3.2
Impact Factor
ISSN: 1449-1907
Int J Med Sci 2018; 15(4):376-384. doi:10.7150/ijms.22823 This issue Cite
Research Paper
SERPINB5 Expression: Association with CCRT Response and Prognostic Value in Rectal Cancer
1. Department of Pathology, E-DA Hospital, I-Shou University, Kaohsiung, Taiwan
2. School of Medicine, I-Shou University, Kaohsiung, Taiwan
3. Department of Pathology, Taipei Medical University Hospital, Taipei, Taiwan
4. Department of Pathology, School of Medicine, College of Medicine, Taipei Medical University, Taipei, Taiwan
5. Department of Pathology, Chi-Mei Medical Center, Tainan, Taiwan
6. Institute of Biomedical Sciences, National Sun Yat-sen University, Kaohsiung, Taiwan
7. Department of Medical Research, Chi-Mei Medical Center, Chiali Branch, Tainan, Taiwan
#These two authors equally contributed to this work.
Received 2017-9-14; Accepted 2018-1-5; Published 2018-2-12
Abstract
Background: Due to the varying characteristics and conflicting outcomes on the overall survival of rectal cancer patients, many studies have been undertaken to determine various prognostic and predictive factors for the mainstay treatment of CCRT followed by surgery. Cancer cell motility contributes to tumor invasion, migration and eventually metastasis. However, the genes associated with cell motility (i.e., GO:0048870) have not been systemically evaluated in rectal cancers.
Methods: A comparative analysis of gene expression profiles was applied to the transcriptomic dataset (GSE35452) with a focus on genes associated with cell motility (GO:0048870), where SERPINB5 was recognized as the most significantly up-regulated gene. Tumor samples from 172 primary rectal cancer patients who underwent neoadjuvant CCRT followed by surgical resection were collected. Immunohistochemistry was used to semi-quantitatively assess the expression level of SERPINB5 protein. Statistical analyses of SERPINB5 expression and various clinicopathological features as well as survival were then performed.
Results: High immunoreactivity of SERPINB5 was significantly linked to pre- and post-CCRT advanced disease, lymphovascular invasion, and poor response to CCRT (all P ≤ 0.015). SERPINB5 overexpression was not only negatively associated with disease-specific survival (DSS), local recurrence-free survival (LRFS) and metastasis-free survival (MeFS) rates in univariate analyses but also was an independent prognostic factor for DSS and MeFS in rectal cancer patients (all P ≤ 0.043).
Conclusion: SERPINB5 may play an important role in rectal cancer progression and response to neoadjuvant CCRT and serve as a novel prognostic factor.
Keywords: CCRT, chemoradiotherapy, Maspin, rectal cancer, SERPINB5
Introduction
Colorectal cancer is the third most common cancer in men (746,000 cases per year, 10.0% of the total number of men with cancer) and the second in women (614,000 cases, 9.2% of the total number of women with cancer) worldwide [1]. In the United States, cancer is the second leading cause of mortality, of which colon and rectal cancer are the second leading cause of cancer death. Rectal cancer also accounts for the second most common cancer in the large intestines [2-4]. In Taiwan throughout the years, there has been a consistent increase in colorectal cancer mortality that has shown a strong association with aging [5].
Many risk factors have been associated with rectal cancer, including family history, physical activity, cigarette smoking, and consumption of red meats, fish, fried foods and oils [6-8]. Adequate management of rectal cancer requires a multidisciplinary approach with preoperative staging to determine the need for neoadjuvant therapy or the type and extent of surgery required. The mainstay treatment for mid and distal rectal cancer is total mesorectal excision surgery and a combination of surgical resection and chemoradiotherapy for the lower two-thirds of the rectum [9-12]. Radical tumor resection following neoadjuvant concurrent chemoradiotherapy (CCRT) is now the gold standard treatment for patients with rectal cancers that invade through the muscularis propria or have regional lymph node metastasis [13].
With the advancement of biological technology, many studies have attempted to identify the effects of neoadjuvant chemoradiation therapy on rectal cancers with different molecular characteristics. These prognostic and predictive biomarkers can facilitate risk stratification according to the genes present in order to plan the best treatment strategy for patients with rectal cancer [14]. Some of the major biomarkers derived from clinical studies in colorectal cancer include EGFR copy number, EGFR ligand expression, activating KRAS mutations in codons 12 and 13, KRAS G13D mutation, NRAS and BRAF mutations, PIK3CA exon 20 mutations, Serpin B5, and mucinous or signet-ring histopathology. Although many of these biomarkers have shown predictive efficacy, they require further clinical validation [15].
Cancer cell motility contributes to tumor invasion, migration and eventually metastasis, which are the fundamental characteristics of cancer [16]. After analyzing the gene expression profiling associated with cell motility (GO:0048870) based on a transcriptomic database on CCRT response in rectal cancer (GSE35452), the gene serpin family B member 5 (SERPINB5) was shown to be the most significantly up-regulated in a non-responder group.
The SERPINB5 gene encodes a 375-amino acid, 42-kDa protein, SERPINB5, also known as Maspin (mammary serine protease inhibitor). SERPINB5 protein was first reported in 1994 as a serine protease inhibitor (serpin) with tumor suppressive properties and has been extensively researched throughout the years [17]. SERPINB5 has been classified as a tumor suppressor that is lost in breast and prostate cancer and can be used as potential diagnostic marker for tumor progression. Strong expression has also been associated with CEA levels and a worse prognosis in colorectal cancer. Studies have shown that SERPINB5 may have a stage-specific function that is possibly related to tumor cell dissemination and/or metastatic outgrowth and may correlate to the aggressiveness of colorectal adenocarcinomas [18-21]. However, no research has investigated the relationship between SERPINB5 expression and the response of neoadjuvant CCRT in rectal cancer or the significance of prognostication in rectal cancer, a special type different from other anatomical counterparts. Therefore, we conducted the current study.
Materials and Methods
Analysis of the expression profiles in rectal cancer
The model established by Watanabe T et al. in 2006 on the prediction of rectal cancer sensitivity to preoperative radiotherapy by DNA microarray analysis of gene expression profiles [22] was applied to the transcriptomic dataset (GSE35452) composed of 46 rectal cancer patients who were treated with neoadjuvant CCRT. A comparative analysis of the raw .cel files of GSE35452 with a focus on the genes associated with cell motility (GO:0048870) was performed using Nexus Expression 3 software (BioDiscovery, El Segundo, CA, United States). Genes with P value<0.01 and log2-transformed expression fold change >±0.1 were selected for further analysis.
Patients and tissue samples
Between 1998 and 2004, patients at Chi Mei Medical Center (Tainan, Taiwan) with histologically verified primary rectal adenocarcinoma and adequate paraffin-embedded tissue blocks were collected first. There were 172 participants enrolled who met the inclusion criteria of primary rectal adenocarcinoma who underwent neoadjuvant CCRT followed by surgical resection with no distant metastasis. All participants were screened by chest X-radiography and/or abdominopelvic computed tomography (CT). All patients with distant metastasis were excluded. Pre-treatment clinical staging was evaluated using rectal endoscopic ultrasound (EUS) with or without abdominopelvic CT scan. All of the participants received radiation therapy at a total dose of 45 Gy in 25 fractions over a 5-week period with a 24-h continuous infusion of 5-fluorouracil concurrently before surgery. Adjuvant systemic chemotherapy was performed for those with either a positive nodal status or a tumor status of T3 to T4 in the pre-treatment (Pre-Tx) or post-treatment (Post-Tx) status. All patients were under regular follow-up after diagnosis until death or until their last appointment. Approval was granted by the Institutional Review Board of Chi Mei Medical Center (IRB10302-014).
Immunohistochemistry and histopathological evaluation
To increase the inter-observer reliability and validity and to reduce bias, two pathologists (He HL and Chang IW) were blinded to the patients' information. They separately evaluated the tumor specimens for histopathological features. Post-treatment tumor samples were assessed and staged based on the 7th American Joint Committee on Cancer (AJCC) TNM staging system [23]. The grading system of tumor regression after preoperative chemoradiotherapy was evaluated using the modified Dworak system described by Rödel [24]. The Dworak/Rödel tumor regression grade (TRG) is a five-tiered quantitative system: grade 0 indicates no regression; grade 1 indicates <25% fibrosis of the tumor mass; grade 2 indicates 25%-50% fibrosis of the tumor mass; grade 3 indicates >50% fibrosis of the tumor mass; grade 4 indicates complete regression. Immunohistochemistry was performed to assess the expression of the SERPINB5 protein. This procedure was performed by cutting 3-mm sections from pre-treatment paraffin-embedded blocks and placing the sections onto pre-coated glass slides. Xylene was used to deparaffinize the slides followed by rehydration with ethanol. Antigen retrieval was carried out in a 10-mM citrate buffer (pH 6) after being heated for 7 min by microwave. Using 3% H2O2, endogenous peroxidases were blocked. Slides were then washed with TRIS-buffered saline (TBS) for 15 min and then incubated with a primary antibody against SERPINB5 (dilution 1:20000, rabbit polyclonal, Abcam, Cambridge, United Kingdom). The SERPINB5 immunostaining was assessed using the H-score by the following equation: H-score = ΣPi (i+1), in which i is the intensity of the stained tumor cells (0 to 3+), and Pi is the percentage of stained tumor cells of various intensities. High expression of SERPINB5 was defined as having an H-score greater than the median of all scored cases.
Statistical analysis
The relationships between SERPINB5 expression and various clinicopathological features were determined using Pearson's chi-squared (χ2) test. The Kaplan-Meier method was applied for survival analysis, including disease-free survival (DFS), local (pelvic) recurrence-free survival (LRFS), and metastasis-free survival (MeFS). Log-rank tests were used for univariate analyses. A Cox proportional hazards model was used to identify independent prognostic factors for the multivariate analysis. Statistical significance was associated when a P value was less than 0.05 under two-sided tests. All statistical analyses were performed with IBM SPSS Statistics 22.0 (IBM Corporation, Armonk, NY, U.S.).
Results
SERPINB5 gene was identified as the most significantly up-regulated gene among those linked to cell motility (GO:0048870)
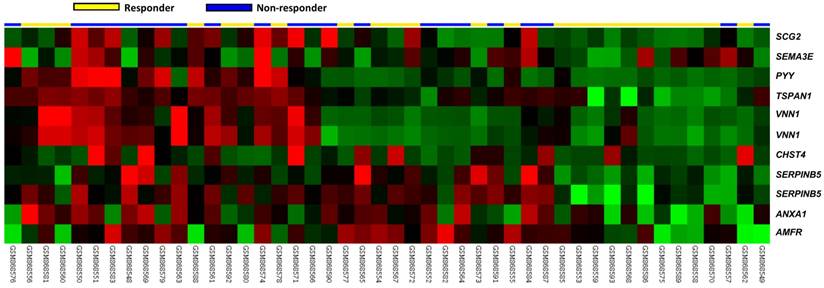
In the downloaded transcriptomic dataset of rectal cancer (GSE35452) from GEO, NCBI, 24 out of 46 (52.2%) patients were classified as responders (having a positive response to preoperative CCRT), and the remaining 22 (47.8%) patients were categorized as non-responders (having a resistance to preoperative CCRT). Eleven probes covering nine transcripts belonging to cell motility (GO:0048870) were significantly up-regulated, including the SERPINB5, VNN1, TSPAN1, AMFR, CHST4, PYY, SCG2, ANXA1 and SEMA3E genes (P ≤ 0.0092, Fig. 1). Of these, the SERPINB5 transcript exhibited the most significant up-regulation in non-responders compared to that in the responders, whose log2 ratios by comparison between the non-responders and responders were 0.2908 and 1.3577, respectively (P ≤ 0.0002, Table 1). The expression of VNN1 and its prognostic significance in rectal cancer was described in our previous study [25].
SERPINB5 expression and the associations with clinicopathological variables
The clinical and pathological features of our rectal cancer patient cohort are shown in Table 2. Among them, the majority was male (M:F = 62.8%:37.2%) and younger than seventy years old (61.6%). Eighty-one tumors (47.1%) were early cancers (T1-2) before preoperative chemoradiotherapy, whereas 91 tumors (52.9%) were advanced (T3-4). Forty-seven patients (27.3%) had lymph node metastasis, and 125 (72.7%) did not have lymph node metastasis before treatment. After neoadjuvant CCRT, half of the tumors (n = 86) were early cancers (yT0-2) while the other half (n = 86) were advanced (yT3-4). Forty-nine patients (28.5%) had pathologically confirmed nodal metastasis, and 123 (71.5%) did not after treatment. Vascular and perineural invasion was observed in 15 (8.7%) and 5 (2.9%) cases, respectively. The post-treatment prostatectomy specimens revealed no or little response to neoadjuvant CCRT in 37 cases (TRG 0-1, 21.5%), moderate response in 118 cases (TRG 2-3, 68.6%) and complete response in 17 cases (TRG 4, 9.9%).
The subcellular localization of SERPINB5 was predominantly in the cytoplasm of cancer cells in low-stage cases and in both the cytoplasm and nuclei in high-stage tumors. As demonstrated in Table 2, low immunoreactivity of SERPINB5 was significantly associated with a less advanced post-CCRT tumor invasive depth (P = 0.001), a negative pre- and post-CCRT lymph node metastasis (P < 0.001 for both), an absence of lymphovascular invasion (P = 0.015) and a better response to neoadjuvant CCRT (higher TRG, P < 0.001, Fig. 2). The expression of SERPINB5 was not significantly correlated to gender, age, pre-CCRT T status, pre-CCRT serum CEA level, or perineural invasion.
Survival analyses and the prognostic impact of SERPINB5 expression
In the univariate analyses (Table 3), a less advanced post-CCRT tumor invasive depth and a higher TRG were positively linked to DSS, LRFS and MeFS (all P ≤ 0.0040). Low pre-CCRT serum CEA and absence of lymphovascular invasion were significantly associated with improved DSS and LRFS rates (all P ≤ 0.0216). Only negative pre-CCRT nodal metastasis was significantly correlated to a higher LRFS rate (all P = 0.0070). In the multivariate analysis (Table 4), TRG was an independent prognostic factor for all survival indices (all P ≤ 0.033). Lymphovascular invasion and pre-CCRT CEA were independent indicators for DSS and LRFS (all P ≤ 0.049).
Significantly deregulated genes associated with cell motility (GO:0048870) based on CCRT response in rectal cancer
| Probe | Comparison log2 ratio | Comparison P-value | Gene Symbol | Gene Name | Biological Process | Molecular Function |
|---|---|---|---|---|---|---|
| 204855_at | 1.3577 | <0.0001 | SERPINB5 | serpin peptidase inhibitor; clade B (ovalbumin); member 5 | cell motility | serine-type endopeptidase inhibitor activity |
| 205844_at | 1.2511 | 0.0002 | VNN1 | vanin 1 | cell motility, nitrogen compound metabolic process | GPI anchor binding, hydrolase activity, hydrolase activity; acting on carbon-nitrogen (but not peptide) bonds, hydrolase activity; acting on carbon-nitrogen (but not peptide) bonds; in linear amides |
| 209114_at | 0.9852 | 0.0002 | TSPAN1 | tetraspanin 1 | cell adhesion, cell motility, cell proliferation | |
| 202203_s_at | 0.9747 | 0.0001 | AMFR | autocrine motility factor receptor | ER-associated protein catabolic process, cell motility, signal transduction, ubiquitin cycle | ligase activity, metal ion binding, protein binding, receptor activity, ubiquitin-protein ligase activity, zinc ion binding |
| 220446_s_at | 0.7544 | 0.0004 | CHST4 | carbohydrate (N-acetylglucosamine 6-O) sulfotransferase 4 | N-acetylglucosamine metabolic process, carbohydrate metabolic process, cell adhesion, cell motility, cell-cell signaling, immune response, inflammatory response, protein amino acid sulfation, sulfur metabolic process | N-acetylglucosamine 6-O-sulfotransferase activity, sulfotransferase activity, transferase activity |
| 207080_s_at | 0.7389 | 0.0092 | PYY | peptide YY | G-protein coupled receptor protein signaling pathway, cell motility, cell proliferation, cell-cell signaling, cytoskeleton organization and biogenesis, digestion, feeding behavior | hormone activity |
| 1558549_s_at | 0.6644 | 0.0022 | VNN1 | vanin 1 | cell motility, nitrogen compound metabolic process | GPI anchor binding, hydrolase activity, hydrolase activity; acting on carbon-nitrogen (but not peptide) bonds, hydrolase activity; acting on carbon-nitrogen (but not peptide) bonds; in linear amides |
| 204035_at | 0.6374 | 0.0004 | SCG2 | secretogranin II (chromogranin C) | MAPKKK cascade, angiogenesis, cell motility, endothelial cell migration, eosinophil chemotaxis, induction of positive chemotaxis, inflammatory response, intracellular signaling cascade, negative regulation of apoptosis, negative regulation of endothelial cell proliferation, positive regulation of endothelial cell proliferation, protein secretion | calcium ion binding, chemoattractant activity, cytokine activity |
| 201012_at | 0.4402 | 0.0035 | ANXA1 | annexin A1 | anti-apoptosis, arachidonic acid secretion, cell cycle, cell motility, cell surface receptor linked signal transduction, inflammatory response, keratinocyte differentiation, lipid metabolic process, peptide cross-linking, regulation of cell proliferation, signal transduction | calcium ion binding, calcium-dependent phospholipid binding, phospholipase A2 inhibitor activity, phospholipase inhibitor activity, phospholipid binding, protein binding, protein binding; bridging, receptor binding, structural molecule activity |
| 1555551_at | 0.2908 | 0.0002 | SERPINB5 | serpin peptidase inhibitor; clade B (ovalbumin); member 5 | cell motility | serine-type endopeptidase inhibitor activity |
| 206941_x_at | 0.1285 | 0.0057 | SEMA3E | sema domain; immunoglobulin domain (Ig); short basic domain; secreted; (semaphorin) 3E | cell differentiation, cell motility, multicellular organismal development, nervous system development | serine-type endopeptidase inhibitor activity |
Analysis of gene expression in rectal cancers with preoperative concurrent chemoradiotherapy using a published transcriptome dataset (GSE35452). A clustering analysis of genes focused on cell motility (GO:0048870) revealed that SERPINB5 is the most significantly up-regulated gene in non-responders compared with responders. Tumors classified as responders (yellow) or non-responders (blue) are illustrated at the top of the heat map, and the up-regulation and down-regulation of gene expression are represented as a continuum of brightness of red or green, respectively. Tumors with an unchanged transcriptional level are in black.
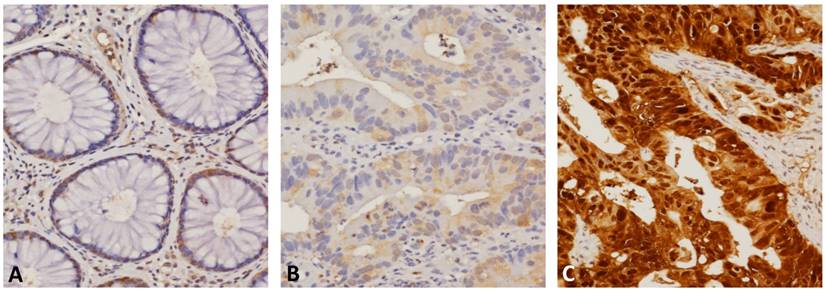
SERPINB5 immunostaining of representative sections revealed (A) low immunoreactivity in normal colonic mucosa, (B) weak cytoplasmic expression in the low-stage tumors that showed a positive response to neoadjuvant CCRT, and (C) strong combined cytoplasmic and nuclear expression in the high-stage tumors that showed resistance to CCRT.
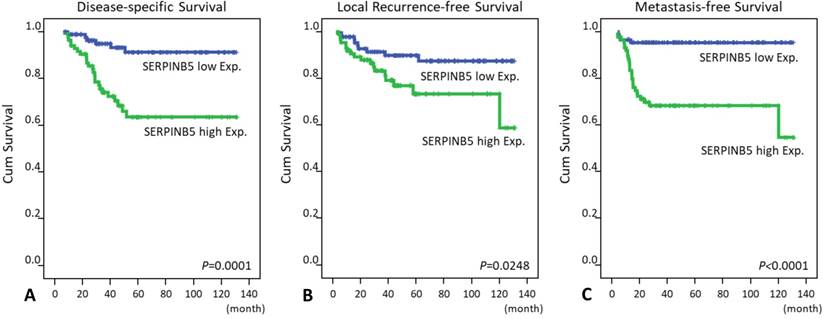
Notably, SERPINB5 overexpression not only predicted worse DSS, LRFS and MeFS outcomes in the univariate log-rank tests (P = 0.0001, P = 0.0248, P < 0.0001, respectively, Table 3 and Fig. 3) but also served as an independent poor prognosticator for DSS (hazard ratio = 2.217, confidence interval = 1.105-7.058) and MeFS (HR = 5.413, CI = 1.848-15.854; Table 4).
Discussion
Neoadjuvant CCRT followed by surgery has been the mainstay treatment for rectal cancer, but the therapeutic outcomes have varied widely among persons. This has led to an increased amount of research with the goal of identifying predictive and prognostic factors for the treatment options. Many studies have focused on identifying genes associated with cell differentiation, cell proliferation and signal transduction, but very few studies have been performed on cell motility [20, 26]. In the current study, we focused on genes associated with cell motility in response to CCRT in rectal cancer and revealed that the SERPINB5 gene yielded the highest level of dysregulated genes compared to other genes. SERPINB5, encoded by the SERPINB5 gene which belongs to the serpin superfamily, is a serine protease inhibitor and has displayed tumor suppressor activity in breast and prostatic cancers along with tumor progressive features in colorectal cancers [20, 27, 28].
Relationships between SERPINB5 expression and clinicopathological factors in rectal cancer patients receiving preoperative CCRT
| Parameter | No. (%) | SERPINB5 Expression | P-value | ||
|---|---|---|---|---|---|
| Low Exp | High Exp. | ||||
| Gender | Male | 108 (62.8) | 55 | 53 | 0.752 |
| Female | 64 (37.2) | 31 | 33 | ||
| Age | <70 | 106 (61.6) | 53 | 53 | 1.000 |
| ≥70 | 66 (38.4) | 33 | 33 | ||
| Pre-CCRT T stage | T1-T2 | 81 (47.1) | 43 | 38 | 0.445 |
| T3-T4 | 91 (52.9) | 43 | 48 | ||
| Pre-CCRT N stage | N0 | 125 (72.7) | 72 | 53 | <0.001* |
| N1-N2 | 47 (27.3) | 14 | 33 | ||
| Pre-CCRT CEA | ≤5 ng/ml | 114 (66.3) | 62 | 52 | 0.107 |
| >5 ng/ml | 58 (33.7) | 24 | 34 | ||
| Post-CCRT T stage | yT0-yT2 | 86 (50.0) | 54 | 32 | 0.001* |
| yT3-yT4 | 86 (50.0) | 32 | 54 | ||
| Post-CCRT N stage | yN0 | 123 (71.5) | 77 | 46 | <0.001* |
| yN1-yN2 | 49 (28.5) | 9 | 40 | ||
| Lymphovascular invasion | Absent | 157 (91.3) | 83 | 74 | 0.015* |
| Present | 15 (8.7) | 3 | 12 | ||
| Perineural invasion | Absent | 167 (97.1) | 85 | 82 | 0.173 |
| Present | 5 (2.9) | 1 | 4 | ||
| Tumor regression grade | Grade 0-1 | 37 (21.5) | 10 | 27 | <0.001* |
| Grade 2-3 | 118 (68.6) | 61 | 57 | ||
| Grade 4 | 17 (9.9) | 15 | 2 | ||
*, statistically significant
Univariate log-rank analysis for important clinicopathological variables and SERPINB5 expression
Parameter | No. of case (%) | Disease-specific survival | Local recurrence-free survival | Metastasis-free survival | ||||
|---|---|---|---|---|---|---|---|---|
| No. of event | P | No. of event | P | No. of event | P | |||
| Gender | Male | 108 (62.8) | 20 | 0.9026 | 7 | 0.2250 | 17 | 0.3520 |
| Female | 64 (37.2) | 11 | 20 | 14 | ||||
| Age | <70 | 106 (61.6) | 19 | 0.8540 | 18 | 0.6615 | 20 | 0.7427 |
| ≥70 | 66 (38.4) | 12 | 9 | 11 | ||||
| Pre-CCRT T stage | T1-T2 | 81 (47.1) | 10 | 0.0776 | 10 | 0.2261 | 11 | 0.1745 |
| T3-T4 | 91 (52.9) | 21 | 17 | 20 | ||||
| Pre-CCRT N stage | N0 | 125 (72.7) | 19 | 0.0711 | 15 | 0.0070* | 19 | 0.0973 |
| N1-N2 | 47 (27.3) | 21 | 12 | 12 | ||||
| Pre-CCRT CEA | ≤5 ng/ml | 114 (66.3) | 15 | 0.0216* | 13 | 0.0179* | 17 | 0.1460 |
| >5 ng/ml | 58 (33.7) | 16 | 14 | 14 | ||||
| Post-CCRT T stage | yT0-yT2 | 86 (50.0) | 7 | 0.0006* | 7 | 0.0040* | 8 | 0.0033* |
| yT3-yT4 | 86 (50.0) | 24 | 20 | 23 | ||||
| Post-CCRT N stage | yN0 | 123 (71.5) | 21 | 0.5998 | 16 | 0.1320 | 20 | 0.4634 |
| yN1-yN2 | 49 (28.5) | 10 | 11 | 11 | ||||
| Lymphovascular invasion | Absent | 157 (91.3) | 25 | 0.0184* | 21 | 0.0028* | 27 | 0.4470 |
| Present | 15 (8.7) | 6 | 6 | 4 | ||||
| Perineural invasion | Absent | 167 (97.1) | 29 | 0.2559 | 25 | 0.0940 | 30 | 0.9083 |
| Present | 5 (2.9) | 2 | 2 | 1 | ||||
| Tumor regression grade | Grade 0-1 | 37 (21.5) | 13 | 0.0038* | 10 | 0.0090* | 14 | 0.0006* |
| Grade 2-3 | 118 (68.6) | 17 | 17 | 16 | ||||
| Grade 4 | 17 (9.9) | 1 | 0 | 1 | ||||
| SERPINB5 expression | Low Exp. | 86 (50.0) | 6 | 0.0001* | 9 | 0.0248* | 4 | <0.0001* |
| High Exp. | 86 (50.0) | 25 | 18 | 27 | ||||
*, statistically significant
Multivariate survival analysis
| Parameter | Disease-specific survival | Local recurrence-free survival | Metastasis-free survival | ||||||
|---|---|---|---|---|---|---|---|---|---|
| HR | 95% CI | P | HR | 95% CI | P | HR | 95% CI | P | |
| Tumor regression grade | 2.217 | 1.066-4.608 | 0.033* | 2.933 | 1.316-6.536 | 0.009* | 1.484 | 1.060-2.075 | 0.021* |
| SERPINB5 expression | 2.792 | 1.105-7.058 | 0.030* | 0.996 | 0.408-2.435 | 0.994 | 5.413 | 1.848-15.854 | 0.002* |
| Lymphovascular invasion | 2.671 | 1.004-7.108 | 0.049* | 4.097 | 1.337-12.551 | 0.014* | - | - | - |
| Pre-CCRT CEA | 2.180 | 1.025-4.639 | 0.043* | 2.653 | 1.090-6.028 | 0.031* | |||
| Post-CCRT T stage | 1.963 | 0.814-4.734 | 0.133 | 1.635 | 0.650-4.109 | 0.296 | 1.700 | 0.747-3.873 | 0.206 |
| Pre-CCRT N stage | - | - | - | 1.581 | 0.653-3.827 | 0.310 | - | - | - |
Kaplan-Meier survival curves demonstrate the significant prognostic impact of SERPINB5 expression on disease-specific survival (P = 0.0001), local recurrence-free survival (P = 0.0248) and metastasis-free survival (P < 0.0001).
SERPINB5 is located on chromosome 18q21.3-q23 and encodes the serpin family B member 5 (SERPINB5) protein, also called Maspin (mammary serine protease inhibitor) [17]. SERPINB5 belongs to serpin (serine protease inhibitor) superfamily, which irreversibly inhibits the target protease via a large conformational change to disorganize the binding or catalytic sites. The specific inhibitory mechanism is referred as the “stressed and relaxed” transition. All proteins of the serpin superfamily contain a reactive center loop (RCL), which is a key substructure to permit the reactive site presentation in an ideal configuration for binding and inhibition of the target protease [26]. Nonetheless, the RCL of SERPINB5 is relatively short, divergent and hydrophobic and is incapable of conducting the transition [29]. Hence, SERPINB5 is considered a non-inhibitory member of the serpin superfamily, and researchers have paid more attention to its tumor suppressive properties. The G-helix and RCL of SERPINB5 mediate the effects of cell migration and cell adhesion [30-33]. A 15-mer G-helix peptide binding to the β1 integrin, RCL-mediated cell adhesion to type I collagen and fibronectin regulate the interaction of cells and the extracellular matrix, which is necessary for tumor invasion, migration and eventually metastasis [34]. The SERPINB5 gene was first identified as a tumor suppressor gene by Zou et al. in 1994 [17]. SERPINB5 was expressed in normal human breast epithelial cells but not in most breast cancer cell lines. SERPINB5-transfected breast cancer cells also showed reduced abilities for invasion and metastasis in vitro and in vivo, respectively. Expression of SERPINB5 was also associated with a better prognosis in prostate cancer [35], bladder cancer [36, 37], non-small cell lung cancer [38, 39] and ovarian cancer [40]. However, SERPINB5 was overexpressed in pancreatic [41], gallbladder [42], thyroid [43], as well as colorectal cancers [44]. Up-regulation of SERPINB5 was also significantly correlated with advanced invasive depth, high Dukes' stage and high-grade tumor budding [45]. In a study by Märkl et al, nuclear expression of SERPINB5 was associated with shorter overall survival intervals compared with cytoplasmic expression in colorectal cancer patients without lymph node metastasis [46]. Nuclear SERPINB5 expression was not only an independent unfavorable prognosticator predicting lower overall survival rate (hazard ratio 2.08; 95% CI, 1.13-3.81) but also an indicator of a positive response to adjuvant 5-FU-based chemotherapy (hazard ratio 0.384; 95% CI, 0.188-0.784) for patients with stage III (nodal positive) colon cancer in another study [47]. In contrast, a recent investigation demonstrated that nuclear localization of SERPINB5 was mandatory for the tumor suppressor properties, where SERPINB5 bound to chromatin and inhibited metastasis of breast and ovarian cancer cells [48]. The discrepancy is still obscure and may be due to different biological functions of the same protein in different cancers. Moreover, in the current study, we illustrated that the expression of SERPINB5 was predominantly in the cytoplasm of low-stage rectal cancer cells and tended to be expressed in both the cytoplasm and nuclei of high-stage rectal cancer cells. However, low immunoreactivity was significantly associated with a positive response to preoperative chemoradiotherapy. The divergence is probably caused by different populations of cohorts (patients with nodal-positive colon cancers with/without adjuvant chemotherapy vs. rectal cancer patients who received neoadjuvant CCRT).
In conclusion, up-regulated expression of SERPINB5 was associated with adverse clinical and pathological features, including neoadjuvant CCRT resistance in rectal cancer patients. SERPINB5 overexpression was also an independent prognostic indicator for predicting worse survival rates (DSS and MeFS). SERPINB5 may play an important role in rectal cancer progression and in the response to neoadjuvant CCRT and serve as a novel prognostic factor. Although many genes have been identified and are used as prognostic biomarkers for rectal cancers, the results of this study add value to the overall management and treatment outcomes of colorectal cancer patients. Clinicians can predict the efficacy of neoadjuvant CCRT in the presence of up-regulated SERPINB5 prior to initiating the treatment. Further investigations to elucidate the comprehensive molecular mechanisms of SERPINB5 in the oncogenesis of rectal cancer are necessary for developing a potential SERPINB5-targeted therapy for high-risk patients, as we have described the promising therapeutic targets for patients with rectal cancer [49, 50].
Abbreviations
AJCC: American Joint Committee on Cancer; CCRT: concurrent chemoradiotherapy; DSS: disease-free survival; LRFS: local recurrence-free survival; MeFS: metastasis-free survival; SERPINB5: serpin family B member 5; TRG: tumor regression grade.
Acknowledgements
This study was supported by grants from E-Da Hospital (EDAHP106038 and EDAHP106055) and Chi-Mei Medical Center, Chiali Branch, (CCFHR10606 and CCFHR10502).
Competing Interests
The authors have declared that no competing interest exists.
References
1. WHO: Geneva, Switzerland. Colorectal Cancer Estimated Incidence, Mortality and Prevalence Worldwide in 2012. 2017. http://globocan.iarc.fr/Pages/fact_sheets_cancer.aspx?cancer=colorectal
2. NIH: Bethesda, MD, USA. Cancer Stat Facts: Colon and Rectum Cancer. http://seer.cancer.gov/statfacts/html/colorect.html
3. Siegel RL, Miller KD, Jemal A. Cancer statistics, 2015. CA: A Cancer Journal for Clinicians. 2015;65:5-29
4. Siegel R, Desantis C, Jemal A. Colorectal cancer statistics, 2014. CA Cancer J Clin. 2014;64:104-17
5. Su SY, Huang JY, Jian ZH, Ho CC, Lung CC, Liaw YP. Mortality of colorectal cancer in Taiwan, 1971-2010: temporal changes and age-period-cohort analysis. International Journal of Colorectal Disease. 2012;27:1665-72
6. Iswarya SK, Premarajan KC, Kar SS, Kumar SS, Kate V. Risk factors for the development of colorectal carcinoma: A case control study from South India. World J Gastrointest Oncol. 2016;8:207-14
7. Cheng J, Chen Y, Wang X. et al. Meta-analysis of prospective cohort studies of cigarette smoking and the incidence of colon and rectal cancers. Eur J Cancer Prev. 2015;24:6-15
8. Wei EK, Giovannucci E, Wu K. et al. Comparison of risk factors for colon and rectal cancer. Int J Cancer. 2004;108:433-42
9. Wiig JN, Giercksky KE, Tveit KM. Intraoperative radiotherapy for locally advanced or locally recurrent rectal cancer: Does it work at all? Acta Oncol. 2014;53:865-76
10. Petrelli F, Coinu A, Lonati V, Barni S. A systematic review and meta-analysis of adjuvant chemotherapy after neoadjuvant treatment and surgery for rectal cancer. Int J Colorectal Dis. 2015;30:447-57
11. Breugom AJ, Swets M, Bosset JF. et al. Adjuvant chemotherapy after preoperative (chemo)radiotherapy and surgery for patients with rectal cancer: a systematic review and meta-analysis of individual patient data. Lancet Oncol. 2015;16:200-7
12. McCarthy K, Pearson K, Fulton R, Hewitt J. Pre-operative chemoradiation for non-metastatic locally advanced rectal cancer. Cochrane Database of Systematic Reviews. 2012
13. Benson AB 3rd, Venook AP, Bekaii-Saab T. et al. Rectal Cancer, Version 2.2015. J Natl Compr Canc Netw. 2015;13:719-28
14. McCourt M, Armitage J, Monson JR. Rectal cancer. Surgeon. 2009;7:162-9
15. De Divitiis C, Nasti G, Montano M, Fisichella R, Iaffaioli RV, Berretta M. Prognostic and predictive response factors in colorectal cancer patients: between hope and reality. World J Gastroenterol. 2014;20:15049-59
16. Brown GT, Murray GI. Current mechanistic insights into the roles of matrix metalloproteinases in tumour invasion and metastasis. J Pathol. 2015;237:273-81
17. Zou Z, Anisowicz A, Hendrix MJ. et al. Maspin, a serpin with tumor-suppressing activity in human mammary epithelial cells. Science. 1994;263:526-9
18. Baek JY, Yeo HY, Chang HJ. et al. Serpin B5 is a CEA-interacting biomarker for colorectal cancer. Int J Cancer. 2014;134:1595-604
19. Bailey CM, Khalkhali-Ellis Z, Seftor EA, Hendrix MJ. Biological functions of maspin. J Cell Physiol. 2006;209:617-24
20. Snoeren N1, Emmink BL, Koerkamp MJ. et al. Maspin is a marker for early recurrence in primary stage III and IV colorectal cancer. Br J Cancer. 2013;109:1636-47
21. Umekita Y, Souda M, Yoshida H. Expression of maspin in colorectal cancer. In Vivo. 2006;20:797-800
22. Watanabe T, Komuro Y, Kiyomatsu T. et al. Prediction of sensitivity of rectal cancer cells in response to preoperative radiotherapy by DNA microarray analysis of gene expression profiles. Cancer Res. 2006;66:3370-4
23. AJCC Cancer Staging Manual. 7th ed. Springer: Berlin. 2011
24. Rödel C, Martus P, Papadoupolos T. et al. Prognostic significance of tumor regression after preoperative chemoradiotherapy for rectal cancer. J Clin Oncol. 2005;23:8688-96
25. Chai CY, Zhang Y, Song J, Lin SC, Sun S, Chang IW. VNN1 overexpression is associated with poor response to preoperative chemoradiotherapy and adverse prognosis in patients with rectal cancers. Am J Transl Res. 2016;8:4455-63
26. Berardi R, Morgese F, Onofri A. et al. Role of maspin in cancer. Clin Transl Med. 2013;2:8
27. Joensuu KM, Leidenius MH, Andersson LC, Heikkilä PS. High expression of maspin is associated with early tumor relapse in breast cancer. Hum Pathol. 2009;40:1143-51
28. Sheng S, Carey J, Seftor EA, Dias L, Hendrix MJ, Sager R. et al. Maspin acts at the cell membrane to inhibit invasion and motility of mammary and prostatic cancer cells. Proc Natl Acad Sci USA. 1996;93:11669-74
29. Pemberton PA, Wong DT, Gibson HL. et al. The tumor suppressor maspin does not undergo the stressed to relaxed transition or inhibit trypsin-like serine proteases. Evidence that maspin is not a protease inhibitory serpin. J Biol Chem. 1995;270:15832-7
30. Cella N, Contreras A, Latha K, Rosen JM, Zhang M. Maspin is physically associated with [beta]1 integrin regulating cell adhesion in mammary epithelial cells. FASEB J. 2006;20:1510-2
31. Ravenhill L, Wagstaff L, Edwards DR, Ellis V, Bass R. G-helix of maspin mediates effects on cell migration and adhesion. J Biol Chem. 2010;285:36285-92
32. Seftor RE, Seftor EA, Sheng S, Pemberton PA, Sager R, Hendrix MJ. Maspin suppresses the invasive phenotype of human breast carcinoma. Cancer Res. 1998;58:5681-5
33. Zhang W, Shi HY, Zhang M. Maspin overexpression modulates tumor cell apoptosis through the regulation of Bcl-2 family proteins. BMC Cancer. 2005;5:50
34. Chang IW, Liang PI, Li CC. et al. HAS3 underexpression as an indicator of poor prognosis in patients with urothelial carcinoma of the upper urinary tract and urinary bladder. Tumour Biol. 2015;36:5441-50
35. Machtens S, Serth J, Bokemeyer C. et al. Expression of the p53 and Maspin protein in primary prostate cancer: correlation with clinical features. Int J Cancer. 2001;95:337-42
36. Kramer MW, Waalkes S, Hennenlotter J. et al. Maspin protein expression correlates with tumor progression in non-muscle invasive bladder cancer. Oncol Lett. 2010;1:621-626
37. Acikalin D, Oner U, Can C, Acikalin MF, Colak E. Predictive value of maspin and Ki-67 expression in transurethral resection specimens in patients with T1 bladder cancer. Tumori. 2012;98:344-50
38. Berardi R, Santinelli A, Onofri A. et al. Maspin expression is a favorable prognostic factor in non-small cell lung cancer. Anal Quant Cytol Histol. 2012;34:72-8
39. Katakura H, Takenaka K, Nakagawa M. et al. Maspin gene expression is a significant prognostic factor in resected non-small cell lung cancer (NSCLC). Maspin in NSCLC. Lung Cancer. 2006;51:323-8
40. Klasa-Mazurkiewicz D, Narkiewicz J, Milczek T, Lipińska B, Emerich J. Maspin overexpression correlates with positive response to primary chemotherapy in ovarian cancer patients. Gynecol Oncol. 2009;113:91-8
41. Liu H, Shi J, Anandan V. et al. Reevaluation and identification of the best immunohistochemical panel (pVHL, Maspin, S100P, IMP-3) for ductal adenocarcinoma of the pancreas. Arch Pathol Lab Med. 2012;136:601-9
42. Kim J, Jang KT, Kim KH. et al. Aberrant maspin expression is involved in early carcinogenesis of gallbladder cancer. Tumour Biol. 2010;5:471-6
43. Ito Y, Yoshida H, Tomoda C. et al. Maspin expression is directly associated with biological aggressiveness of thyroid carcinoma. Thyroid. 2004;14:13-8
44. Zheng H, Tsuneyama K, Cheng C. et al. Maspin expression was involved in colorectal adenoma-adenocarcinoma sequence and liver metastasis of tumors. Anticancer Res. 2007;27:259-65
45. Umekita Y, Souda M, Yoshida H. Expression of maspin in colorectal cancer. In Vivo. 2006;20:797-800
46. Märkl B, Arnholdt HM, Jähnig H. et al. Shift from cytoplasmic to nuclear maspin expression correlates with shorter overall survival in node-negative colorectal cancer. Hum Pathol. 2010;41:1024-33
47. Dietmaier W1, Bettstetter M, Wild PJ. et al. Nuclear Maspin expression is associated with response to adjuvant 5-fluorouracil based chemotherapy in patients with stage III colon cancer. Int J Cancer. 2006;118:2247-54
48. Goulet B, Chan G, Chambers AF, Lewis JD. An emerging role for the nuclear localization of maspin in the suppression of tumor progression and metastasis. Biochem Cell Biol. 2012;90:22-38
49. Tian YF, Hsieh PL, Lin CY. et al. High Expression of Aldolase B Confers a Poor Prognosis for Rectal Cancer Patients Receiving Neoadjuvant Chemoradiotherapy. J Cancer. 2017;8:1197-204
50. Lin SC, Chang IW, Hsieh PL. et al. High Immunoreactivity of DUOX2 Is Associated With Poor Response to Preoperative Chemoradiation Therapy and Worse Prognosis in Rectal Cancers. J Cancer. 2017;8:2756-64
Author contact
![]() Corresponding author: Shou-Chun Yu, Department of Medical Research, Chi Mei Medical Center, Chiali Branch, Tainan, Taiwan, No.606, Jialising, Xinghua Vil., Jiali Dist., Tainan City 722, Taiwan, E-mail: sherry0517com, TEL: 866-6-7263333 ext. 9, FAX: 866-6-7264612
Corresponding author: Shou-Chun Yu, Department of Medical Research, Chi Mei Medical Center, Chiali Branch, Tainan, Taiwan, No.606, Jialising, Xinghua Vil., Jiali Dist., Tainan City 722, Taiwan, E-mail: sherry0517com, TEL: 866-6-7263333 ext. 9, FAX: 866-6-7264612

 Global reach, higher impact
Global reach, higher impact