Impact Factor
ISSN: 1449-1907
Int J Med Sci 2018; 15(5):436-446. doi:10.7150/ijms.22894 This issue Cite
Research Paper
A New Look at Etiological Factors of Idiopathic Scoliosis: Neural Crest Cells
1. Novosibirsk Research Institute of Traumatology and Orthopaedics n.a. Ya.L. Tsivyan, Novosibirsk, Russia
2. Institute of Cytology and Genetics, Russian Academy of Science, Novosibirsk, Russia
3. Institute of Chemical Biology and Fundamental Medicine, Russian Academy of Science, and Meshalkin National Medical Research Center, Ministry of Health of the Russian Federation, Novosibirsk, Russia
4. Arrowhead Pharmaceuticals, Madison WI, and University of Pittsburgh, Pittsburgh PA, USA.
Received 2017-9-19; Accepted 2017-12-18; Published 2018-3-6
Abstract
Idiopathic scoliosis is one of the most common disabling pathologies of children and adolescents. Etiology and pathogenesis of idiopathic scoliosis remain unknown. To study the etiology of this disease we identified the cells' phenotypes in the vertebral body growth plates in patients with idiopathic scoliosis.
Materials and methods: The cells were isolated from vertebral body growth plates of the convex and concave sides of the deformity harvested intraoperatively in 50 patients with scoliosis. Cells were cultured and identified by methods of common morphology, neuromorphology, electron microscopy, immunohistochemistry and PCR analysis.
Results: Cultured cells of convex side of deformation were identified as chondroblasts. Cells isolated from the growth plates of the concave side of the deformation showed numerous features of neuro- and glioblasts. These cells formed synapses, contain neurofilaments, and expressed neural and glial proteins.
Conclusion: For the first time we demonstrated the presence of cells with neural/glial phenotype in the concave side of the vertebral body growth plate in scoliotic deformity. We hypothesized that neural and glial cells observed in the growth plates of the vertebral bodies represent derivatives of neural crest cells deposited in somites due to alterations in their migratory pathway during embryogenesis. We also propose that ectopic localization of cells derived from neural crest in the growth plate of the vertebral bodies is the main etiological factor of the scoliotic disease.
Keywords: scoliosis, growth plate, neural crest, the expression of proteins, neurofilaments, synapses, chondrocyte
Introduction
For centuries, the etiological factor of idiopathic scoliosis attracts attention of scientists. In spite of different numerous studies, the causes of scoliosis remain unknown [1-5]. It is now widely discussed genetic nature of idiopathic scoliosis [6-10]. However, gene or genes determining idiopathic scoliosis are still not identified [11].
Comprehensive studies of idiopathic scoliosis from its earliest to severe forms [12, 13] showed that the asymmetric growth of the spine is associated with impaired functioning of chondroblasts in the growth plates on the concave side of the spinal deformity. It was found that chondroblasts on the convex side have appropriate differentiation and subsequent osteogenesis, while those on the concave side remain in the early stages of histogenesis, becoming а factor of the asymmetric growth. In a previous study, expression analyses of key chondroblast genes were performed to determine the possible genetic causes of violation in histogenesis on the concave side of the deformation [14]. The genes assessed by real-time PCR were associated with growth (GHR, EGFR, IGFIR, TGFBRI), synthesis and structure of the extracellular matrix (COL2AI, HAPLNI; ACAN, LUM, VCAN, COL1AI), as well as with the sulfation and transmembrane sulfate transport (DTDST, CHSTI, CHTST3). The data showed that cells in the scoliotic growth plate maintain the level of synthesis of matrix and proteoglycan core proteins, however they have abnormalities in sulfation, link protein, growth and transcription factors. Statistical analysis revealed pronounced differences between chondroblasts in normal patients and in those with idiopathic scoliosis. It was assumed that the detected gene expression profile might be associated with the presence of different cell phenotypes in the growth plates of the vertebral bodies of patients with idiopathic scoliosis [14].
The present study was performed to identify cell phenotypes in vertebral body growth plates of scoliotic patients. Written patient/legal guardian consent to use the removed specimens for research purposes was obtained for all cases.
Materials and methods
Cell isolation and cultivation
The identification of cell phenotype in the vertebral growth plate was based on the findings that these cells in in vitro conditions still preserve patterns of gene expression and morphological features typical for orthotopic localization [15, 16].
Cells from the convex and concave sides of the vertebral growth plate deformation were isolated and cultured separately. The growth plates of the vertebral bodies were harvested during surgery for severe forms of idiopathic scoliosis in 50 children aged 11-15 years performed in pediatric clinic of Research Institute of Traumatology and Orthopaedics. Samples were collected in sterile tubes containing 0.9% physiological saline solution and antibiotics gentamicin at a concentration of 20 ug/ml.
Hyaline cartilage of the growth plates was washed in saline, crushed to size of 1-2 mm in a petri dish with a minimal volume of RPMI medium (Biolot), and then it was placed in a 1.5% collagenase solution (Gibco) into CO2 incubator at 37 ° C for 22-24 hours. The resulting cell suspension was passed through a nylon filter (Nylon) for removing bits of tissue. Cells were pelleted by centrifugation for 10 minutes at 2000 rpm. The isolated cells were cultured in DMEM F12 medium (Invitrogen) supplemented with 15% FBS (Gibco) , 50 U/ml penicillin/streptomycincs/amphotericin B (Biolot) in a CO2 incubator at 37 ° C. Cells were cultured without replating during 21 days. Change of medium was performed each 3 days. Morphological studies of the cells were carried out in a period of from 5 to 21 days. One day before analyses the cultured cells were detached by 0.25% trypsin and passaged in the fresh growth medium to coverslips, films, chips and four-well plates.
Scanning electron microscopy
Cells on the chips were fixed in growth medium containing 2.5% glutaraldehyde for 15 minutes and then transferred to a solution of 2.5% glutaraldehyde in 0.1 M cacodylate buffer for one hour. Thereafter the chips successively underwent two washes in 0.1 M cacodylate buffer, fixation in 1% aqueous solution of osmium tetroxide, two washes in water and dehydration by incubation in solutions of increasing concentration of ethanol (30%, 50%, 70%, 100%) for 10 minutes each. The dehydrated samples were dried by the critical point protocol in the Critical Point Dryer (BAL-TEC, Liechtenstein) and then examined in a scanning electron microscope (Zeiss, Germany) before and after spraying 1 nm chromium layer under argon atmosphere (Coating Unit, Leica Microsystems, Austria). The samples were observed under magnifications ranging from 1000 to 30000 and an accelerating voltage of 30kV.
Transmission electron microscopy
Cells on special plastic films were fixed by a 2.5% glutaraldehyde solution in 0.1 M Na-cacodylate buffer (pH 7.4) for 1 hour. Then the films were washed three times with 0.1 M Na-cacodylate buffer (pH 7.4) and post-fixed in 1% osmium tetroxide solution supplemented with 0.8% potassium ferrocyanide in the same buffer for 1 hour. After three washes in distilled water, the cells were left overnight in a 1% aqueous solution of uranylacetate at 4 °C. The next day the samples were washed with water and dehydrated in alcohols of increasing concentration (for 5 min in 30% and 50% ethanol, and for 10 min in 70%, 96% and 100% ethanol). The cells were further dehydrated in acetone (2 x 20 min). After that the samples were impregnated with a resin mixture consisting of 4 components (Epon 812, DDSA, MNA and DMP-30) as follows: for 1 hour in a resin: acetone mix 1:2 (V/V); for 2 hours in a resin: acetone mix 1:1; for 2 hours in a resin: acetone mix 2:1; for 2 hours in pure resin; and one hour more in fresh portion of pure resin. Further the samples were filled in the foil forms and incubated overnight in a desiccator with CaCl2 (to remove air bubbles from the resin). For polymerization the samples were placed in a thermostat at +60°C for 3 days.
Cell staining
Cells attached to the coverslip were fixed in 70° ethanol and further stained with hematoxylin-eosin, Alcian blue, as well as in accordance with the protocols of Ramon-Kahal and Nissl [17].
Immunohistochemical staining was performed according to the manufacturer's recommendations to the antibodies. Dewaxing and antigen retrieval of tissue sections were carried out using RT Link module (Dako, Denmark) in citrate buffer (pH 9,0) at 95°C for 1 hour. Endogenous peroxidase was blocked with 3% solution of H2O2. For protein blocking FBS was used. Tissue sections were incubated with antibodies at 22°C for 30 minutes. The antibodies were anti GFAP (clone N1506, rabbit polyclonal, «DAKO»); anti S100 (clone IR504, rabbit polyclonal, "DAKO"); anti Synaptophysin (clone DAK-SYNAP, mouse monoclonal, "DAKO"); anti Neurofilament Protein (NF, clone 2F11, mouse monoclonal, "DAKO"). To visualize immunohistochemical reaction a polymer detection system EnVision FLEX Systems (Dako, Denmark) was used. The cell nuclei were stained with hematoxylin.
For inderect immunofluorescene staining, the cells were fixed in 4% formalin solution for 10 minutes and permeabilized in 0.2% Triton X-100 for 15 minutes. Samples were washed twice in PBS for 10 minutes and blocked with 1% BSA for 30 minutes. Cells were incubated with antibodies to Aggrecan (Abcam, mouse monoclonal, dilution 1:100), Sox 9 (Abcam, mouse monoclonal, dilution 1:100), Chondroitin sulfates A and C (Abcam, mouse monoclonal, dilution 1:200), Collagen I (Abcam, rabbit polyclonal, dilution 1:200), Collagen II (Abcam, mouse monoclonal, dilution 1:100), Neuronal Class III β-Tubulin TUJ1 (Covance, mouse monoclonal, dilution 1:1000) and Anti-Neurofilament 200 (Sigma, rabbit polyclonal, dilution 1:500) at 4°C overnight. First antibody localization was visualized using appropriate secondary antibodies raised against rabbit or mouse IgG and conjugated to fluorescent dyes Alexa 488 or Alexa 568 (Life Technologies). Nuclei were stained with DAPI (Vector Laboratories). Stained slides were imaged on a Nikon TE microscope.
Results
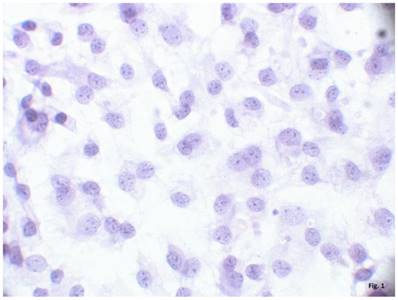
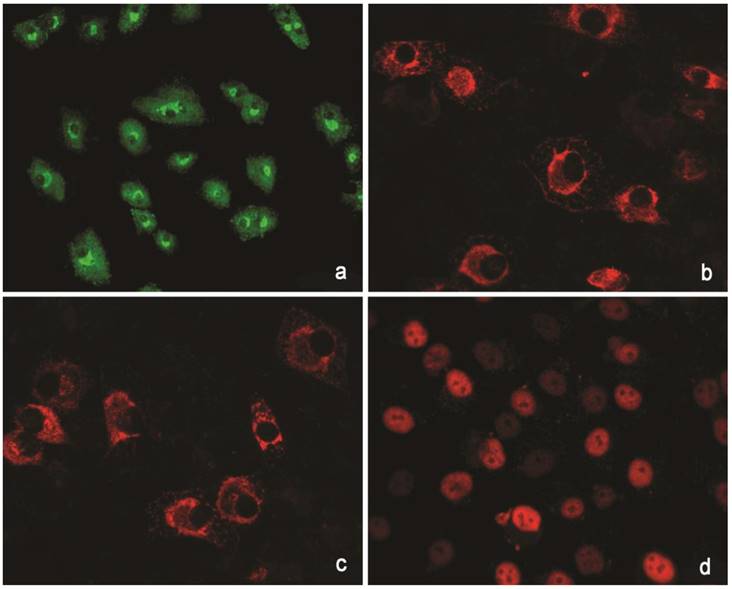
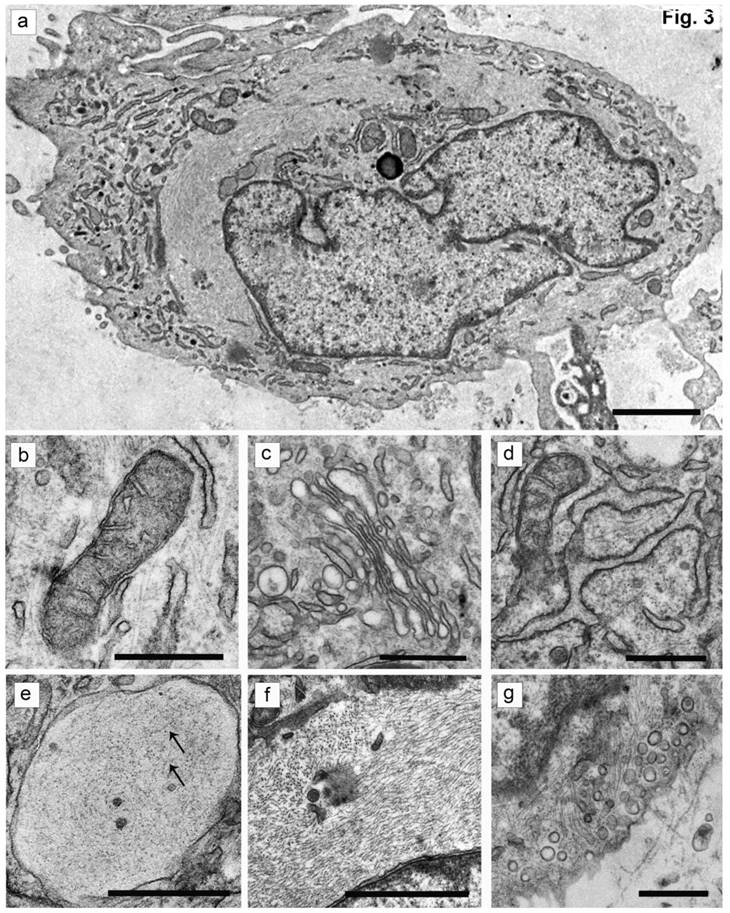
The cells isolated from the external parts of the growth plates, located above and below scoliotic deformity, under culture condition formed a monolayer consisting of a bright round cells adjoining to each other (Figure 1). Nuclei with 1-2 nucleoli and dispersed chromatin were located in the center of the cells. Chondroitin sulfates A and C and glycogen granules were detected in their cytoplasm. Immunohistochemical staining of the cells revealed expression of aggrecan, Sox 9, chondroitin sulfates A and C, as well as collagen types I and II (Figure 2). At the level of the electron microscopy the cells had a round shape with a large bright nucleus with invaginations. The cytoplasm was rich in organelles and intermediate microfilaments. Mitochondria were large, mostly oval in shape with short transverse cristae. The endoplasmic reticulum showed extended tanks. The Golgi apparatus was represented by dictyosomes and numerous vesicles located near the plasma membrane. Accumulation of intermediate filaments was observed near to the nuclei (Figure 3).
A monolayer of cultured chondroblasts (convex side of the spinal deformity). Hematoxylin-eosin staining, magnification x600.
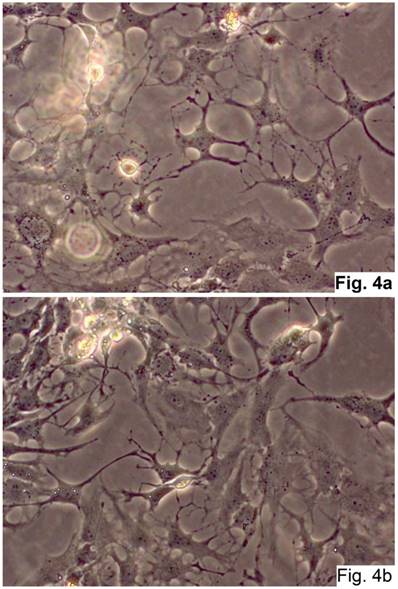
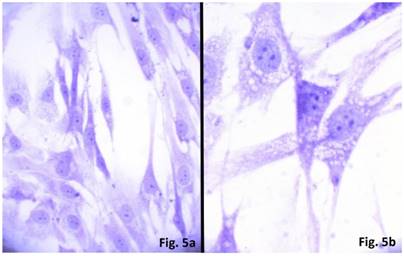
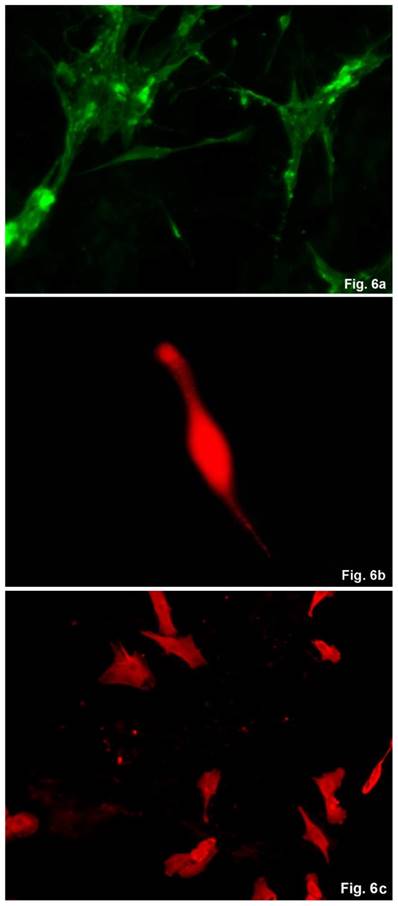
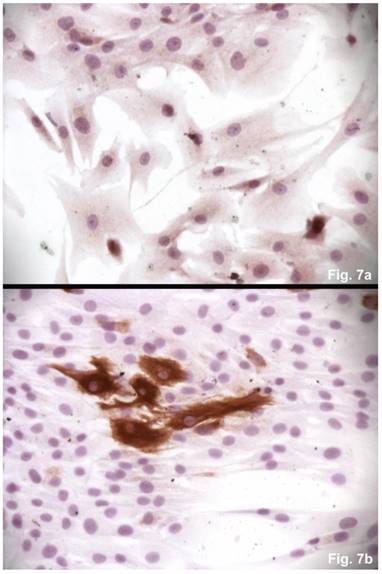
The cells isolated from the concave side growth plates of the deformity (scoliosis apex) were represented by several types (Figures 4a and 4b). The first type cells were large multipolar cells with one long axon and numerous branching short processes (Figure 5a). At the center of these cells, the spherical nucleus with 1-2 nucleoli was located. Granular network (Nissl substance) was detected in the cytoplasm of the cells including processes. We found the same cells with a large oval nucleus located centrally and a narrow rim of Nissl-positive cytoplasm, turning into long processes on both ends of the cell. We have recognized uni-, bi- and pseudounipolar cells, which had Nissl substance in their cytoplasm and processes (Figure 5b). Immunohistochemical staining of the cells revealed expression of high molecular weight neurofilament NF-200 and neural βIII-tubulin as well as NF1 gene (Figures 6 a, b, c). The second type cells were large cells with round nuclei and numerous branched cytoplasmic processes. These cells expressed astrocyte protein S-100 (Figure 7a). We observed cells with clear boundaries, round or oval shape with a few processes. At the center, the cell had a large round nucleus bordered with bright rim of cytoplasm. These cells including their processes were positively stained by Cajal (Fig. 8). These cells expressed glial acidic protein GFAP, as shown in Figure 7b. We also detected the third type cells, which matched cells derived from the convex side of the deformation (data not shown).
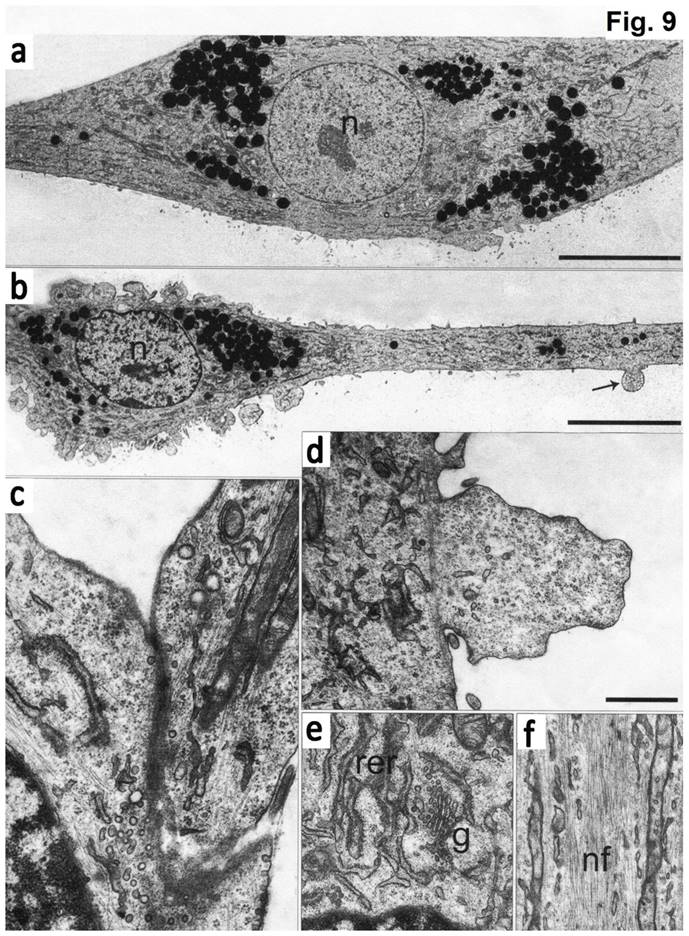
Electron microscopy data showed that at the ultrastructure level the first type cells had elongated morphology with a round nucleus and 1-2 electron-dense nucleoli (Fig. 9). The cell body was continued to a long process (axon), which contained the membrane protrusions (axon hillocks). Similar membrane protrusions ("spines") were also found on the cell bodies. Inside the "spines" a large number of small bright bubbles (vacuoles) were observed. Axons of the same cells contact to the bodies of the other cells forming synapses. On both sides of the sinus membrane, we detected small and large vacuoles. The cytoplasm of cells was enriched with organelles. The endoplasmic reticulum (EPR) consisted of long narrow tanks with extended regions. The Golgi apparatus was highly developed and represented by numerous dictyosome and bubbles. There were exocytic vesicles in the cells. Mitochondria had thin stretched branched cristae and dense matrix. The processes of the cells showed neurofilaments. In the cytoplasm and processes we detected granules, which likely had a protein nature. Furthermore, we found the stacks of rough EPR resembling Nissl's bodies. The cells of the second type had numerous processes, which form contacts. These cells contained a bright nucleus with 1-2 nucleoli and clumps of heterochromatin. EPR was presented as branched channels. The Golgi apparatus had multiple vesicles and exocytic bubbles. The mitochondria had different shapes with extended cristae. In the cytoplasm, large number of intermediate filaments were accumulated. Neurofilaments in these cells were not detected (Figure 9).
Immunohistochemical reactions to proteins. (a) Collagen type I (green). (b) Collagen type II (red). (C) Aggrecan (red). ( d) Sox 9 (red).
The ultrastructure of chondroblasts from the convex side of the growth plate of the vertebral body at the top of deformation in a patient with idiopathic scoliosis. (a) General view of cells containing a nucleus with invaginations and a large number of intermediate filaments in the cytoplasm. (b, c) Mitochondria with short transverse cristae. (b) Narrow and (c) extended tanks of rough EPR. (d) The dictyosomes of the Golgi apparatus. (e) Vacuole with short thin filaments inside (pointed by arrows). (f) Accumulation of the intermediate filaments near the nucleus. (g) - Numerous vesicles near the plasma membrane of cells. The scale bars are 2 microns for (a) and 0.5 microns for (b-g).
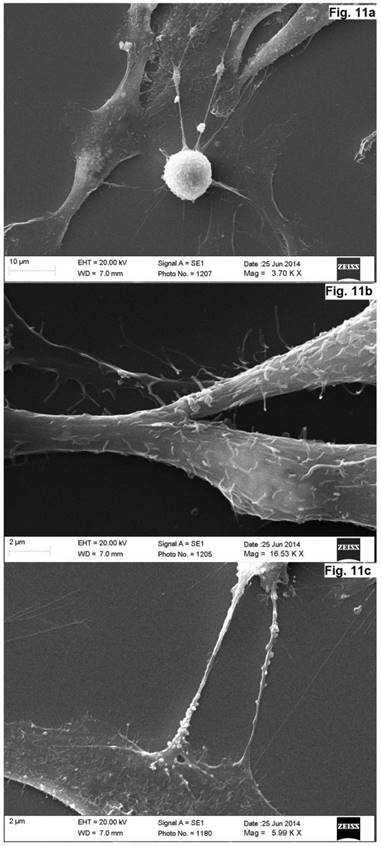
As was evident from the scanning electron microscopy that the cells derived from the concave side growth plate of the deformation had an elongated shape and the long processes forming contacts. We have seen synapse and the large "spinule" between the cells. Dense granules ranging from 300 to 500 nm in diameter were present in the cytoplasm and processes of the cells (Figures. 10, 11). The cells displayed numerous "spines" on their processes.
Cells of neural origin in the culture of the concave side of the scoliotic deformation (Native preparation, two different fields, x400).
Cultured cells from idiopathic scoliosis concave side (the top of the deformation) stained according to Nissl. (a) x 200, (b) x 400.
Immunohistochemical detection of neural proteins in cultured cells of the concave side of spinal deformity obtained from idiopathic scoliosis patients. (a) - NF-1 (green); (b) β- III tubuline (red), c) NF200 (red).
Discussion
The obtained data demonstrated that cells of different phenotypes exist in different locations of the vertebral body growth plates in patients with idiopathic scoliosis. The cultured cells of the convex side growth plates of the deformation were identified as chondroblasts. Their chondrogenic nature was confirmed by their morphological structure, including ultrastructural organization, the presence of tissue-specific proteoglycans and expression of genes associated with the cartilage growth [14].
Immunohistochemical detections of glial proteins. (a) S-100, an astrocytic protein, x 200. (b) GFAP, a glial protein, x200.
Cultured cells from idiopathic scoliosis concave side (the top of the deformation) stained according to Cajal. x200.

The cultured cells, isolated from the concave side growth plates of idiopathic scoliotic deformity, were identified as neuro- and glioblasts. Morphologically neuroblasts represented multi-, uni-, and psevdounipolar cells forming multiple contacts with both the processes and cell bodies. The Nissl substance was identified and neurospecific proteins NF-200, βIII-tubulin as well as NF1 gene were expressed in the cells. Electron microscopy data also revealed some properties attributable to neural cells, namely an extended network of neurofilaments, mature and developing sinuses with vesicle and specific elongated mitochondria. The processes and the cell bodies had numerous axon hillocks ("spines") containing vesicles. Multiple contacts were revealed between the processes and the cells. The cells of the second type were round-shaped and contained a large number of processes that formed numerous contacts. Glial proteins are expressed in the processes and cells positively stained by Ramon-Kahal. According to morphological and ultrastructural data, this type of cells was referred to glioblasts.
Naturally, the question arises how the cells of neural and glial origin could be localized in the growth plate of a patient with idiopathic scoliosis. To answer this question we should refer to the early stages of embryogenesis. It is known that the spine is formed from mesenchyme [18-20]. However, developing neural tube gives rise to the neural crest cells, which migrate along three pathways [21, 22]. One of the pathways of migration of neural crest cells is a truncal path that passes through the anterior (rostral) section of somites, which leads to the sensory ganglia formation [23]. Migrating cells undergo epithelial- mesenchymal transition, by switching over the expression from neural proteins of cell adhesion to proteins of mesenchymal adhesion [24].
During the transition, the neural crest cells become round and acquire mesenchymal phenotype indistinguishable from those of surrounding cells [25]. This facilitates migration of neural crest cells along mesenchymal extracellular matrix lining the path from the neural tube to the somites. Formation of the matrix is associated with the expression of Pax3 gene resulting in synthesis of two versican isoforms (V1 and V0) [26]. The direction of the neural crest cell migration is determined by the asymmetric distribution of versican isoforms and aggrecan [27]. The versican isoforms V0, V1 acts as inductors of neural crest cells migration, while aggrecan which is a high-molecular weight proteoglycan, has an inhibitory effect on distribution of the cells [28]. Chondrogenic differentiation of mesenchymal cells in the somites occurs during the migration of neural crest cells. These data demonstrate that the chondrogenesis and gangliagenesis are mutually dependent [20, 29]. Removal of somite results in inability to form sensory ganglia, while violation in somite segmentation leads to the formation of ugly ganglia [30]. Therefore, the migration of neural crest cells through somites is a normal course of events when regulation of spine morphogenesis and sensory ganglia occur at the same time [31, 32].
The microphotography of cultured cells isolated from concave side of vertebral body growth plate of patient with IS (a, b) Neuron-like elongated cells with a long process (axon). Numerous electron-dense granules present around the nuclei (N) and in the cytoplasm of the cells. Spines on the axons are pointed by arrow. (c) Cytoplasm segment of a cell in culture, which contains mature rough endoplasmic reticulum (shEPR) and the Golgi complex (G). (d) A fragment of axon with elongated mitochondria (pointed by arrow M) and a wide network of neurofilaments (NF) ongoing into process. (d) A contact which is formed between the process of a cell and the body of the other cell. Numerous vesicles located in the contact (pointed by arrows). (f, g) Spines present on the cell body and processes. The scale bars are 10 micron for (a, b) and 1 micron for (c-g).
Scanning electron microscope images of neurons in cell culture derived from concave side of vertebral body growth plate of patient with IS (samples without chromium layer). (a, b) A general view of the cells at the low magnification. The cells have an elongated shape and long processes. (c) The contacts between two cells. The synapse and the large spines are visible. In the cytoplasm of neurons, as well as in their processes are revealed small dense granules of 300 to 500 nm in diameter. The scale bars are 200 microns for (a, b), and 50 microns for (c).

Scanning electron microscope image of neurons in cell culture derived from concave side of vertebral body growth plate of patient with IS (samples with chromium layer). (a) A general view of the cells. (b) A fragment of a process of neuron at high magnification. (c) Process of neuron (axon) with spines. (d) A fragment of a neuron at high magnification. Its surface contains short processes and cytoplasm has granules which shine through the plasma membrane. The scale bars are 1 micron for (a), and 2 microns for (b-d).
According to this, we can assume that because of deviation in the spatial and temporal patterns of migration of neural crest cells some of them may “settle” and deposit in the somite. This may occur because of Pax3 gene mutation followed by impaired synthesis of versican isoforms along the migration route [26]. These data were additionally confirmed later [31, 32]. Violation in versican secretion and inhibition of their sulfation led to stop the migration of neural crest cells. It is known [33, 34] that the interaction of neural crest cells with interstitial matrix is carried out on the basis of cell - cell - matrix rule. Any impairment in the synthesis of receptors (integrins) and / or in their interaction with the migration substrate of the neural crest cells may be the cause of further violations of morphogenetic events in the somite [30].
Thus, we can assume that neural and glial cells we observed in the growth plates of the vertebral bodies of patients with idiopathic scoliosis are derived from neural crest cells deposited in the somite. It is known that neural crest cells acquire phenotype of surrounding cells at the site of their final destination retaining many properties of the original cells [35, 36]. This may explain why the cells in the concave side of the growth plate in idiopathic scoliotic spinal deformity were previously defined as poorly differentiated chondroblasts.
It is understandable that neurogenic cells within the growth plates of the vertebral bodies are not determined to the proper growth and chondrogenic differentiation resulting in asymmetry and local dysplasia with subsequent formation of spinal deformity. Thus, abnormalities in spine morphogenesis during embryonic development result in scoliosis with all its clinical and morphological attributes.
Thus, our separate analysis of cells from convex and concave sides of the vertebral growth plate of 50 patients with idiopathic scoliosis allowed us to suggest the causes of dysplasia and spinal deformity and to assume the variability of clinical manifestations of idiopathic scoliosis depending on the degree of violation in morphogenetic processes within the growth plate of vertebral bodies. We undertake further studies to get more details on how neural crest cells might be involved in formation and manifestation of idiopathic scoliosis.
Conclusions
The study of cell phenotype in the growth plates of the vertebral bodies on the convex and concave sides of the deformity in 50 patients revealed the presence of neurogenic cells. Although deviation of neural crest cells from physiologic migration/signaling was shown to induce pathology in cranial skeleton [37], such pathogenesis was never suggested for scoliotic deformity. We hypothesized those neural and glial cells we observed in the growth plate concave side represent derivatives of neural crest cells deposited in somites due to violation in their migration during embryogenesis. These cells are not genetically determined to the growth process that might lead to impaired growth on the concave side with continuation of growth on the convex side causing the formation of deformity. Thus, the deviation in morphogenesis of the spine development might result in the development of idiopathic scoliosis.
Abbreviations
GA: the Golgi apparatus; GAG: glycosaminoglycans; IS: idiopathic scoliosis; NF: neurofilaments; GP: growth plate; EPR: the endoplasmic reticulum.
Acknowledgements
This work was supported by Grant 17-75-30009 from the Russian Science Foundation (Pavel P. Laktionov).
Author Contribution
All authors contributed equally to this work. All authors discussed the results and implications and commented on the manuscript at all stages.
Conflict of Interest
The authors declare that they have no conflict of interest.
References
1. Stokes IA, Burwell RG, Dangerfield PH. Biomechanical spinal growth modulation and progressive adolescent scoliosis-a test of the'vicious cycle'pathogenetic hypothesis: Summary of an electronic focus group debate of the IBSE. Scoliosis. 2006;1(1):16
2. Veldhuizen A, Wever D, Webb P. The aetiology of idiopathic scoliosis: biomechanical and neuromuscular factors. European Spine Journal. 2000;9(3):178-84
3. Porter RW. The pathogenesis of idiopathic scoliosis: uncoupled neuro-osseous growth? European Spine Journal. 2001;10(6):473-81
4. Machida M, Dubousset J, Yamada T. et al. Serum melatonin levels in adolescent idiopathic scoliosis prediction and prevention for curve progression-a prospective study. Journal of pineal research. 2009;46(3):344-8
5. Dayer R, Haumont T, Belaieff W. et al. Idiopathic scoliosis: etiological concepts and hypotheses. Journal of children's orthopaedics. 2013;7(1):11-6
6. Ogilvie JW. Update on prognostic genetic testing in adolescent idiopathic scoliosis (AIS). Journal of Pediatric Orthopaedics. 2011;31:S46-S8
7. Miller NH. Idiopathic scoliosis: cracking the genetic code and what does it mean? Journal of Pediatric Orthopaedics. 2011;31:S49-S52
8. Wise CA, Gao X, Shoemaker S. et al. Understanding genetic factors in idiopathic scoliosis, a complex disease of childhood. Current genomics. 2008;9(1):51-9
9. Guo X, Chau WW, Hui-Chan CW. et al. Balance control in adolescents with idiopathic scoliosis and disturbed somatosensory function. Spine. 2006;31(14):E437-E40
10. Salehi L, Mangino M, De Serio S. et al. Assignment of a locus for autosomal dominant idiopathic scoliosis (IS) to human chromosome 17p11. Human genetics. 2002;111(4):401-4
11. Gorman KF, Julien C, Moreau A. The genetic epidemiology of idiopathic scoliosis. European Spine Journal. 2012;21(10):1905-19
12. Zaydman AM. Neurofibromatosis and scoliosis. Novosibirsk, Russia: Science Publising. 2011
13. Zaydman AM. Idiopathic scoliosis. The morphology, biochemistry and genetics. Novosibirsk, Russia: Novosibirsk University Press. 1994
14. Zaydman AM, Strokova EL, Novikov VV. et al. Gene expression in chondrocytes from growth plate of patients with idiopathic scoliosis. Surgery of spine. 2014;4:88-98
15. Melrose J, Smith S, Knox S. et al. Perlecan, the multidomain HS-proteoglycan of basement membranes, is a prominent pericellular component of ovine hypertrophic vertebral growth plate and cartilaginous endplate chondrocytes. Histochemistry and cell biology. 2002;118(4):269-80
16. Wang S, Qiu Y, Ma Z. et al. Expression of Runx2 and type X collagen in vertebral growth plate of patients with adolescent idiopathic scoliosis. Connective tissue research. 2010;51(3):188-96
17. Merchán MA, DeFelipe J, De Castro F. Cajal and de Castro's neurohistological methods. Oxford University Press. 2016
18. Peters H, Wilm B, Sakai N. et al. Pax1 and Pax9 synergistically regulate vertebral column development. Development. 1999;126(23):5399-408
19. Kaplan KM, Spivak JM, Bendo JA. Embryology of the spine and associated congenital abnormalities. The Spine Journal. 2005;5(5):564-76
20. Bronner ME, Simões-Costa M. Chapter Seven-The Neural Crest Migrating into the Twenty-First Century. Current topics in developmental biology. 2016;116:115-34
21. Scaal M. Early development of the vertebral column. Seminars in Cell & Developmental Biology: Elsevier. 2016:83-91
22. Moore KL, Persaud TVN. The Developing Human: Clinically Oriented Embryology. Saunders, Elsevier. 2008
23. Roffers-Agarwal J, Gammill LS. Neuropilin receptors guide distinct phases of sensory and motor neuronal segmentation. Development. 2009;136(11):1879-88
24. Bronner-Fraser M, García-Castro M. Manipulations of neural crest cells or their migratory pathways. Methods in cell biology. 2008;87:75-96
25. Perris R, Perissinotto D. Role of the extracellular matrix during neural crest cell migration. Mechanisms of development. 2000;95(1):3-21
26. Henderson DJ, Ybot-Gonzalez P, Copp AJ. Over-expression of the chondroitin sulphate proteoglycan versican is associated with defective neural crest migration in the Pax3 mutant mouse (splotch). Mechanisms of development. 1997;69(1):39-51
27. McGonnell IM, Graham A. Trunk neural crest has skeletogenic potential. Current biology. 2002;12(9):767-71
28. Pettway Z, Domowicz M, Schwartz NB. et al. Age-dependent inhibition of neural crest migration by the notochord correlates with alterations in the S103L chondroitin sulfate proteoglycan. Experimental cell research. 1996;225(1):195-206
29. Le Douarin NM, Teillet M-AM. Experimental analysis of the migration and differentiation of neuroblasts of the autonomic nervous system and of neurectodermal mesenchymal derivatives, using a biological cell marking technique. Developmental biology. 1974;41(1):162-84
30. Bundy J, Rogers R, Hoffman S. et al. Segmental expression of aggrecan in the non-segmented perinotochordal sheath underlies normal segmentation of the vertebral column. Mechanisms of development. 1998;79(1):213-7
31. Krull CE. Inhibitory interactions in the patterning of trunk neural crest migration. Annals of the New York Academy of Sciences. 1998;857(1):13-22
32. Harris ML, Erickson CA. Lineage specification in neural crest cell pathfinding. Developmental Dynamics. 2007;236(1):1-19
33. Erickson CA, Perris R. The role of cell-cell and cell-matrix interactions in the morphogenesis of the neural crest. Developmental biology. 1993;159(1):60-74
34. Duband JL. Neural crest delamination and migration: integrating regulations of cell interactions, locomotion, survival and fate. Advances in experimental medicine and biology. 2006;589:45-77
35. Logan M, Martin JF, Nagy A. et al. Expression of Cre Recombinase in the developing mouse limb bud driven by a Prxl enhancer. Genesis. 2002;33(2):77-80
36. Firulli BA, Milliar H, Toolan KP. et al. Defective Hand1 phosphoregulation uncovers essential roles for Hand1 in limb morphogenesis. Development. 2017 dev149963
37. Mishina Y, Snider TN. Neural crest cell signaling pathways critical to cranial bone development and pathology. Experimental cell research. 2014;325(2):138-47
Author contact
![]() Corresponding author: Alla M. Zaydman, AZaydmanru and Vladimir M. Subbotin, vsubbotincom, vsbbtinedu. Office: 608-316-3924; Fax: 608-441-0741
Corresponding author: Alla M. Zaydman, AZaydmanru and Vladimir M. Subbotin, vsubbotincom, vsbbtinedu. Office: 608-316-3924; Fax: 608-441-0741

 Global reach, higher impact
Global reach, higher impact