Impact Factor
ISSN: 1449-1907
Int J Med Sci 2018; 15(7):703-712. doi:10.7150/ijms.21660 This issue Cite
Research Paper
Differential Expression of CircRNAs in Embryonic Heart Tissue Associated with Ventricular Septal Defect
1. Department of Pediatrics, The Affiliated Obstetrics and Gynecology Hospital of Nanjing Medical University, No. 123 Tian Fei Xiang, Mo Chou Road, Nanjing 210004, Jiangsu Province, China.
2. Department of Pediatrics, Nanjing Maternity and Child Health Care Hospital, No. 123 Tian Fei Xiang, Mo Chou Road, Nanjing 210004, Jiangsu Province, China.
#These authors contributed equally to this work.
Received 2017-6-28; Accepted 2018-2-4; Published 2018-5-14
Abstract
Objectives: To explore and validate the differential expression of circRNAs in the myocardium of congenital ventricular septal defect (VSD) and to explore a new avenue of research regarding the pathological mechanisms of VSD.
Methods: We detected circRNAs expression profiles in heart tissues taken from six aborted fetuses with VSD and normal group using circRNA microarray. Some differentially expressed circRNAs were studied by bioinformatics analysis. Finally, quantitative reverse transcription polymerase chain reaction (qRT-PCR) was performed to confirm these results.
Results: This study found abundant circRNAs in the myocardium taken from individuals in the normal group and the VSD group. After that, totally 6234 differentially expressed circRNAs between the normal group and the VSD group were confirmed (Fold change ≥ 2.0; p < 0.05). Then, this research carried out bioinformatics analysis and predicted the potential biological functions of circRNAs. Finally, the over-expression of hsa_circRNA_002086 and under-expression of hsa_circRNA_007878, hsa_circRNA_100709, hsa_circRNA_101965, hsa_circRNA_402565 were further validated by qRT-PCR.
Conclusions: There is a significant difference in expression of the circRNA in cardiac tissue from VSD group compared to the normal group. Combined with the microarray results and previous researches, circRNAs may contribute to the occurrence of VSD by acting as miRNA sponges or by binding proteins, these possible roles for circRNAs in VSD require elucidation in additional studies.
Keywords: Congenital Heart Disease (CHD), fetation, heart development, miRNA sponges
Introduction
Congenital heart disease (CHD) is the most common birth defect and a leading cause of morbidity and mortality in patients with congenital malformations[1]. Moreover, the VSDs are the most common congenital cardiac abnormalities. The isolated incidence of VSD was 2.62 in 1000 births[2]. In addition to imaging, there was little laboratory tests were performed to confirm the diagnosis of VSD. Present interventions show little effect in early prevention or treatment of VSD, for its unclear pathogenesis.
With the developing of transcriptomic, previous studies have demonstrated that non-coding RNAs, such as miRNAs and lncRNAs play important roles in cardiac development [3]. As early as 1980s, the circRNA had been discovered[4], but the circRNA did not receive much attention when receiving the research technology at that time. With the development of the study, the researchers found that, unlike the previously studied linear RNA, circRNA forms a covalently closed continuous loop, there is no linear RNA molecule in the 3'and 5' ends connected together[5]. This feature confers the insensitivity of cyclic RNA to nuclease[4], and thus is more stable than linear RNA, which makes circRNAs more obvious advantage in the development and application of potential new clinical diagnostic markers than other types of RNA[6-8].
With the development of high-throughput sequencing technology, a large number of circRNA have now been identified spanning a wide variety of organisms. In 1993, it was discovered that there is a circular transcript in the mouse sperm-determining gene Sry[9]; this indicated that circRNAs are not a product of false shear in the transcription process. With the deepening of the research, circular RNAs (circRNAs) are discovered class of evolutionarily conserved endogenous non-coding RNA that play important roles in the regulation of gene expression[10-12], so as to participate in the occurrence and development of various diseases. In subsequent studies, it was reported that cirs-7 acts as a miRNA sponge in brain cells [12], and circTCF-25 was shown to inhibit the miRNA function[13]. Recently abundant circRNAs were found in the amphicytulas before implantation[14]. It is suggested that the circRNA plays an important role not only in the process of disease, but also in the process of embryonic development.
Detection of all circRNAs by microarray. A. Volcano Plot: The red points in the plot represent differentially expressed circRNAs that were statistically significant between the two groups. B. Scatter Plot: The values for the X and Y axes are normalized signal values (log2 scaled). The green lines represent fold change. CircRNAs above the top line and below the bottom green line exhibit more than a 2.0-fold change of circRNAs between the two groups. C. The histogram and fan diagram shows the distribution of all the differentially expressed circRNAs on human chromosomes: red represents over-expressed and blue represents under-expressed circRNAs. D. The thermal map revealed that the top 20 over-expressed circRNAs in CHD group. E. The thermal map revealed that the top 20 under-expressed circRNA in CHD group.
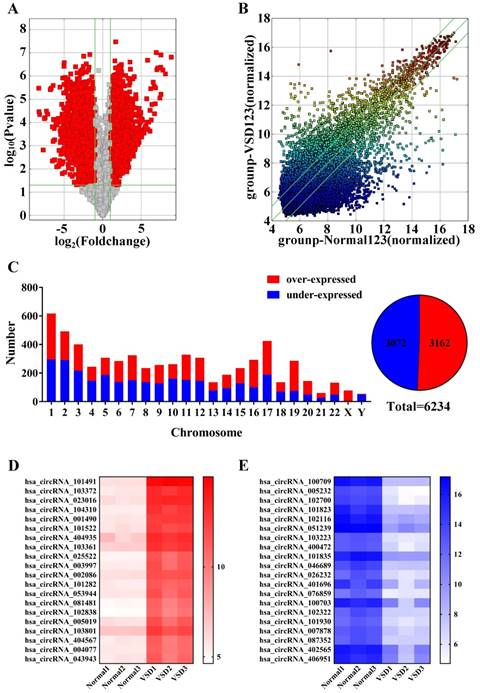
In previous studies, little research is about the role of circRNAs in the regulation of congenital heart disease. In this study, through the application of microarray, we determine the differences expression of circRNAs between the VSD group and the control group in the embryo abortion. Specifically, we identified a total of 6234 differentially expressed circRNAs, then predicted the functions of circRNA. At length, this work provided a new direction which can access to the pathological mechanisms of VSD.
Results
Identification of differentially expressed circRNA profiles
We performed microarray assays on circRNA to identify circRNA expression signatures in CHD. A total of 12842 circRNA targets were detected by microarray probes in three pairs of samples. Comparing VSD cardiac tissue (n=3) to normal cardiac tissue (n=3), we identified 6234 differentially expressed circRNAs: 3162 circRNAs were over-expressed, and 3072 circRNAs were under-expressed at fold change (Fold change ≥ 2.0 and p < 0.05, Figure 2c right, Supplementary Table 1).
Volcanic maps (Figure 2a) and scatter plots (Figure 2b) show all of the detected circRNAs. Moreover, in volcano plot filtering identified significantly differentially expressed circRNAs between the two groups. The left red points mean the under-expressed circRNAs and the right red points represent the over-expressed circRNAs. The points out of two green lines near the middle of the scatter plots mean over 2.0-fold change. The histogram depicts the distribution of circRNAs on human chromosomes (Figure 2C left). Furthermore, the thermal map revealed that the top 20 over-expressed and under-expressed circRNAs between the VSD group and normal group (Figure 2D and 2E), and the top 20 differentially expressed circRNAs are shown in Table 1.
Top 20 Over- and under- expressed circRNAs derived from VSD group
| CircRNA | Host Gene Name | Fold Change | p-value |
|---|---|---|---|
| Over-expressed | |||
| hsa_circRNA_101491 | MAPKBP1 | 469.87 | 1.52110E-07 |
| hsa_circRNA_103372 | IP6K2 | 186.88 | 6.12402E-06 |
| hsa_circRNA_023016 | RBM4 | 185.63 | 1.22810E-07 |
| hsa_circRNA_104310 | ZDHHC4 | 168.79 | 8.76757E-06 |
| hsa_circRNA_001490 | KIF2A | 122.80 | 9.69000E-08 |
| hsa_circRNA_101522 | DMXL2 | 82.44 | 1.66974E-05 |
| hsa_circRNA_404935 | ZBTB16 | 78.26 | 7.78875E-05 |
| hsa_circRNA_103361 | SMARCC1 | 74.09 | 3.47999E-06 |
| hsa_circRNA_025522 | ARHGDIB | 62.48 | 9.51537E-05 |
| hsa_circRNA_003997 | CLMP | 58.13 | 1.96476E-04 |
| hsa_circRNA_002086 | LOC401320 | 55.98 | 2.08650E-07 |
| hsa_circRNA_101282 | ABCC4 | 51.74 | 9.45923E-06 |
| hsa_circRNA_053944 | FAM98A | 48.27 | 1.56638E-04 |
| hsa_circRNA_081481 | FBXO24 | 47.01 | 1.42946E-04 |
| hsa_circRNA_102838 | ITGB6 | 46.62 | 1.86864E-04 |
| hsa_circRNA_005019 | CHSY1 | 40.75 | 2.87632E-04 |
| hsa_circRNA_103801 | MYO10 | 35.02 | 7.82817E-05 |
| hsa_circRNA_404567 | PHTF1 | 33.33 | 3.20992E-04 |
| hsa_circRNA_004077 | VAT1L | 33.02 | 6.97139E-05 |
| hsa_circRNA_043943 | VAT1 | 32.63 | 4.21995E-04 |
| Under-expressed | |||
| hsa_circRNA_100709 | FAM53B | 312.08 | 3.33090E-06 |
| hsa_circRNA_005232 | SLC8A1 | 305.53 | 3.43120E-05 |
| hsa_circRNA_102700 | SLC8A1 | 291.22 | 4.54877E-05 |
| hsa_circRNA_101823 | CNOT1 | 177.22 | 6.10264E-05 |
| hsa_circRNA_102116 | ZNF652 | 158.09 | 3.24855E-05 |
| hsa_circRNA_051239 | ATP5SL | 140.01 | 4.31932E-05 |
| hsa_circRNA_103223 | DDX17 | 110.65 | 3.81112E-06 |
| hsa_circRNA_400472 | RYR2 | 93.16 | 1.54288E-04 |
| hsa_circRNA_101835 | NFATC3 | 83.73 | 7.31852E-05 |
| hsa_circRNA_046689 | ENOSF1 | 76.66 | 7.56315E-05 |
| hsa_circRNA_026232 | LARP4 | 76.64 | 1.78835E-04 |
| hsa_circRNA_401696 | ANKFY1 | 74.42 | 4.03502E-03 |
| hsa_circRNA_076859 | DST | 67.12 | 9.68140E-05 |
| hsa_circRNA_100703 | CHST15 | 62.61 | 8.07226E-03 |
| hsa_circRNA_102322 | TMEM241 | 61.19 | 1.29360E-04 |
| hsa_circRNA_101930 | YWHAE | 59.74 | 9.22908E-05 |
| hsa_circRNA_007878 | MAP4 | 56.33 | 9.76382E-05 |
| hsa_circRNA_087352 | UBQLN1 | 54.63 | 6.71974E-05 |
| hsa_circRNA_402565 | EDEM2 | 53.33 | 7.63520E-03 |
| hsa_circRNA_406951 | LOC493754 | 50.48 | 6.65697E-03 |
Validation of differentially expressed circRNAs
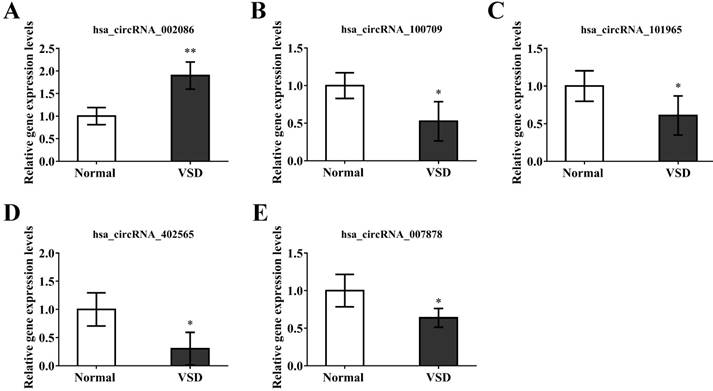
In order to verify the reliability of our microarray results. we randomly selected 5 circRNAs, hsa_circRNA_002086, hsa_circRNA_007878, hsa_circRNA_100709, hsa_circRNA_101965, hsa_circRNA_402565, from the top 20 differentially expressed circRNAs. GAPDH was used as a normalization control for qRT-PCR analysis. The results of the qRT-PCR indicated significant over-expression of hsa_circRNA_002086 and under-expression of hsa_circRNA_007878, hsa_circRNA_100709, hsa_circRNA_101965, hsa_circRNA_402565 (Figure 4). Because microarray and qRT-PCR belong to two difference genetic tests, there are some errors between them. We confirmed the trend of 5 circRNAs differential expression were the same with chip, though the results of fold changes form qRT-PCR were different from those of microarray. This indicated that the results of qRT-PCR were well consistent with microarray results, demonstrating the high reliability of the microarray expression results.
Prediction of the function for the circRNAs host genes
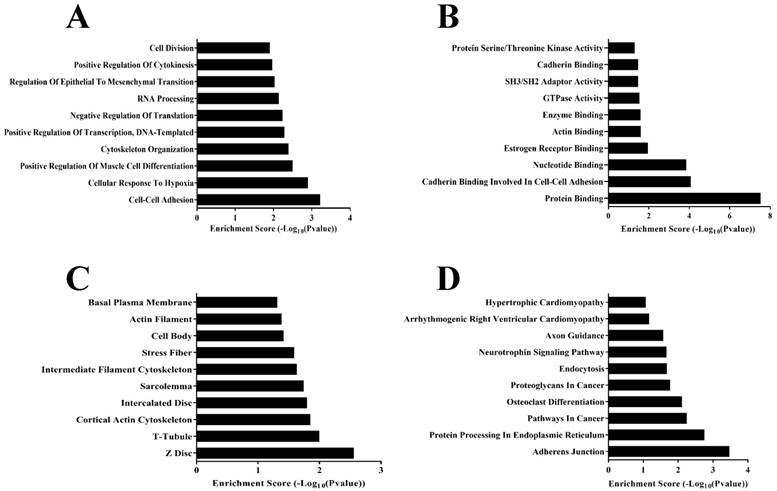
According to previous researches, many circRNAs functions are related to their host genes[15, 16]. To eliminate some low variance multiples which may belongs to interference information, and convenient data analysis. We selected 282 differentially expressed circRNAs (Fold change ≥ 15.0; p < 0.05), of which 88 were over-expressed and 194 were under-expressed in CHD cardiac tissue were analyzed (Supplementary Table 1). The host genes of 282 differentially expressed circRNAs were input into DAVID (https://david.ncifcrf.gov), an online gene ontology (GO) analysis tool, and the number of target genes in each GO term was counted. Enrichment score was used to test and calculate the significance of the target gene enrichment in each GO term, and a p-value was acquired to describe the significance of the target gene GO term. Target genes were classified and analyzed according to cellular component, molecular function, biological process, and KEGG pathway (Figure 4).
We can find out from bioinformatics analysis, in biological process category (Figure 4a) parts of these genes are involved in the transcriptional regulation of RNA and cell differentiation. Then we also found the Protein Serine, Threonine Kinase Activity and Actin Binding that are involved in the regulation of cardiac cell activity and function in the molecular functions category (Figure 4b). In the cellular component category (Figure 4c), there are same peculiar structures of heart cells, such as Z Disc, T-Tubule, or some of the cellular structures, Actin Filament, Intercalated Disc, that are involved in cardiac cell function. In the final KEGG-pathway analysis (Figure 4d), we also found some signal pathways similar to Hypertrophic Cardiomyopathy, Arrhythmogenic Right Ventricular Cardiomyopathy, the heart disease related diseases. Therefore, from the results of bioinformatics analysis, these circRNAs we screened were largely related to the development of the heart.
Validation of circRNA microarray data using real-time quantitative-PCR. The real-time RT-PCR reactions were repeated three times for hsa_circRNA_002086, hsa_circRNA_007878, hsa_circRNA_100709, hsa_circRNA_101965, hsa_circRNA_402565.
Gene ontology and Pathway-Express analysis about 282 differentially expressed circRNAs. a. Predicted target genes ontology terms in the biological process category. b. Predicted target genes ontology terms in the molecular functions category. c. Predicted target genes ontology terms in the cellular component category. d. Predicted target genes identified by Pathway-Express analysis using the DAVID online analysis tools.
Construction of the circRNA/miRNA interaction network
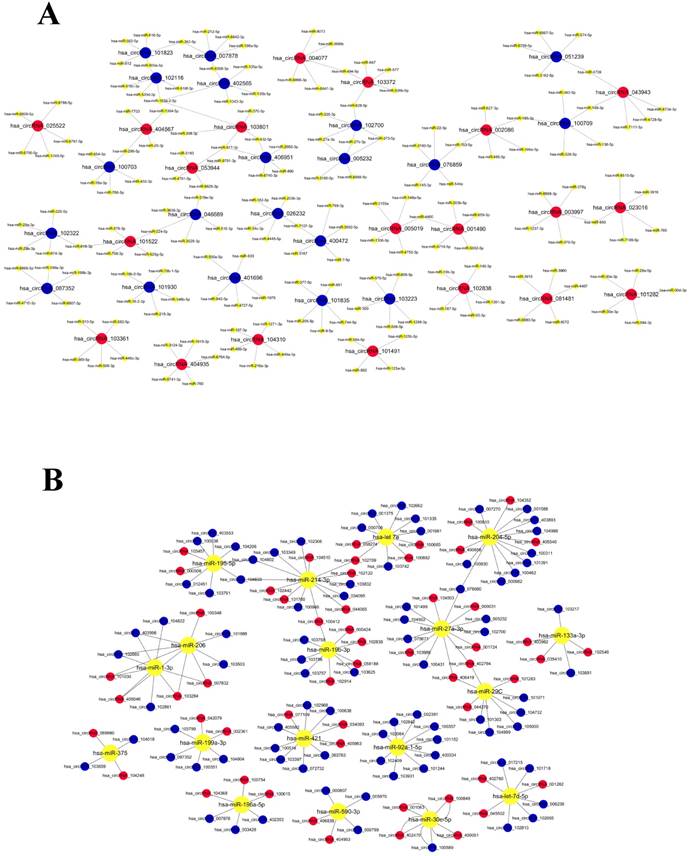
In the early researches for circRNA functions, most investigator paid close attention to the function of miRNA sponges[12, 13, 17-19]. In order to evaluate the target miRNAs of circRNAs, this study used TargetScan and miRanda database to theoretically predict, based on conserved seed-matching sequence. The 6234 differentially expressed circRNAs are theoretically bound to miRNAs (Supplementary Table 1). The relationship between circRNA and miRNA in the first 20 sites of differential expression has been sorted out as network (Figure 5a). Reviewed previous studies, we have identified a number of miRNA which can take participation in regulation of cardiac development. The relationship between the miRNA, which had been researched, and their potentially combined circRNAs (Figure 5b). The relationship between them deserves further exploration.
The network of the relationship between circRNAs and miRNAs. A. The relationship between Top 20 circRNAs and their potentially combined miRNAs B. The relationship between the miRNA, which had been researched, and their potentially combined circRNAs
Discussion
Ventricular septal defect (VSD) is the most common major congenital malformation, accounting for approximately 20% of neonatal deaths[2]. Although investigators have been throw themselves to explore the development and progression of congenital heart disease, including the identification of mutations in genes associated with VSD abnormalities, the detailed mechanisms of VSD remain a mystery[20]. In our study, it investigated the role of circRNAs in the development of human embryonic heart through miRNA sponges, based on high-throughput microarray screening.
It will be contributed to the diagnosis and rescue of abnormal embryos during pregnancy, for studying the circRNAs involved in embryonic development and the functional mechanisms of circRNAs. The researchers used high-throughput sequencing to explore the expression changes of circRNAs at different time points in the development of the rat retina, and tried to combine apoptosis to explore the role of circRNA in the development of neural cells. Han, J., et al. have researched that affluent differential expression of circRNAs among 3 different developmental time points, and 15 of which are related to apoptosis of circRNA[21]. From their study, we concluded that circRNA also plays a very important role in the process of embryonic development. Thus, the role of circRNAs in the development of embryonic heart is a direction worthy of further study.
Presently, circRNAs are at the forefront of research in cancer and cardiovascular disease, and these studies will further explore the mechanisms of development and progression of these diseases[22, 23]. MiRNA sponges function is the main research direction of investigators in existing researched. A latest study in 2017, circRNA-MYLK could serve as a sponge for miR-29a to abolish the endogenous suppressive effect on target gene VEGFA which promoted bladder cancer growth[24]. Through the complete pathway axis of the circRNA-miRNA-downstream target, the researchers elaborated the mechanism of circRNAs in the course of bladder cancer. Legnini, I. et al provided us with the latest research model of miRNA sponges. In 2016, the first study about angiocardiopathy found that the circRNA HRCR can regulate miR-223 by inhibiting the expression of ARC, which inhibits the development of cardiac hypertrophy and heart failure, thus confirming that circRNAs participate in the regulation of protein expression by effecting the biological function of miRNA[25]. Thus, in the development of cardiovascular system, circRNA may also affect the biological function of miRNA, which has an impact on the downstream target, and this mode of action is worthy of further exploration.
In the past studies, miRNA has been authenticated to be involved in the regulation of cardiac development. These preliminary work laid the foundation for us to further explore the function of miRNA sponges. In this study, through the software analysis and prediction, it found many miRNA binding sites in the circRNAs. As shown in Table 3, many circRNAs contain miR-30c binding sites; the high differential expression suggests that these circRNAs might be involved in the onset and development of CHD by regulating miRNA expression. Liu et al. reported that up-regulated miR-30c can act through the sonic hedgehog signal pathway, a signaling pathway associated with embryonic development and differentiation of P19 cells, and influences the balance between proliferation and apoptosis[26]. In another early research, miR-29c regulates the proliferation and apoptosis of P19 cells by regulating WNT 4 signaling molecules and regulates its differentiation into cardio myocytes[27]. Combined with the above researches suggest that one role for these differentially expressed circRNAs may be to function as a miR-30c or miR-29c sponge, which may affect heart development. We can further study the regulatory mechanism of upstream circRNAs through the discovered miRNAs. Of course, we can also further analyze and tap our chip results, then select the circRNA that we're interested in, and discover a new circRNA-miRNA-Target regulatory signaling pathway for heart development.
Network analysis between miRNAs and circRNAs
| MiRNA Binding Sites | Fold change | Regulation | CircRNA |
|---|---|---|---|
| hsa-let-7d-5p[31, 32] | 38.3150488 | down | hsa_circRNA_006235 |
| 18.6883554 | down | hsa_circRNA_101718 | |
| 7.6873022 | down | hsa_circRNA_017215 | |
| 6.649988 | down | hsa_circRNA_102655 | |
| 5.662462 | down | hsa_circRNA_102813 | |
| 3.362295 | up | hsa_circRNA_001282 | |
| 3.207422 | up | hsa_circRNA_045502 | |
| 3.040495 | up | hsa_circRNA_402780 | |
| hsa-let-7e[31, 32] | 11.1323253 | down | hsa_circRNA_101335 |
| 6.9339121 | down | hsa_circRNA_102662 | |
| 6.626608 | down | hsa_circRNA_001375 | |
| 6.139229 | down | hsa_circRNA_103742 | |
| 5.694939 | down | hsa_circRNA_000708 | |
| 5.286563 | down | hsa_circRNA_001961 | |
| 12.48349 | up | hsa_circRNA_100882 | |
| 7.884907 | up | hsa_circRNA_102709 | |
| 5.058363 | up | hsa_circRNA_100685 | |
| 4.504873 | up | hsa_circRNA_058274 | |
| hsa-miR-1-3p[33, 34] | 41.9001898 | down | hsa_circRNA_403996 |
| 6.9060521 | down | hsa_circRNA_102865 | |
| 4.007695 | down | hsa_circRNA_102861 | |
| 3.283393 | down | hsa_circRNA_104822 | |
| 6.540579 | up | hsa_circRNA_101030 | |
| 3.882837 | up | hsa_circRNA_405046 | |
| 3.864211 | up | hsa_circRNA_103284 | |
| 3.21382 | up | hsa_circRNA_007832 | |
| hsa-miR-133a-3p[33, 34] | 10.9918076 | down | hsa_circRNA_103881 |
| 8.0107769 | down | hsa_circRNA_103217 | |
| 7.158794 | up | hsa_circRNA_102546 | |
| 4.499071 | up | hsa_circRNA_403982 | |
| 4.148428 | up | hsa_circRNA_035410 | |
| hsa-miR-19b-3p[33, 35] | 6.3395835 | down | hsa_circRNA_103625 |
| 5.2972091 | down | hsa_circRNA_103756 | |
| 5.1675127 | down | hsa_circRNA_103757 | |
| 4.5267012 | down | hsa_circRNA_103758 | |
| 46.61967 | up | hsa_circRNA_102838 | |
| 9.424552 | up | hsa_circRNA_100412 | |
| 6.843182 | up | hsa_circRNA_000424 | |
| 5.656437 | up | hsa_circRNA_102914 | |
| 5.311273 | up | hsa_circRNA_058188 | |
| hsa-miR-195-5p[33, 34] | 12.1003825 | down | hsa_circRNA_012451 |
| 11.4621834 | down | hsa_circRNA_104603 | |
| 9.1727498 | down | hsa_circRNA_103791 | |
| 8.3094564 | down | hsa_circRNA_104602 | |
| 5.2359807 | down | hsa_circRNA_104206 | |
| 4.9777722 | down | hsa_circRNA_100038 | |
| 4.4409499 | down | hsa_circRNA_403553 | |
| 7.55831 | up | hsa_circRNA_103457 | |
| 6.324974 | up | hsa_circRNA_000508 | |
| hsa-miR-196a-5p[3] | 56.3330834 | down | hsa_circRNA_007878 |
| 4.3106251 | down | hsa_circRNA_402353 | |
| 3.0475854 | down | hsa_circRNA_003428 | |
| 7.735672 | up | hsa_circRNA_104368 | |
| 4.679547 | up | hsa_circRNA_100754 | |
| 3.397228 | up | hsa_circRNA_100615 | |
| hsa-miR-199a-3p[33] | 54.63029 | down | hsa_circRNA_087352 |
| 11.80673 | down | hsa_circRNA_104804 | |
| 7.568176 | down | hsa_circRNA_100351 | |
| 5.618868 | down | hsa_circRNA_103799 | |
| 4.55836 | up | hsa_circRNA_042079 | |
| 3.594391 | up | hsa_circRNA_002361 | |
| hsa-miR-204-5p[36] | 15.59543 | down | hsa_circRNA_001588 |
| 11.19295 | down | hsa_circRNA_000982 | |
| 7.635485 | down | hsa_circRNA_007270 | |
| 7.612147 | down | hsa_circRNA_104988 | |
| 7.220489 | down | hsa_circRNA_100462 | |
| 7.004912 | down | hsa_circRNA_100311 | |
| 5.825728 | down | hsa_circRNA_403893 | |
| 5.723085 | down | hsa_circRNA_100830 | |
| 5.461927 | down | hsa_circRNA_078680 | |
| 4.036292 | down | hsa_circRNA_101391 | |
| 4.163626 | up | hsa_circRNA_100810 | |
| 3.937716 | up | hsa_circRNA_104352 | |
| 3.810963 | up | hsa_circRNA_400658 | |
| 3.283699 | up | hsa_circRNA_405540 | |
| hsa-miR-206[37] | 41.90019 | down | hsa_circRNA_403996 |
| 14.206515 | down | hsa_circRNA_103503 | |
| 6.9060521 | down | hsa_circRNA_102865 | |
| 5.44311 | down | hsa_circRNA_101688 | |
| 6.540579 | up | hsa_circRNA_101030 | |
| 4.634693 | up | hsa_circRNA_100348 | |
| 3.882837 | up | hsa_circRNA_405046 | |
| 3.864211 | up | hsa_circRNA_103284 | |
| 3.21382 | up | hsa_circRNA_007832 | |
| hsa-miR-214-3p[33] | 12.52012 | down | hsa_circRNA_103349 |
| 11.46218 | down | hsa_circRNA_104603 | |
| 10.3626 | down | hsa_circRNA_034095 | |
| 8.309456 | down | hsa_circRNA_104602 | |
| 5.554202 | down | hsa_circRNA_100986 | |
| 5.368699 | down | hsa_circRNA_103832 | |
| 4.821045 | down | hsa_circRNA_102306 | |
| 18.58771 | up | hsa_circRNA_102442 | |
| 9.424552 | up | hsa_circRNA_100412 | |
| 7.884907 | up | hsa_circRNA_102709 | |
| 7.453939 | up | hsa_circRNA_104510 | |
| 6.999214 | up | hsa_circRNA_102122 | |
| 6.680745 | up | hsa_circRNA_044065 | |
| 5.141421 | up | hsa_circRNA_101760 | |
| hsa-miR-27a-3p[38, 39] | 305.5269 | down | hsa_circRNA_005232 |
| 291.2235 | down | hsa_circRNA_102700 | |
| 14.14891 | down | hsa_circRNA_075671 | |
| 10.77599 | down | hsa_circRNA_104502 | |
| 10.18955 | down | hsa_circRNA_100431 | |
| 6.716819 | down | hsa_circRNA_101499 | |
| 5.461927 | down | hsa_circRNA_078680 | |
| 8.634449 | up | hsa_circRNA_000031 | |
| 6.265159 | up | hsa_circRNA_104503 | |
| 5.785755 | up | hsa_circRNA_001724 | |
| 5.440395 | up | hsa_circRNA_103986 | |
| 5.099158 | up | hsa_circRNA_402794 | |
| 4.149106 | up | hsa_circRNA_406419 | |
| hsa-miR-29c[27] | 13.852547 | down | hsa_circRNA_101071 |
| 10.17929 | down | hsa_circRNA_104999 | |
| 6.6941486 | down | hsa_circRNA_101303 | |
| 4.6987668 | down | hsa_circRNA_104732 | |
| 3.2086528 | down | hsa_circRNA_105000 | |
| 5.099158 | up | hsa_circRNA_402794 | |
| 4.280459 | up | hsa_circRNA_101283 | |
| 4.149106 | up | hsa_circRNA_406419 | |
| 4.008815 | up | hsa_circRNA_044370 | |
| hsa-miR-30c-5p[26, 33] | 14.99199 | down | hsa_circRNA_100589 |
| 25.32313 | up | hsa_circRNA_001063 | |
| 3.875839 | up | hsa_circRNA_402470 | |
| 3.640062 | up | hsa_circRNA_100849 | |
| 3.079875 | up | hsa_circRNA_400051 | |
| hsa-miR-375[40] | 3.1354057 | down | hsa_circRNA_104018 |
| 2.0539568 | down | hsa_circRNA_103659 | |
| 3.688859 | up | hsa_circRNA_104248 | |
| 2.205257 | up | hsa_circRNA_069980 | |
| hsa-miR-421[33] | 11.004075 | down | hsa_circRNA_063763 |
| 8.3824519 | down | hsa_circRNA_103397 | |
| 7.5803979 | down | hsa_circRNA_100638 | |
| 7.3394522 | down | hsa_circRNA_072732 | |
| 5.2343335 | down | hsa_circRNA_102968 | |
| 3.9867779 | down | hsa_circRNA_405582 | |
| 3.7947983 | down | hsa_circRNA_100514 | |
| 31.05648 | up | hsa_circRNA_034093 | |
| 26.61819 | up | hsa_circRNA_405963 | |
| 3.100921 | up | hsa_circRNA_077109 | |
| hsa-miR-590-3p[35] | 77.197715 | down | hsa_circRNA_000799 |
| 11.605572 | down | hsa_circRNA_000807 | |
| 5.6559582 | down | hsa_circRNA_005970 | |
| 4.906641 | up | hsa_circRNA_406838 | |
| 3.586507 | up | hsa_circRNA_404953 | |
| hsa-miR-92a-1-5p[33] | 16.27706 | down | hsa_circRNA_102842 |
| 8.83981 | down | hsa_circRNA_103931 | |
| 5.648434 | down | hsa_circRNA_100357 | |
| 5.005185 | down | hsa_circRNA_002391 | |
| 4.601707 | down | hsa_circRNA_102409 | |
| 3.924155 | down | hsa_circRNA_400334 | |
| 3.521439 | down | hsa_circRNA_101244 | |
| 3.465761 | down | hsa_circRNA_102064 | |
| 3.369293 | down | hsa_circRNA_101152 |
Furthermore, circRNAs are involved in many important regulatory functions, not just through the miRNAs sponges function. RNA binding motifs (RBM), and even the translation of synthetic proteins may be the pathway for circRNA to function[9, 28, 29]. As in bioinformatics analysis, molecular functions (Figure 4c) show that the circRNAs we detected may be similar to its host genes, give full play to its functions through protein binding. For example, it has been found that circ-Foxo3 can affect protein cell localization by binding proteins. Circ-Foxo3 is expressed mainly in the cytoplasm, where it is associated with aging related proteins Id1 and E2F1, as well as the stress proteins HIF1 alpha and FAK. Circ-Foxo3 can reduce the expression of Id1 and E2F1 in the nucleus, but also reduce the stress response by regulating the expression of FAK and HIF1 alpha in mitochondria, and accelerating myocardial aging[30]. This suggests that in our microarray results, there may be a non-miRNA-sponges involved in the regulation of cardiac development, but this mode of action needs further analysis and screening.
This study still has some limitations. We had neither proved these circRNAs could directly regulate heart development, nor detected the dynamic expression of these circRNAs during heart development. Furthermore, we have no expression pattern analysis of the host genes of candidate circRNAs and their effects on mRNAs or miRNAs. It calls for further validations. Additionally, combined with the bioinformatics analysis and miRNA target prediction, prediction function, provides fertile areas for further research.
In conclusion, this study demonstrated the significant differentially circRNAs in myocardial tissue between VSD and normal group. These circRNAs might involve in the regulation of myocardial development. Our study provides some fundamental data for the follow-up studies of diagnostic markers and potential mechanisms of heart development. To our knowledge, this study is the preliminary exploration of circRNAs as a mechanism for heart development. Our data suggest that circRNAs might play an important role in heart development, and establish rationale to investigate the role of circRNA involved in heart development in additional studies that will elucidate mechanisms of heart development and development of VSD.
Materials and methods
Ethical statement
All human fetal heart tissues were obtained from Obstetrics and Gynecology Hospital affiliated of Nanjing Medical University from deceased donors as approved by the medical ethics committee. And it complies with The Population and Family Planning Law of the People's Republic of China. We followed established procedures for written informed parental consent. We conducted basic research in accordance with national institutes of health guidelines.
Experimental design
This experiment adopts a case-control study design. To examine the different expression of circRNA, we conducted high-throughput microarray technology to detect heart tissue divided into two different groups: VSD and normal (n=3 tissues per groups). We collected cardiac tissue from aborted fetus at 24-28 weeks of gestation depending on embryos diagnosed by ultrasonography (Figure 1). In order to exclude the interference of non-research purposes related factors, we excluded the tissues collected from whose mother had other diseases, and the embryos had genetic disorders, such as 21- trisomy syndrome. To validate the microarray, we randomly selected 5 circRNAs (hsa_circRNA_002086, hsa_circRNA_007878, hsa_circRNA_100709, hsa_circRNA_101965, hsa_circRNA_402565) and examined its expression in 12 pairs of fetal heart tissue samples at 24-28 week of gestation by quantitative reverse transcription-polymerase chain reaction (qRT-PCR).
Patients and sample collection
Enrollment occurred from January to June 2016 at the Obstetrics and Gynecology Hospital affiliated of Nanjing Medical University Department of Family Planning. Prenatal ultrasound diagnosis of VSD for aborted fetuses, and fetal abortion with VSD were confirmed by anatomy, and are not associated with other malformations. Controls included aborted fetuses whose prenatal diagnosis were no abnormal genotype and were confirmed to lack VSD or other cardiac malformation. The results of imaging diagnosis are shown in Figure 1.
Microarray analysis
Arraystar circRNA Microarray Technology (KANGCHEN, Shanghai, China) was used to analyze the differential expression of circRNAs.
Total RNA extraction and reverse transcription
Total RNA was extracted from the samples using TRIzol Reagent (Invitrogen, Carlsbad CA, USA), according to manufacturer's instructions. The RNA prep pure tissue kit (TIANGEN, DP431) was also used for subsequent RNA preparation. Based on the concentration of each sample, 1000 ng total RNA was input into the 20ul reverse transcription reaction. cDNA synthesis was performed on each sample using reverse transcription with random primers following the recommendations of the TaKaRa Prime ScriptTM RT Master Mix kit.
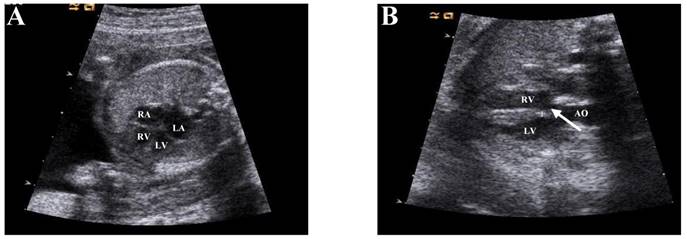
Echocardiographic diagnosis of fetal heart. a. The four chambers of the heart of a normal fetus (Arrow position). b: Resting diagram of Ventricular Septal Defect (VSD) (Arrow position). Note RV, right ventricle; LV, left ventricle; RA, right atrium; LA, left atrium; AO, aorta.
qRT-PCR detection of target genes
We used SYBR for qRT-PCR to evaluation results of chip. The experimental data were analyzed using the 2-ΔΔCT method. All data are the average of three independent experiments. Primer sequences are shown in Table 2.
GO analysis and Bioinformatics pathway
We retrieved the genes encoded by the circRNAs source region from the circBase (http://www.circbase.org) and predicted their target genes. Target genes were input into the DAVID (https://david.ncifcrf.gov) online GO analysis tool.
Statistical analysis
SPSS statistical software was used for data analysis. Data are given as mean ± standard deviation. Significant differences between groups were evaluated by the t test. A difference with p < 0.05 was considered statistically significant.
Primers used in present study
| Primer name | Primer sequences |
|---|---|
| Gapdh-F | TCGACAGTCAGCCGCATCTTCTTT |
| Gapdh-R | ACCAAATCCGTTGACTCCGACCTT |
| hsa_circRNA_402565-F | CAATCCCTCACATTCTCCA |
| hsa_circRNA_402565-R | GTTGCCACAGTAACCACATC |
| hsa_circRNA_101965-F | TAGAGGGTCGGCAGCA |
| hsa_circRNA_101965-R | TGTGGATAGTCCGTTCGT |
| hsa_circRNA_100709-F | GTGACACCTGGAGCCCT |
| hsa_circRNA_100709-R | CCTTGACTCATCTTCTTTGG |
| hsa_circRNA_007878-F | AGCCAAAGATGTTCCACC |
| hsa_circRNA_007878-R | GCTTCCACAGACCACCC |
| hsa_circRNA_002086-F | CTGGTGTCTGTCCTCCTTAC |
| hsa_circRNA_002086-R | GGGTGACCTGGTTGTGA |
Supplementary Material
Supplementary table S1.
Acknowledgements
This study was supported by grants from the National Natural Science Foundation of China (Grant No. 81470376, 81370200), the National Natural Science Foundation of Jiangsu Province of China (No. BK20141077), and the Nanjing Medical Science and Technique Development Foundation (No. 201605052).
Competing Interests
The authors have declared that no competing interest exists.
References
1. Kaltman JR, Burns KM, Pearson GD. Perspective on Congenital Heart Disease Research. Circulation research. 2017;120:898-900
2. van der Linde D, Konings EE, Slager MA, Witsenburg M, Helbing WA, Takkenberg JJ. et al. Birth prevalence of congenital heart disease worldwide: a systematic review and meta-analysis. Journal of the American College of Cardiology. 2011;58:2241-7
3. Tian J, An X, Niu L. Role of microRNAs in cardiac development and disease. Exp Ther Med. 2017;13:3-8
4. Jeck WR, Sharpless NE. Detecting and characterizing circular RNAs. Nature biotechnology. 2014;32:453-61
5. Salzman J, Gawad C, Wang PL, Lacayo N, Brown PO. Circular RNAs are the predominant transcript isoform from hundreds of human genes in diverse cell types. PloS one. 2012;7:e30733
6. Li H, Hao X, Wang H, Liu Z, He Y, Pu M. et al. Circular RNA Expression Profile of Pancreatic Ductal Adenocarcinoma Revealed by Microarray. Cellular physiology and biochemistry: international journal of experimental cellular physiology, biochemistry, and pharmacology. 2016;40:1334-44
7. Qian Y, Lu Y, Rui C, Qian Y, Cai M, Jia R. Potential Significance of Circular RNA in Human Placental Tissue for Patients with Preeclampsia. Cellular physiology and biochemistry: international journal of experimental cellular physiology, biochemistry, and pharmacology. 2016;39:1380-90
8. Chen LL. The biogenesis and emerging roles of circular RNAs. Nature reviews Molecular cell biology. 2016;17:205-11
9. Barrett SP, Salzman J. Circular RNAs: analysis, expression and potential functions. Development (Cambridge, England). 2016;143:1838-47
10. Hansen TB, Jensen TI, Clausen BH, Bramsen JB, Finsen B, Damgaard CK. et al. Natural RNA circles function as efficient microRNA sponges. Nature. 2013;495:384-8
11. Memczak S, Jens M, Elefsinioti A, Torti F, Krueger J, Rybak A. et al. Circular RNAs are a large class of animal RNAs with regulatory potency. Nature. 2013;495:333-8
12. Zhao Y, Alexandrov PN, Jaber V, Lukiw WJ. Deficiency in the Ubiquitin Conjugating Enzyme UBE2A in Alzheimer's Disease (AD) is Linked to Deficits in a Natural Circular miRNA-7 Sponge (circRNA; ciRS-7). Genes. 2016:7
13. Zhong Z, Lv M, Chen J. Screening differential circular RNA expression profiles reveals the regulatory role of circTCF25-miR-103a-3p/miR-107-CDK6 pathway in bladder carcinoma. Scientific reports. 2016;6:30919
14. Dang Y, Yan L, Hu B, Fan X, Ren Y, Li R. et al. Tracing the expression of circular RNAs in human pre-implantation embryos. Genome biology. 2016;17:130
15. Legnini I, Di Timoteo G, Rossi F, Morlando M, Briganti F, Sthandier O. et al. Circ-ZNF609 Is a Circular RNA that Can Be Translated and Functions in Myogenesis. Molecular cell. 2017;66:22-37.e9
16. Zeng Y, Du WW, Wu Y, Yang Z, Awan FM, Li X. et al. A Circular RNA Binds To and Activates AKT Phosphorylation and Nuclear Localization Reducing Apoptosis and Enhancing Cardiac Repair. Theranostics. 2017;7:3842-55
17. Zheng XB, Zhang M, X MQ. Detection and characterization of ciRS-7: a potential promoter of the development of cancer. Neoplasma. 2017:64
18. Zheng Q, Bao C, Guo W, Li S, Chen J, Chen B. et al. Circular RNA profiling reveals an abundant circHIPK3 that regulates cell growth by sponging multiple miRNAs. Nature communications. 2016;7:11215
19. Bai Y, Zhang Y, Han B, Yang L, Chen X, Huang R. et al. Circular RNA DLGAP4 ameliorates ischemic stroke outcomes by targeting miR-143 to regulate endothelial-mesenchymal transition associated with blood-brain barrier integrity. The Journal of neuroscience: the official journal of the Society for Neuroscience. 2017
20. Bruneau BG. The developmental genetics of congenital heart disease. Nature. 2008;451:943-8
21. Han J, Gao L, Dong J, Bai J, Zhang M, Zheng J. The expression profile of developmental stage-dependent circular RNA in the immature rat retina. Mol Vis. 2017;23:457-69
22. Devaux Y, Creemers EE, Boon RA, Werfel S, Thum T, Engelhardt S. et al. Circular RNAs in heart failure. European journal of heart failure. 2017
23. Kristensen LS, Hansen TB, Veno MT, Kjems J. Circular RNAs in cancer: opportunities and challenges in the field. Oncogene. 2017
24. Legnini I, Di Timoteo G, Rossi F, Morlando M, Briganti F, Sthandier O. et al. Circ-ZNF609 Is a Circular RNA that Can Be Translated and Functions in Myogenesis. Molecular cell. 2017;66(e9):22-37
25. Wang K, Long B, Liu F, Wang JX, Liu CY, Zhao B. et al. A circular RNA protects the heart from pathological hypertrophy and heart failure by targeting miR-223. European heart journal. 2016;37:2602-11
26. Liu X, Li M, Peng Y, Hu X, Xu J, Zhu S. et al. miR-30c regulates proliferation, apoptosis and differentiation via the Shh signaling pathway in P19 cells. Exp Mol Med. 2016;48:e248
27. Liu M, Chen Y, Song G, Chen B, Wang L, Li X. et al. MicroRNA-29c overexpression inhibits proliferation and promotes apoptosis and differentiation in P19 embryonal carcinoma cells. Gene. 2016;576:304-11
28. Pamudurti NR, Bartok O, Jens M, Ashwal-Fluss R, Stottmeister C, Ruhe L. et al. Translation of CircRNAs. Molecular cell. 2017;66(e7):9-21
29. Khan MA, Reckman YJ, Aufiero S, van den Hoogenhof MM, van der Made I, Beqqali A. et al. RBM20 Regulates Circular RNA Production From the Titin Gene. Circulation research. 2016;119:996-1003
30. Du WW, Yang W, Chen Y, Wu ZK, Foster FS, Yang Z. et al. Foxo3 circular RNA promotes cardiac senescence by modulating multiple factors associated with stress and senescence responses. European heart journal. 2016
31. Yu ZB, Han SP, Bai YF, Zhu C, Pan Y, Guo XR. microRNA expression profiling in fetal single ventricle malformation identified by deep sequencing. International journal of molecular medicine. 2012;29:53-60
32. Rustagi Y, Jaiswal HK, Rawal K, Kundu GC, Rani V. Comparative Characterization of Cardiac Development Specific microRNAs: Fetal Regulators for Future. PloS one. 2015;10:e0139359
33. Dong Y, Liu C, Zhao Y, Ponnusamy M, Li P, Wang K. Role of noncoding RNAs in regulation of cardiac cell death and cardiovascular diseases. Cellular and molecular life sciences: CMLS. 2017
34. Shah P, Bristow MR, Port JD. MicroRNAs in Heart Failure, Cardiac Transplantation, and Myocardial Recovery: Biomarkers with Therapeutic Potential. Current heart failure reports. 2017;14:454-64
35. Wojciechowska A, Braniewska A, Kozar-Kaminska K. MicroRNA in cardiovascular biology and disease. Adv Clin Exp Med. 2017;26:865-74
36. Yester JW, Kuhn B. Mechanisms of Cardiomyocyte Proliferation and Differentiation in Development and Regeneration. Current cardiology reports. 2017;19:13
37. Shan ZX, Lin QX, Fu YH, Deng CY, Zhou ZL, Zhu JN. et al. Upregulated expression of miR-1/miR-206 in a rat model of myocardial infarction. Biochemical and biophysical research communications. 2009;381:597-601
38. Yu K, Ji Y, Wang H, Xuan QK, Li BB, Xiao JJ. et al. Association of miR-196a2, miR-27a, and miR-499 polymorphisms with isolated congenital heart disease in a Chinese population. Genetics and molecular research: GMR. 2016:15
39. Vegter EL, Ovchinnikova ES, van Veldhuisen DJ, Jaarsma T, Berezikov E, van der Meer P. et al. Low circulating microRNA levels in heart failure patients are associated with atherosclerotic disease and cardiovascular-related rehospitalizations. Clinical research in cardiology: official journal of the German Cardiac Society. 2017;106:598-609
40. Garikipati VNS, Verma SK, Jolardarashi D, Cheng Z, Ibetti J, Cimini M. et al. Therapeutic inhibition of miR-375 attenuates post-myocardial infarction inflammatory response and left ventricular dysfunction via PDK-1-AKT signalling axis. Cardiovascular research. 2017;113:938-49
Author contact
![]() Corresponding authors: Zhang-Bin Yu, Ph.D. Department of Pediatrics, Obstetrics and Gynecology Hospital Affiliated to Nanjing Medical University, Nanjing, Jiangsu 210004, China, Phone: 86-25-52226561, Fax: 86-25-52226561. E-mail address: yuzhangbincom. and Shu-Ping Han, Ph.D. Department of Pediatrics, Obstetrics and Gynecology Hospital Affiliated to Nanjing Medical University, Nanjing, Jiangsu 210004, China, Phone: 86-25-52226561, Fax: 86-25-52226561. E-mail address: shupinghanedu.cn.
Corresponding authors: Zhang-Bin Yu, Ph.D. Department of Pediatrics, Obstetrics and Gynecology Hospital Affiliated to Nanjing Medical University, Nanjing, Jiangsu 210004, China, Phone: 86-25-52226561, Fax: 86-25-52226561. E-mail address: yuzhangbincom. and Shu-Ping Han, Ph.D. Department of Pediatrics, Obstetrics and Gynecology Hospital Affiliated to Nanjing Medical University, Nanjing, Jiangsu 210004, China, Phone: 86-25-52226561, Fax: 86-25-52226561. E-mail address: shupinghanedu.cn.

 Global reach, higher impact
Global reach, higher impact