Impact Factor
ISSN: 1449-1907
Int J Med Sci 2018; 15(8):808-815. doi:10.7150/ijms.25140 This issue Cite
Review
Iroquois Homeodomain transcription factors in ventricular conduction system and arrhythmia
1. Department of Cardiology, the First Affiliated Hospital of China Medical University, Shenyang, Liaoning 110001, China
2. Department of Cardiology, West China Hospital of Sichuan University, Chengdu, Sichuan 610041, China
3. Department of Cardiology, Jinqiu Hosipital Of Liaoning Province, Shenyang, Liaoning110001, China
4. Department of Neurology, the First Affiliated Hospital of China Medical University, Shenyang, Liaoning 110001, China
*These two authors contributed equally to this work.
Received 2018-1-25; Accepted 2018-4-29; Published 2018-5-22
Abstract
Iroquois homeobox genes, Irx, encode cardiac transcription factors, Irx1-6 in most mammals. These six transcription factors are expressed in different patterns mainly in the ventricular part of the heart. Existing researches show that Irx genes play key roles in the differentiation and development of ventricular conduction system and the establishment and maintenance of gradient expression of potassium channels, Kv4.2. Our main focus of this review is on the recent advances in the discovery of above-mentioned genes and the function of the encoding products, how Irx genes establish ventricular conduction system and regulate ventricular repolarization, how the individual and complementary functions can be verified to complement our cognition and leads to novel therapeutic approaches.
Keywords: Arrhythmia, Ionic channels, Iroquois homeobox, Ventricular conduction system, Ventricular repolarization
Introduction
The conduction of electrocardiosignals and the synchronous contraction between ventricles with rhythmicity are essentially important for forming effective cardiac beating cycles and thus ensuring unidirectional blood flow circulation in whole bodies. As an indispensable part of the heart, ventricular conduction system (VCS) plays a vital role in coordinating the heartbeat to activate contraction from the apex and maximize efficiency of expulsion of the blood through aorta (systemic circulation) and pulmonary artery (pulmonary circulation) at the base of the heart. VCS is majorly composed of the following parts: atrioventricular bundle (AVB), left and right bundle branches (BB), and Purkinje fiber (PF) network. The deficiency or damage of this system and genetic loci involved in the pathogenesis will lead to slowed conduction velocities, abnormal activation patterns of the ventricles, different kinds of ventricular arrhythmia and even sudden cardiac death [1-3].
Homeobox genes encode various kinds of transcription factors containing homeodomains, and mutations of these genes or transcription factors can lead to dysembryoplasia, or even early embryonic death. Among which, recent studies have revealed Iroquois homeobox (Irx) family of cardiac transcription factors are important for the development of VCS[4] and heterogeneity of the ventricular repolarization of rodents and humans[5, 6]. This review focuses on the regulatory effects of these Irx transcription factors in VCS and grants a summarization of some unanswered questions about their developmental and molecular functions.
Iroquois Homeodomain transcription factors
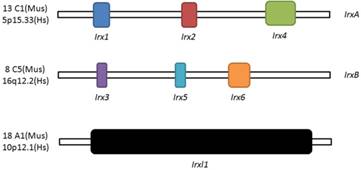
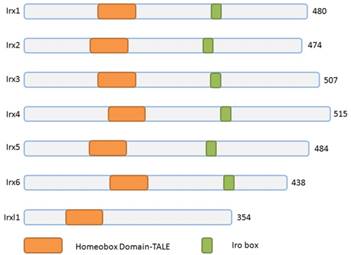
Three homeobox genes of the Irx family, caupolican, araucan and mirror were first discovered in Drosophila which play a critical role in establishing an organizing center that is essential for formation of follicle[7], head[8], eyes[9, 10], muscle[11], mesothorax[12], lateral notum[13], bristles on the thorax[14] and heart development[15] and specification of the veins in the wing and associated sensory organs[16-19]. Irx gene family consists of Irx1, Irx2 and Irx4 in the IrxA cluster, and Irx3, Irx5 and Irx6 in the IrxB cluster in mouse and human[20] (Figure 1), 11 genes, Ziro1a, 2a, 4a of IrxAa, Ziro1b, 4b of IrxAb, Ziro3a, 5a, 6a of IrxBa, Ziro3b, 5b of IrxBb and Ziro7 in Zebrafish[21-24]. In vertebrates, Irx gene family members encode highly conserved homeodomain-containing transcription factors of a 13 amino-acid domain (the Iro box) and the 3-amino-acid-loop-extension (TALE) family[25] (Figure 2), which are required for the opportune patterning and formation of the nervous system[26-31] and the heart[32], specification of the membranous labyrinth[33] and nephron segment fate during kidney development [34-36], lung development and maturation[37], pancreatic, retinal and microvascular endothelial cell development and function[38-40], female gonad[41] and early limb development[42] and also give an advantage to morbid obesity, type 2 diabetes mellitus [43], fused toes[44] and tumor cells[45]. In the heart, all the Irx transcription factors are expressed in unique and overlapping patterns, abnormality or absence of encoding Irx may cause congenital structural heart disease[46], cardiomyopathy[47, 48] and a variety of arrhythmia[49]. The present article focuses on their roles in VCS development, ionic channels maintenance and thus involvement in arrhythmia as below (Table 1).
Formation of ventricular conduction system during cardiac development
Before heart becomes developmentally mature, the electric signals generated from pacemaker tissue can be slowly transmitted through the ventral and dorsal parts of atrioventricular canal (AVC) between embryonic atrium and ventricle[50]. After the cardiac chambers have been divided, the fibrotic nonconductors consist mainly of atrioventricular cushion and extraepicardial mesenchymal tissues shield electric conduction between atrium and ventricle, with the exclusive atrioventricular electric channel, AVB. AVB is part of the VCS, deriving from interventricular septum crista and physically connecting with AVC and atrioventricular node (AVN). With the further development of interventricular septum, both left and right BB develop inch by inch and form from AVB branches under endocardium[51]. The cells forming AVB and BB continue to feature in primitive cardiomyocytes and are more like the genetic and electrophysiological phenotypes of AVN. This specific lineage of cardiomyocytes can substitute for the upper positioned pacemaker tissues to discharge rhythmic electric impulses, junctional rhythms, during atrioventricular block. Different than AVN, Cx40 is enriched between the cells from these tissues, leading to relatively fast conduction velocity.
Within the maturely developed heart, the distal BB and PF network are the tissues containing only several layers of cells, and they constitute the rapid intraventricular conduction system with the maturation of endocardial trabecula[52]. Even in the lower vertebrates, similar conduction characteristics can also be recorded[53]. During the developmental process of embryonic ventricles, the regions specifically expressing Cx40, Nppa and TASK-1 in the heart are gradually restricted within the trabecula tissues, and form peripheral ventricular conduction system simultaneously with the cardiac pericardium linking layer not expressing the above-mentioned ventricular chamber and conduction system specific molecular markers. With the Nkx2.5 regulating the cardiac developmental maturation, the trabecular region of myocardium finally composes the peripheral ventricular conduction system in terms of structure and function.
Schematic illustration of Irx and Irxl1 genes. Location and organization of Irx and Irxl1 genes in mouse (Mus) and human (Hs).
Schematic representation of Irx and Irxl1 proteins. The Irx, Irxl1 proteins and homeobox domains are depicted according to the size. TALE, 3-amino acid-loop-extension.
Expression, function and associated phenotype of Irx genes in conduction system.
| Gene | Cluster | Chromosomal position | Expression ranges | Physiological function | Associated phenotype |
|---|---|---|---|---|---|
| Irx1 | A | 13 C1(Mus) 5p15.33(Hs) | E10.5, interventricular septum trabecular E11.5, ventricular septum E14.5, regions of the developing VCS, including AVB and BB | Potentially involved the development of VCS | Irx1b-MO in zebrafish, bradycardia |
| Irx2 | A | 13 C1(Mus) 5p15.33(Hs) | identical to Irx1 | Not been verified | None |
| Irx3 | B | 8 C5(Mus) 16q12.2(Hs) | From E9.5, trabeculated component of the ventricles | Establish fast conduction in the VCS | Irx3-KO mice, prolonged QRS, notched R waves and RBBB on ECG |
| Irx4 | A | 13 C1(Mus) 5p15.33(Hs) | E7.5, anterior domain of the linear heart; E9.5, atrioventricular canal, inner curvature, and proximal outflow tract; E11.5, ventricular myocardium. | Regulate chamber-specific gene expression, act as a multifunctional co-factor | No defect in cardiacelectricalactivity |
| Irx5 | B | 8 C5(Mus) 16q12.2(Hs) | E9, heart tube ventricle; E9.5, endocardial chamber myocardium (atrioventricular canal, inner curvature, and outflow track); E14.5, ventricular trabeculae, AVB and BB. | Establish and maintain the gradients of Ito,f in ventricles | Irx5-/- mice, T-wave alterations |
| Irx6 | B | 8 C5(Mus) 16q12.2(Hs) | E10.5, endocardial lining of atrial and ventricular chambers | Not been verified | None |
| Irxl1 | 18 A1(Mus) 10p12.1(Hs) | No expression found in the heart | Not been verified | No defect in cardiacelectricalactivity |
Expression of Irx and roles in the VCS
During the development process of heart, Irx1 expression could be detected as early as E10.5 in the ventricular septal trabecular myocardium, which is restricted to the ventricular septum at E11.5, and expands to the developing VCS regions from E14.5 onward, including the AVB and BB [54] (Figure 3), which implies that Irx1 may be involved in the development of VCS. Zebrafish, after injection of a morpholino oligonucleotide designed to knockdown Ziro1b, a gene homologous to Irx1, exhibits profound dose-dependent bradycardia, with slower heart rate reduced by 35% of the normal control[55]. Further studies should be carried out to evaluate the functions of Irx1 knockout (KO) in conduction system.
Expression distribution of Irx transcription factors in the heart. Schematic illustration of Irx transcription factors expression in the embryonic and mature hearts.
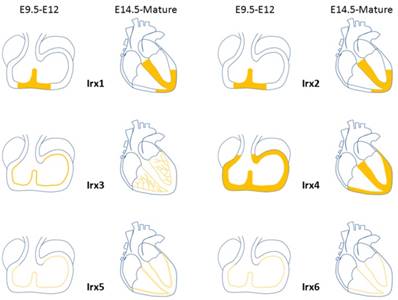
Expression pattern of Irx2 is nearly identical to that of Irx1 [54] (Figure 3). Despite the high expression in the developing heart, Irx2 KO mice are fertile and well-grown, and show no phenotype. And Electrocardiographic analysis of 8- to 10-week-old mice revealed no difference between Irx2-/- mice and wildtype controls[56], implying that Irx2 is unessential to the cardiac conduction system, probably as a result of functional redundancy of other Irx genes.
In embryonic hearts, Irx3 can be detected in regions contributing to the VCS, such as the ventricular septum and trabeculae at E9.5 [54, 57]. Irx3 expression is overlapped in the mature heart with Cx40, a major functional gap junction protein of the AVB, BB, and PF of the VCS[51, 58-60], and accurately labels the mature VCS[4] (Figure 3). Irx3-null mice show QRS complex duration prolongation, notched R waves (R') on an electrocardiogram and an increase of HV interval (the conduction time between the AVB and ventricles) in electrophysiological examination, all indicative of prolonged conduction times in the VCS, by 2 weeks of age[4]. And telemetry ECG displays right axis deviation in 70% of Irx3 KO mice, consistent with right bundle-branch block (RBBB) or abnormal impulse conduction in the right ventricular free wall as published in dog and mouse studies previously[59-61]. The impaired conduction in the VCS is identified with decreased expression of Cx40, which forms gap junction channels with a high conductance and is expressed in all compartments of the fast conduction system, from the AVB, left and right BB, to the whole PF network[60, 62]. Meanwhile, ectopic expression of Cx43, characterize the nodal conduction structures with slow velocity[63], was also verified in the proximal VCS with a direct coupling to the septal myocardium. In accordance with the results that Irx3 activates Cx40 expression while inhibits Cx43. These results suggest that transcription of Cx40 is activated by Irx3 repressing an unidentified repressor indirectly, nevertheless, expression of Cx43 is likely to be suppressed by Irx3 directly through binding to the Cx43 promoter and antagonizing Nkx2.5 dependent activation, which is required for the postnatal differentiation of VCS[64, 65]. Thus, Irx3 plays a key role for maintaining rapid electric conduction through the VCS as well as proper ventricular activation. It is still unclear that whether deficiency in the VCS lacking Irx3 would induce the changes of the expression of gap junction expression located in the VCS. Additionally, optical mapping utilizing intact Zebrafish expressing the in vivo cardiac-specific fluorescent calcium indicator Tg(cmlc2:gCaMP)s878 showed that knockdown of Ziro3a led to low conduction velocity and asynchronization[66, 67]. Aforementioned results illuminate that Irx3 is required for normal ventricular activation and impulse conduction.
In early stage of heart development at E7.5, Irx4 starts to be found in the anterior cardiac primordia, and in atrioventricular canal, inner curvature, ventricles, and proximal outflow tract at E9.5. Irx4 does not show any expression until E11.5 in the myocardial but not in non-myocardial cushion cells located in the atrioventricular canal. After E11.5, expression of Irx4 is restricted to the ventricular myocardium while absent from both atria and the outflow tract [54] (Figure 3). Unlike its involvement in establishing chamber-specific gene expression and differentiation of atrial and ventricular myocytes in the developmental heart [68-70], there is no direct evidence that Irx4 participates in the differentiation and development of VCS.
Irx5 expression was first detected at E9 at the ventral side of the looping heart tube. At E9.5, Irx5 started to be observed in the endocardial lining of the atrial and ventricular chamber myocardium, conversely, it is absent from the endocardium of the atrioventricular canal, inflow tract, inner curvature, outflow tract, and endothelium of the aortic arch arteries [54]. By E14.5, expression region of Irx5 in ventricles is confined to the ventricular trabeculae, AVB and BB[57] (Figure 3). The pattern of expression remained unchanged during expansion of the chambers [54]. Irx5 expresses in the region of immature AVB and BB, which opens the possibility of the sealed regulatory effect of the developing VCS.
Irx6's level of expression is much lower compared to the other five Irx genes and is detected first at E10.5 in the heart[71]. Expression pattern of Irx6 is restricted to the endocardial lining of atrial and ventricular chambers, similar to Irx5 (Figure 3). The function of Irx6 in VCS has not been verified.
Actually, there is another divergent Irx family transcription factor gene Irxl1, Iroquois homeobox-like 1, or Mkx, Mohawk, which also belongs to the TALE (three-amino-acid loop extension) superclass of the atypical homeobox gene family[72] (Figure 1). Irxl1/Mkx encodes transcription factor Irxl1 (Figure 2) that regulates the expression of matrix molecule genes and differentiation in developing tendons[73-76], including type 1 collagen and Tenomodulin, volar plates[77], homeostasis of the periodontal ligament [78], MyoD expression and myoblast differentiation [79], brain and pharyngeal arch morphogenesis[80] and participates in some pathological process, such as, diabetic tendinopathy[81], anterior cruciate ligament degradation[82], serous ovarian cancer[83], and Rett Syndrome[84]. Mouse Mkx shares 56% homology with Irx2 over the entire homeodomain, though, no expression found in the heart.
Irx in cardiacelectricalactivity and ionic channel
In mammalian mature heart, both Irx3 and Irx5 are expressed in the ventricles with transmural, endocardium to epicardium gradients [5, 85-87]. Irx5-/- mice exhibit abnormal ECG characterized by T-wave alterations, whereas heart rate, atrioventricular (PR interval) and intraventricular conduction (QRS duration) are not affected, consistent with accelerated repolarization and reduced ventricular refractoriness [5]. Depression of T-wave originated from ventricular repolarization abnormity which in mice is attributed mainly to gradients of the fast transient outward K+ currents, Ito,f[88-91]. Ito,f in the rodent heart is initiated mainly by two voltage-gated potassium (Kv) channels, Kv4.2 and Kv4.3[92], along with KChIP2, an accessory subunit [93-95], and cardiovascular disease-related[96, 97]. In Irx5 KO mice, the Ito,f gradient in ventricular walls flattens as a result of selective increases in Ito,f and Kv4.2 along with shorter action potential duration in endocardium[5, 87]. The inverse relationship between Irx5 and Kv4.2 expression suggests that an Irx5 repressor gradient negatively regulates Kv4.2 gene expression, forms an opposite gradient of Ito,f and coordinates repolarization in the heart. Also, we should note that Irx5 can act as either an activator or repressor in a cell-type-dependent manner. For instance, the C-terminal portion of Irx5 physically binds to the N-terminal region of Irx4, through which Irx4 could suppress Kv4.2 gene promoter activation mediated by Irx5 in non-cardiomyocytes, like 10T1/2 fibroblastic and PC12 cells. Further tests demonstrate endogenous Irx4 inhibits the function of transfected Irx5 to increase Kv4.2 promoter activity which degree may be determined by the expression ratio of Irx5/Irx4 [6]. This contradictorily activated or repressive impact of Irx5 in cardiomyocytes or non-cardiomyocytes is gained from recruitment of mBop (Smyd1), which has been verified as a cardiac corepressor essential for cardiomyocyte differentiation and cardiac morphogenesis[98, 99]. However, physical interaction between Irx5 and mBop might not exist, nor affect Irx5-mediated Kcnd2 promoter activation [6], this is just contrary to previous conclusion that Irx5 represses Kv4.2 expression through mBop recruitment [5]. Therefore, further studies will be necessary to examine whether and how Irx5, Irx4 and mBop are interacting to regulate Kv4.2 expression.
Recent studies have revealed crucial roles of miRNAs as regulators of the development and function of the heart [100-103]. Irx gene expression is also regulated by miRNAs in the heart, including miR-1,which is highly conserved and enriched in heart and encoded by two separate genes, miR-1-1 and miR-1-2[104, 105], and has important pathophysiological functions (regulates cardiac arrhythmogenic potential) in the heart[106]. There is a miR-1 binding site in the 3`UTR of Irx5 , and in miR-1-2-/- mice, Irx4 and Irx5 expression increase in the heart, which exhibit lower heart rate, prolonged PR interval and QRS complex, consistent with the observations in Irx5 KO mice, a decrease in Kcnd2 along with increased Irx5 expression[107].
Irx and arrhythmia
As the Irx genes are critical for efficient conduction in the VCS development, defects of Irx genes may be associated with arrhythmia. Loss of Irx3 leads to demolition of the rapid concerted spread of excitation in ventricles, prolonged QRS, notched R waves (R') and an increase of HV interval, right bundle branch block to be specific, which is considered to be associated with increase of mortality in patients with acute myocardial infarction[108], and a reasonable candidate of cardiac resynchronization therapy for heart failure patients with evidence of either electrical or mechanical left-sided delay[109].
Among the many factors can impact cardiac rhythms and cause arrhythmias, ion channel abnormality and defect in repolarization could contribute [2]. Establishment and maintenance of electric transmural gradients play a key role in ventricular repolarization. Irregular myocardium repolarization is associated with many kinds of malignant arrhythmia, including torsades de pointes (in long-QT, short-QT and Brugada syndromes), ventricular tachycardia and fibrillation. The Ito,f gradient in ventricular walls reduces and ventricular tachyarrhythmia could be reproducibly induced by intracardiac programmed stimulation in Irx5 KO mice, consistent with alterations in ventricular repolarization to induce arrhythmia, so there is a fair speculation that defects of IRX5 may be related to cardiac arrhythmia in human. [5] These findings suggest that loss of the ventricular transmural gradient of Ito,f in Irx5-/- hearts is the major cause of enhanced arrhythmia susceptibility, or simply the shorten of ventricular refractory period. It will clearly be of interest to further explore the increased pathogenesis of arrhythmias in the Irx5 KO mice.
Irx as new therapy for clinical applications
Recently, Ban et al. developed a method of profound significance to isolate pure ventricular cardiomyocytes by targeting the mRNA of Irx4, with high specificity using molecular beacons from differentiating human and mouse pluripotent stem cells[110]. This approach of producing a pure population of working cardiomyocytes offers a new option for cell therapy of heart failure and arrhythmia caused by it, and can be used to model the abnormal functional phenotype of genetic or idiopathic cardiac diseases, such as long QT syndrome, and to identify potential new therapeutic agents. For ischemic heart disease, the proangiogenic function of Irx3 in human microvascular endothelial cells opens new avenues for basic research and clinical applications[40].
Discussion
The six Irx genes of mammals are mainly distributed in the ventricles during embryonic development, including the parts that develop into the conduction system. Although only Irx3 has been confirmed to participate in the establishment of the ventricular conduction system, other Irx genes have not been shown any abnormal phenotypes in the conduction system after the individual knockout (normal ECG characteristics), given the overlap and functional redundancy of these six genes, more often function as the form of two clusters, A and B[111], wherein Irx genes are closely related to each other, and regulate certain physiological and pathological processes in contradictory ways, so it's very likely that all these six genes play certain roles in the development of the VCS. Even if it is difficult to identify individual genes alone, it is still necessary to conduct further experiments through multiple gene editing methods to find specific effects of some or certain cluster of Irx genes.
In the same way, like the previous experiment, Irx3 indirectly promotes Cx40 expression by combining with an unknown factor in the development of conduction system, Irx4 plays an important role in the process of cardiac compartmentation by combination with Vitamin D, the interaction between Irx4, Irx5 and mBop has yet to be clarified, and in addition, in recent years, the understanding of the three-dimensional structure of Irx gene clusters has been further deepened. In the process of verifying the function of individual Irx genes, it also needs to verify the function of the complexes and their structures.
In an interesting study, the gender-dependent effects of Irx genes on ionic channels have been verified[86], it may be possible to explain that female gender is associated with a higher risk of torsades de pointes than the opposite sex[112]. And further research in such gender difference could blaze new trails on the basis of that Irx3 gene was confirmed to be involved in the development of the female sex gland[41] and several sex hormones, including progesterone, estradiol and testosterone, rained influence on ionic currents, ventricular action potential and QTc interval duration[112, 113].
Irx genes play critical roles in the differentiation development of VCS and maintenance of normal cardiacelectricalactivity. With the in depth development of genome-editing and stem cell technologies, people will gain in-depth understanding of the development process of VCS, the mechanisms of various cardiac arrhythmia and regulation pathways involved, the clinical application of Irx gene family as a genomic target of arrhythmia will develop ideas for initiating new therapeutic methods.
Abbreviations
VCS: ventricular conduction system; AVB: atrioventricular bundle; BB: bundle branches; PF: Purkinje fiber; Irx: Iroquois homeobox; TALE: 3-amino-acid-loop-extension; AVC: atrioventricular canal; AVN: atrioventricular node; KO: knockout; RBBB: right bundle-branch block; Kv: voltage-gated potassium channel.
Acknowledgements
The authors gratefully acknowledge financial support from China Scholarship Council. We thank Ms. Yumeng Hu for her inspiration and being cute.
Competing Interests
The authors have declared that no competing interest exists.
References
1. Priori SG, Napolitano C. Genetics of cardiac arrhythmias and sudden cardiac death. Ann N Y Acad Sci. 2004;1015:96-110
2. Antzelevitch C. Role of spatial dispersion of repolarization in inherited and acquired sudden cardiac death syndromes. Am J Physiol Heart Circ Physiol. 2007;293:H2024-38
3. Sarkozy A, Brugada P. Sudden cardiac death and inherited arrhythmia syndromes. J Cardiovasc Electrophysiol. 2005;16(Suppl 1):S8-20
4. Zhang SS, Kim KH, Rosen A, Smyth JW, Sakuma R, Delgado-Olguin P. et al. Iroquois homeobox gene 3 establishes fast conduction in the cardiac His-Purkinje network. Proc Natl Acad Sci U S A. 2011;108:13576-81
5. Costantini DL, Arruda EP, Agarwal P, Kim KH, Zhu Y, Zhu W. et al. The homeodomain transcription factor Irx5 establishes the mouse cardiac ventricular repolarization gradient. Cell. 2005;123:347-58
6. He W, Jia Y, Takimoto K. Interaction between transcription factors Iroquois proteins 4 and 5 controls cardiac potassium channel Kv4.2 gene transcription. Cardiovasc Res. 2009;81:64-71
7. Jordan KC, Clegg NJ, Blasi JA, Morimoto AM, Sen J, Stein D. et al. The homeobox gene mirror links EGF signalling to embryonic dorso-ventral axis formation through Notch activation. Nat Genet. 2000;24:429-33
8. Kiritooshi N, Yorimitsu T, Shirai T, Puli OR, Singh A, Nakagoshi H. A vertex specific dorsal selector Dve represses the ventral appendage identity in Drosophila head. Mech Dev. 2014;133:54-63
9. Dominguez M, de Celis JF. A dorsal/ventral boundary established by Notch controls growth and polarity in the Drosophila eye. Nature. 1998;396:276-8
10. McNeill H, Yang CH, Brodsky M, Ungos J, Simon MA. Mirror encodes a novel PBX-class homeoprotein that functions in the definition of the dorsal-ventral border in the Drosophila eye. Genes Dev. 1997;11:1073-82
11. Carrasco-Rando M, Tutor AS, Prieto-Sanchez S, Gonzalez-Perez E, Barrios N, Letizia A. et al. Drosophila araucan and caupolican integrate intrinsic and signalling inputs for the acquisition by muscle progenitors of the lateral transverse fate. PLoS Genet. 2011;7:e1002186
12. Cavodeassi F, Modolell J, Gomez-Skarmeta JL. The Iroquois family of genes: from body building to neural patterning. Development. 2001;128:2847-55
13. Ikmi A, Netter S, Coen D. Prepatterning the Drosophila notum: the three genes of the iroquois complex play intrinsically distinct roles. Dev Biol. 2008;317:634-48
14. Leyns L, GomezSkarmeta JL, DamblyChaudiere C. iroquois: A prepattern gene that controls the formation of bristles on the thorax of Drosophila. Mech Dev. 1996;59:63-72
15. Singh A, Mirzoyan Z, Pandur P. The Iroquois Complex Is Required in the Dorsal Mesoderm to Ensure Normal Heart Development in Drosophila. PLoS One. 2013;8:e76498
16. GomezSkarmeta JL, Modolell J. araucan and caupolican provide a link between compartment subdivisions and patterning of sensory organs and veins in the Drosophila wing. Genes Dev. 1996;10:2935-45
17. GomezSkarmeta JL, Diez del Corral R, delaCalleMustienes E, FerresMarco D, Modolell J. araucan and caupolican, two members of the novel iroquois complex, encode homeoproteins that control proneural and vein-forming genes. Cell. 1996;85:95-105
18. Bilioni A, Craig G, Hill C, McNeill H. Iroquois transcription factors recognize a unique motif to mediate transcriptional repression in vivo. Proc Natl Acad Sci U S A. 2005;102:14671-6
19. Kehl BT, Cho KO, Choi KW. Mirror, a Drosophila homeobox gene in the iroquois complex, is required for sensory organ and alula formation. Development. 1998;125:1217-27
20. Peters T, Dildrop R, Ausmeier K, Ruther U. Organization of mouse Iroquois homeobox genes in two clusters suggests a conserved regulation and function in vertebrate development. Genome Res. 2000;10:1453-62
21. Kerner P, Ikmi A, Coen D, Vervoort M. Evolutionary history of the iroquois/Irx genes in metazoans. BMC Evol Biol. 2009;9:74
22. Feijoo CG, Manzanares M, de la Calle-Mustienes E, Gomez-Skarmeta JL, Allende ML. The Irx gene family in zebrafish: genomic structure, evolution and initial characterization of irx5b. Dev Genes Evol. 2004;214:277-84
23. Dildrop R, Ruther U. Organization of Iroquois genes in fish. Dev Genes Evol. 2004;214:267-76
24. de la Calle-Mustienes E, Feijoo CG, Manzanares M, Tena JJ, Rodriguez-Seguel E, Letizia A. et al. A functional survey of the enhancer activity of conserved non-coding sequences from vertebrate Iroquois cluster gene deserts. Genome Res. 2005;15:1061-72
25. Burglin TR. Analysis of TALE superclass homeobox genes (MEIS, PBC, KNOX, Iroquois, TGIF) reveals a novel domain conserved between plants and animals. Nucleic Acids Res. 1997;25:4173-80
26. Lecaudey V, Anselme I, Dildrop R, Ruther U, Schneider-Maunoury S. Expression of the zebrafish Iroquois genes during early nervous system formation and patterning. J Comp Neurol. 2005;492:289-302
27. Bosse A, Stoykova A, Nieselt-Struwe K, Chowdhury K, Copeland NG, Jenkins NA. et al. Identification of a novel mouse iroquois homeobox gene, Irx5, and chromosomal localisation of all members of the mouse iroquois gene family. Dev Dyn. 2000;218:160-74
28. Gomez-Skarmeta JL, Modolell J. Iroquois genes: genomic organization and function in vertebrate neural development. Curr Opin Genet Dev. 2002;12:403-8
29. Lecaudey V, Anselme I, Rosa F, Schneider-Maunoury S. The zebrafish Iroquois gene iro7 positions the r4/r5 boundary and controls neurogenesis in the rostral hindbrain. Development. 2004;131:3121-31
30. de la Calle-Mustienes E, Glavic A, Modolell J, Gomez-Skarmeta JL. Xiro homeoproteins coordinate cell cycle exit and primary neuron formation by upregulating neuronal-fate repressors and downregulating the cell-cycle inhibitor XGadd45-gamma. Mech Dev. 2002;119:69-80
31. Matsumoto K, Nishihara S, Kamimura M, Shiraishi T, Otoguro T, Uehara M. et al. The prepattern transcription factor Irx2, a target of the FGF8/MAP kinase cascade, is involved in cerebellum formation. Nat Neurosci. 2004;7:605-12
32. Bruneau BG, Bao ZZ, Tanaka M, Schott JJ, Izumo S, Cepko CL. et al. Cardiac expression of the ventricle-specific homeobox gene Irx4 is modulated by Nkx2-5 and dHand. Dev Biol. 2000;217:266-77
33. Cardena-Nunez S, Sanchez-Guardado LO, Corral-San-Miguel R, Rodriguez-Gallardo L, Marin F, Puelles L. et al. Expression patterns of Irx genes in the developing chick inner ear. Brain Struct Funct. 2017;222:2071-92
34. Schwab K, Hartman HA, Liang HC, Aronow BJ, Patterson LT, Potter SS. Comprehensive microarray analysis of Hoxa11/Hoxd11 mutant kidney development. Dev Biol. 2006;293:540-54
35. Alarcon P, Rodriguez-Seguel E, Fernandez-Gonzalez A, Rubio R, Gomez-Skarmeta JL. A dual requirement for Iroquois genes during Xenopus kidney development. Development. 2008;135:3197-207
36. Reggiani L, Raciti D, Airik R, Kispert A, Brandli AW. The prepattern transcription factor Irx3 directs nephron segment identity. Genes Dev. 2007;21:2358-70
37. van Tuyl M, Liu J, Groenman F, Ridsdale R, Han RN, Venkatesh V. et al. Iroquois genes influence proximo-distal morphogenesis during rat lung development. Am J Physiol Lung Cell Mol Physiol. 2006;290:L777-L89
38. Cheng CW, Chow RL, Lebel M, Sakuma R, Cheung HO, Thanabalasingham V. et al. The Iroquois homeobox gene, Irx5, is required for retinal cone bipolar cell development. Dev Biol. 2005;287:48-60
39. Cheng CW, Yan CH, Hui CC, Strahle U, Cheng SH. The homeobox gene irx1a is required for the propagation of the neurogenic waves in the zebrafish retina. Mech Dev. 2006;123:252-63
40. Scarlett K, Pattabiraman V, Barnett P, Liu D, Anderson LM. The proangiogenic effect of iroquois homeobox transcription factor Irx3 in human microvascular endothelial cells. J Biol Chem. 2015;290:6303-15
41. Jorgensen JS, Gao L. Irx3 is differentially up-regulated in female gonads during sex determination. Gene Expr Patterns. 2005;5:756-62
42. McDonald LA, Gerrelli D, Fok Y, Hurst LD, Tickle C. Comparison of Iroquois gene expression in limbs/fins of vertebrate embryos. J Anat. 2010;216:683-91
43. Ragvin A, Moro E, Fredman D, Navratilova P, Drivenes O, Engstrom PG. et al. Long-range gene regulation links genomic type 2 diabetes and obesity risk regions to HHEX, SOX4, and IRX3. Proc Natl Acad Sci U S A. 2010;107:775-80
44. Peters T, Ausmeier K, Dildrop R, Ruther U. The mouse Fused toes (Ft) mutation is the result of a 1.6-Mb deletion including the entire Iroquois B gene cluster. Mamm Genome. 2002;13:186-8
45. Martorell O, Barriga FM, Merlos-Suarez A, Stephan-Otto Attolini C, Casanova J, Batlle E. et al. Iro/IRX transcription factors negatively regulate Dpp/TGF-beta pathway activity during intestinal tumorigenesis. EMBO Rep. 2014;15:1210-8
46. Wang GF, Nikovits W Jr, Bao ZZ, Stockdale FE. Irx4 forms an inhibitory complex with the vitamin D and retinoic X receptors to regulate cardiac chamber-specific slow MyHC3 expression. J Biol Chem. 2001;276:28835-41
47. Bruneau BG, Bao ZZ, Fatkin D, Xavier-Neto J, Georgakopoulos D, Maguire CT. et al. Cardiomyopathy in Irx4-deficient mice is preceded by abnormal ventricular gene expression. Mol Cell Biol. 2001;21:1730-6
48. Beisvag V, Lehre PK, Midelfart H, Aass H, Geiran O, Sandvik AK. et al. Aetiology-specific patterns in end-stage heart failure patients identified by functional annotation and classification of microarray data. Eur J Heart Fail. 2006;8:381-9
49. Koopmann TT, Beekman L, Alders M, Meregalli PG, Mannens MM, Moorman AF. et al. Exclusion of multiple candidate genes and large genomic rearrangements in SCN5A in a Dutch Brugada syndrome cohort. Heart Rhythm. 2007;4:752-5
50. Valderrabano M, Chen F, Dave AS, Lamp ST, Klitzner TS, Weiss JN. Atrioventricular ring reentry in embryonic mouse hearts. Circulation. 2006;114:543-9
51. Miquerol L, Beyer S, Kelly RG. Establishment of the mouse ventricular conduction system. Cardiovasc Res. 2011;91:232-42
52. Rentschler S, Vaidya DM, Tamaddon H, Degenhardt K, Sassoon D, Morley GE. et al. Visualization and functional characterization of the developing murine cardiac conduction system. Development. 2001;128:1785-92
53. Sedmera D, Reckova M, deAlmeida A, Sedmerova M, Biermann M, Volejnik J. et al. Functional and morphological evidence for a ventricular conduction system in zebrafish and Xenopus hearts. Am J Physiol Heart Circ Physiol. 2003;284:H1152-60
54. Christoffels VM, Keijser AG, Houweling AC, Clout DE, Moorman AF. Patterning the embryonic heart: identification of five mouse Iroquois homeobox genes in the developing heart. Dev Biol. 2000;224:263-74
55. Joseph EM. Zebrafish IRX1b in the embryonic cardiac ventricle. Dev Dyn. 2004;231:720-6
56. Lebel M, Agarwal P, Cheng CW, Kabir MG, Chan TY, Thanabalasingham V. et al. The Iroquois Homeobox Gene Irx2 Is Not Essential for Normal Development of the Heart and Midbrain-Hindbrain Boundary in Mice. Mol Cell Biol. 2003;23:8216-25
57. Gaborit N, Sakuma R, Wylie JN, Kim KH, Zhang SS, Hui CC. et al. Cooperative and antagonistic roles for Irx3 and Irx5 in cardiac morphogenesis and postnatal physiology. Development. 2012;139:4007-19
58. Van Veen TAB. Differences in the Pattern of Ventricular Activation in Small Rodents Determined by Morphological Organization of the Cardiac Ventricular Conduction System. The Chinese Journal of Physiology. 2010;53:52-61
59. Tamaddon HS, Vaidya D, Simon AM, Paul DL, Jalife J, Morley GE. High-resolution optical mapping of the right bundle branch in connexin40 knockout mice reveals slow conduction in the specialized conduction system. Circ Res. 2000;87:929-36
60. van Rijen HV, van Veen TA, van Kempen MJ, Wilms-Schopman FJ, Potse M, Krueger O. et al. Impaired conduction in the bundle branches of mouse hearts lacking the gap junction protein connexin40. Circulation. 2001;103:1591-8
61. Kawasuji M, Iwa T. Spread of the epicardial excitation in right bundle branch block pattern. Jpn Circ J. 1978;42:1041-56
62. Gros D, Dupays L, Alcolea S, Meysen S, Miquerol L, Theveniau-Ruissy M. Genetically modified mice: tools to decode the functions of connexins in the heart-new models for cardiovascular research. Cardiovasc Res. 2004;62:299-308
63. Franco D, Icardo JM. Molecular characterization of the ventricular conduction system in the developing mouse heart: topographical correlation in normal and congenitally malformed hearts. Cardiovasc Res. 2001;49:417-29
64. Meysen S, Marger L, Hewett KW, Jarry-Guichard T, Agarkova I, Chauvin JP. et al. Nkx2.5 cell-autonomous gene function is required for the postnatal formation of the peripheral ventricular conduction system. Dev Biol. 2007;303:740-53
65. Kasahara H. Nkx2.5 homeoprotein regulates expression of gap junction protein connexin 43 and sarcomere organization in postnatal cardiomyocytes. J Mol Cell Cardiol. 2003;35:243-56
66. Kim KH, Rosen A, Bruneau BG, Hui CC, Backx PH. Iroquois homeodomain transcription factors in heart development and function. Circ Res. 2012;110:1513-24
67. Chi NC, Shaw RM, Jungblut B, Huisken J, Ferrer T, Arnaout R. et al. Genetic and physiologic dissection of the vertebrate cardiac conduction system. PLoS Biol. 2008;6:e109
68. Bao ZZ, Bruneau BG, Seidman JG, Seidman CE, Cepko CL. Regulation of chamber-specific gene expression in the developing heart by Irx4. Science. 1999;283:1161-4
69. Cheng Z, Wang J, Su D, Pan H, Huang G, Li X. et al. Two novel mutations of the IRX4 gene in patients with congenital heart disease. Hum Genet. 2011;130:657-62
70. Zhang Q, Jiang J, Han P, Yuan Q, Zhang J, Zhang X. et al. Direct differentiation of atrial and ventricular myocytes from human embryonic stem cells by alternating retinoid signals. Cell Res. 2011;21:579-87
71. Mummenhoff J, Houweling AC, Peters T, Christoffels VM, Ruther U. Expression of Irx6 during mouse morphogenesis. Mech Dev. 2001;103:193-5
72. Mukherjee K, Burglin TR. Comprehensive analysis of animal TALE homeobox genes: new conserved motifs and cases of accelerated evolution. J Mol Evol. 2007;65:137-53
73. Suzuki H, Ito Y, Shinohara M, Yamashita S, Ichinose S, Kishida A. et al. Gene targeting of the transcription factor Mohawk in rats causes heterotopic ossification of Achilles tendon via failed tenogenesis. Proc Natl Acad Sci U S A. 2016;113:7840-5
74. Liu W, Watson SS, Lan Y, Keene DR, Ovitt CE, Liu H. et al. The atypical homeodomain transcription factor Mohawk controls tendon morphogenesis. Mol Cell Biol. 2010;30:4797-807
75. Liu H, Zhang C, Zhu S, Lu P, Zhu T, Gong X. et al. Mohawk promotes the tenogenesis of mesenchymal stem cells through activation of the TGFbeta signaling pathway. Stem Cells. 2015;33:443-55
76. Ito Y, Toriuchi N, Yoshitaka T, Ueno-Kudoh H, Sato T, Yokoyama S. et al. The Mohawk homeobox gene is a critical regulator of tendon differentiation. Proc Natl Acad Sci U S A. 2010;107:10538-42
77. Onizuka N, Ito Y, Inagawa M, Nakahara H, Takada S, Lotz M. et al. The Mohawk homeobox transcription factor regulates the differentiation of tendons and volar plates. J Orthop Sci. 2014;19:172-80
78. Koda N, Sato T, Shinohara M, Ichinose S, Ito Y, Nakamichi R. et al. The transcription factor mohawk homeobox regulates homeostasis of the periodontal ligament. Development. 2017;144:313-20
79. Chuang HN, Hsiao KM, Chang HY, Wu CC, Pan H. The homeobox transcription factor Irxl1 negatively regulates MyoD expression and myoblast differentiation. FEBS J. 2014;281:2990-3003
80. Chuang HN, Cheng HY, Hsiao KM, Lin CW, Lin ML, Pan H. The zebrafish homeobox gene irxl1 is required for brain and pharyngeal arch morphogenesis. Dev Dyn. 2010;239:639-50
81. Wu YF, Wang HK, Chang HW, Sun J, Sun JS, Chao YH. High glucose alters tendon homeostasis through downregulation of the AMPK/Egr1 pathway. Sci Rep. 2017;7:44199
82. Nakahara H, Hasegawa A, Otabe K, Ayabe F, Matsukawa T, Onizuka N. et al. Transcription factor Mohawk and the pathogenesis of human anterior cruciate ligament degradation. Arthritis Rheum. 2013;65:2081-9
83. Tomar T, Alkema NG, Schreuder L, Meersma GJ, de Meyer T, van Criekinge W. et al. Methylome analysis of extreme chemoresponsive patients identifies novel markers of platinum sensitivity in high-grade serous ovarian cancer. BMC Med. 2017;15:116
84. Miyake K, Yang C, Minakuchi Y, Ohori K, Soutome M, Hirasawa T. et al. Comparison of Genomic and Epigenomic Expression in Monozygotic Twins Discordant for Rett Syndrome. PLoS One. 2013;8:e66729
85. Rosati B, Grau F, McKinnon D. Regional variation in mRNA transcript abundance within the ventricular wall. J Mol Cell Cardiol. 2006;40:295-302
86. Gaborit N, Varro A, Le Bouter S, Szuts V, Escande D, Nattel S. et al. Gender-related differences in ion-channel and transporter subunit expression in non-diseased human hearts. J Mol Cell Cardiol. 2010;49:639-46
87. Stones R, Billeter R, Zhang H, Harrison S, White E. The role of transient outward K+ current in electrical remodelling induced by voluntary exercise in female rat hearts. Basic Res Cardiol. 2009;104:643-52
88. Liu J, Kim KH, London B, Morales MJ, Backx PH. Dissection of the voltage-activated potassium outward currents in adult mouse ventricular myocytes: I(to,f), I(to,s), I(K,slow1), I(K,slow2), and I(ss). Basic Res Cardiol. 2011;106:189-204
89. Guo W, Xu H, London B, Nerbonne JM. Molecular basis of transient outward K+ current diversity in mouse ventricular myocytes. The Journal of physiology. 1999;521(Pt 3):587-99
90. Gussak I, Chaitman BR, Kopecky SL, Nerbonne JM. Rapid ventricular repolarization in rodents: electrocardiographic manifestations, molecular mechanisms, and clinical insights. J Electrocardiol. 2000;33:159-70
91. Yan G-X, Lankipalli RS, Burke JF, Musco S, Kowey PR. Ventricular repolarization components on the electrocardiogram. J Am Coll Cardiol. 2003;42:401-9
92. Dixon JE, Shi W, Wang HS, McDonald C, Yu H, Wymore RS. et al. Role of the Kv4.3 K+ channel in ventricular muscle. A molecular correlate for the transient outward current. Circ Res. 1996;79:659-68
93. Rosati B, Grau F, Rodriguez S, Li H, Nerbonne JM, McKinnon D. Concordant expression of KChIP2 mRNA, protein and transient outward current throughout the canine ventricle. J Physiol. 2003;548:815-22
94. Kuo HC, Cheng CF, Clark RB, Lin JJ, Lin JL, Hoshijima M. et al. A defect in the Kv channel-interacting protein 2 (KChIP2) gene leads to a complete loss of I(to) and confers susceptibility to ventricular tachycardia. Cell. 2001;107:801-13
95. Rosati B, Pan Z, Lypen S, Wang HS, Cohen I, Dixon JE. et al. Regulation of KChIP2 potassium channel beta subunit gene expression underlies the gradient of transient outward current in canine and human ventricle. J Physiol. 2001;533:119-25
96. Oudit GY, Kassiri Z, Sah R, Ramirez RJ, Zobel C, Backx PH. The molecular physiology of the cardiac transient outward potassium current (I(to)) in normal and diseased myocardium. J Mol Cell Cardiol. 2001;33:851-72
97. Michael G, Xiao L, Qi XY, Dobrev D, Nattel S. Remodelling of cardiac repolarization: how homeostatic responses can lead to arrhythmogenesis. Cardiovasc Res. 2009;81:491-9
98. Gottlieb PD, Pierce SA, Sims RJ, Yamagishi H, Weihe EK, Harriss JV. et al. Bop encodes a muscle-restricted protein containing MYND and SET domains and is essential for cardiac differentiation and morphogenesis. Nat Genet. 2002;31:25-32
99. Sims RJ, Weihe EK, Zhu L, O'Malley S, Harriss JV, Gottlieb PD. m-Bop, a Repressor Protein Essential for Cardiogenesis, Interacts with skNAC, a Heart- and Muscle-specific Transcription Factor. J Biol Chem. 2002;277:26524-9
100. van Rooij E, Olson EN. MicroRNAs: powerful new regulators of heart disease and provocative therapeutic targets. J Clin Invest. 2007;117:2369-76
101. King IN, Qian L, Liang J, Huang Y, Shieh JT, Kwon C. et al. A genome-wide screen reveals a role for microRNA-1 in modulating cardiac cell polarity. Dev Cell. 2011;20:497-510
102. Glass C, Singla DK. MicroRNA-1 transfected embryonic stem cells enhance cardiac myocyte differentiation and inhibit apoptosis by modulating the PTEN/Akt pathway in the infarcted heart. Am J Physiol Heart Circ Physiol. 2011;301:H2038-49
103. Sayed D, Hong C, Chen IY, Lypowy J, Abdellatif M. MicroRNAs play an essential role in the development of cardiac hypertrophy. Circ Res. 2007;100:416-24
104. Bruneau BG. The developmental genetics of congenital heart disease. Nature. 2008;451:943-8
105. Ikeda S, He A, Kong SW, Lu J, Bejar R, Bodyak N. et al. MicroRNA-1 negatively regulates expression of the hypertrophy-associated calmodulin and Mef2a genes. Mol Cell Biol. 2009;29:2193-204
106. Yang B, Lin H, Xiao J, Lu Y, Luo X, Li B. et al. The muscle-specific microRNA miR-1 regulates cardiac arrhythmogenic potential by targeting GJA1 and KCNJ2. Nat Med. 2007;13:486-91
107. Zhao Y, Ransom JF, Li A, Vedantham V, von Drehle M, Muth AN. et al. Dysregulation of cardiogenesis, cardiac conduction, and cell cycle in mice lacking miRNA-1-2. Cell. 2007;129:303-17
108. Xiang L, Zhong A, You T, Chen J, Xu W, Shi M. Prognostic Significance of Right Bundle Branch Block for Patients with Acute Myocardial Infarction: A Systematic Review and Meta-Analysis. Med Sci Monit. 2016;22:998-1004
109. Belkin MN, Upadhyay GA. Does Cardiac Resynchronization Therapy Benefit Patients with Non-Left Bundle Branch Block Prolonged QRS Patterns? Curr Cardiol Rep. 2017;19:125
110. Ban K, Wile B, Cho KW, Kim S, Song MK, Kim SY. et al. Non-genetic Purification of Ventricular Cardiomyocytes from Differentiating Embryonic Stem Cells through Molecular Beacons Targeting IRX-4. Stem Cell Reports. 2015;5:1239-49
111. Houweling AC, Dildrop R, Peters T, Mummenhoff J, Moorman AF, Ruther U. et al. Gene and cluster-specific expression of the Iroquois family members during mouse development. Mech Dev. 2001;107:169-74
112. Salem JE, Alexandre J, Bachelot A, Funck-Brentano C. Influence of steroid hormones on ventricular repolarization. Pharmacol Ther. 2016;167:38-47
113. Kurokawa J, Furukawa T. Non-genomic action of sex steroid hormones and cardiac repolarization. Biol Pharm Bull. 2013;36:8-12
Author contact
![]() Corresponding author: Yinan Zhao, huwycom, Department of Neurology, First Affiliated Hospital of China Medical University, Shenyang, Liaoning 110001, China
Corresponding author: Yinan Zhao, huwycom, Department of Neurology, First Affiliated Hospital of China Medical University, Shenyang, Liaoning 110001, China

 Global reach, higher impact
Global reach, higher impact