-host metabolic interactions. Sci Transl Med. 2012;4:137rv6-137rv6
25. Marteau P, Guyonnet D, Lafaye de Micheaux P, Gelu SA. randomized, double-blind, controlled study and pooled analysis of two identical trials of fermented milk containing probiotic Bifidobacterium lactis CNCM I-2494 in healthy women reporting minor digestive symptoms. Neurogastroenterol Motil. 2013;25:331-e252
26. Mater DD, Bretigny L, Firmesse O, Flores MJ, Mogenet A, Bresson JL. et al. Streptococcus thermophilus and Lactobacillus delbrueckii subsp. bulgaricus survive gastrointestinal transit of healthy volunteers consuming yogurt. FEMS Microbiol Lett. 2005;250:185-87
27. Kebouchi M, Galia W, Genay M, Soligot C, Lecomte X, Awussi AA. et al. Implication of sortase-dependent proteins of Streptococcus thermophilus in adhesion to human intestinal epithelial cell lines and bile salt tolerance. Appl Environ Microbiol. 2016;100:3667-79
28. Mohamadzadeh M, Duong T, Sandwick SJ, Hoover T, Klaenhammer TR. Dendritic cell targeting of Bacillus anthracis protective antigen expressed by Lactobacillus acidophilus protects mice from lethal challenge. P Natl Acad Sci USA. 2009;106:4331-36
29. Kajikawa A, Zhang L, Long J, Nordone S, Stoeker L, LaVoy A. et al. Construction and immunological evaluation of dual cell surface display of HIV-1 Gag and Salmonella enterica serovar Typhimurium FliC in Lactobacillus acidophilus for vaccine delivery. Clin Vaccine Immunol. 2012;19:1374-81
30. Corcoran BM, Stanton C, Fitzgerald GF, Ross RP. Growth of probiotic lactobacilli in the presence of oleic acid enhances subsequent survival in gastric juice. Microbiology. 2007;153:291-99
31. Corcoran BM, Stanton C, Fitzgerald GF, Ross RP. Survival of probiotic lactobacilli in acidic environments is enhanced in the presence of metabolizable sugars. Appl Environ Microbiol. 2005;71:3060-67
32. Chae JP, Valeriano VD, Kim GB, Kang DK. Molecular cloning, characterization and comparison of bile salt hydrolases from Lactobacillus johnsonii PF01. J Appl Microbiol. 2013;114:121-33
33. Kim GB, Brochet M, Lee BH. Cloning and characterization of a bile salt hydrolase (bsh) from Bifidobacterium adolescentis. Biotechnol Lett. 2005;27:817-22
34. Kim GB, Yi SH, Lee BH. Purification and characterization of three different types of bile salt hydrolases from Bifidobacterium strains. J Dairy Sci. 2004;87:258-66
35. Jiang J, Hang X, Zhang M, Liu X, Li D, Yang H. Diversity of bile salt hydrolase activities in different lactobacilli toward human bile salts. Ann Microbiol. 2010;60:81-88
36. Saxami G, Karapetsas A, Lamprianidou E, Kotsianidis I, Chlichlia A, Tassou C. et al. Two potential probiotic lactobacillus strains isolated from olive microbiota exhibit adhesion and anti-proliferative effects in cancer cell lines. J Funct Foods. 2016;24:461-71
37. Kainulainen V, Tang Y, Spillmann T, Kilpinen S, Reunanen J, Saris PE. et al. The canine isolate Lactobacillus acidophilus LAB20 adheres to intestinal epithelium and attenuates LPS-induced IL-8 secretion of enterocytes in vitro. BMC Microbiol. 2015;15:4
38. Bu XD, Li N, Tian XQ, Huang PL. Caco-2 and LS174T cell lines provide different models for studying mucin expression in colon cancer. Tissue and Cell. 2011;43:201-6
39. Bennett KM, Walker SL, Lo DD. Epithelial microvilli establish an electrostatic barrier to microbial adhesion. Infect Immun. 2014;82:2860-71
40. Gerasimov SV, Vasjuta VV, Myhovych OO, Bondarchuk LI. Probiotic supplement reduces atopic dermatitis in preschool children: a randomized, double-blind, placebo-controlled, clinical trial. Am J Clin Dermatol. 2010;11:351-61
41. Gerasimov SV, Ivantsiv VA, Bobryk LM, Tsitsura OO, Dedyshin LP, Guta NV. et al. Role of short-term use of L. acidophilus DDS-1 and B. lactis UABLA-12 in acute respiratory infections in children: a randomized controlled trials. Eur J Clin Nutr. 2016;70:463-69
42. Tejero-Sariñena S, Barlowb J, Costabile A, Gibson GR, Rowland I. In vitro evaluation of the antimicrobial activity of a range of probiotics against pathogens: evidence for the effects of organic acids. Anaerobe. 2012;18:530-38
43. Schoster A, Kokotovic B, Permin A, Pedersen PD, Dal Bello F, Guardabassi L. In vitro inhibition of Clostridium difficile and Clostridium perfringens by commercial probiotic strains. Anaerobe. 2013;20:36-41
44. Ferreira dos Santos T, Alves Melo T, Almeida ME, Passos Rezende R, Romano CC. Immunomodulatory Effects of Lactobacillus plantarum Lp62 on Intestinal Epithelial and Mononuclear Cells. Biomed Res Int. 2016;2016:8404156
45. Ohkusa T, Yoshida T, Sato N, Watanabe S, Tajiri H, Okayasu I. Commensal bacteria can enter colonic epithelial cells and induce proinflammatory cytokine secretion: a possible pathogenic mechanism of ulcerative colitis. J Med Microbiol. 2009;58:535-45
46. Resta-Lenert S, Barrett KE. Probiotics and commensals reverse TNF-α-and IFN-γ-induced dysfunction in human intestinal epithelial cells. Gastroenterology. 2006;130:731-46
47. Candela M, Perna F, Carnevali P, Vitali B, Ciati R, Gionchetti P. et al. Interaction of probiotic Lactobacillus and Bifidobacterium strains with human intestinal epithelial cells: adhesion properties, competition against enteropathogens and modulation of IL-8 production. Int J Food Microbiol. 2008;125:286-92
Author contact
![]() Corresponding authors: School of Health Sciences, College of Health and Medicine, University of Tasmania, Launceston, Tasmania 7250 Australia. Tel: +61 3 6324 5467/0430029656 Fax: +61 3 6324 5555; Email address: ravichandra.vemuriedu.au; rajaraman.eriedu.au
Corresponding authors: School of Health Sciences, College of Health and Medicine, University of Tasmania, Launceston, Tasmania 7250 Australia. Tel: +61 3 6324 5467/0430029656 Fax: +61 3 6324 5555; Email address: ravichandra.vemuriedu.au; rajaraman.eriedu.au

 Global reach, higher impact
Global reach, higher impact