3.2
Impact Factor
ISSN: 1449-1907
Int J Med Sci 2018; 15(2):142-152. doi:10.7150/ijms.21645 This issue Cite
Research Paper
MiR-22-3p Regulates Cell Proliferation and Inhibits Cell Apoptosis through Targeting the eIF4EBP3 Gene in Human Cervical Squamous Carcinoma Cells
1. Department of Gynaecology and Obstetrics, Qixia District Maternity and Child Health Care Hospital, Nanjing, 210028, China;
2. Department of Gynaecology and Obstetrics, Huangdao District of Traditional Chinese Medicine, Qingdao, 266500, China;
3. State Key Laboratory of Reproductive Medicine, Department of Clinical Laboratory, Nanjing Maternity and Child Health Care Hospital affiliated to Nanjing Medical University, Nanjing, 210004, China.
# Kang-tai Lv and Zhu-Liu contributed equally to this work.
* Jing-ping Chu and Ling-juan Gao contributed equally to this work.
Received 2017-6-27; Accepted 2017-10-31; Published 2018-1-1
Abstract
Background: MicroRNAs (miRNAs) are non-coding small RNAs that function as negative regulators of gene expression and are involved in tumour biology. The eIF4E-binding proteins (eIF4EBPs) play essential roles in preventing translation initiation and inhibiting protein synthesis at a global or message-specific level in a variety of tumours. Methods: According to comparative miRNA profiles of clinical cervical cancer and non-cancerous cervical tissue specimens, several miRNAs were aberrantly expressed in the cervical cancer samples. C33a and SiHa cell proliferation and apoptosis were detected using methyl thiazolyl tetrazolium (MTT) and flow cytometry assays, respectively. Results: Among the aberrantly expressed miRNAs, miR-22-3p was significantly differentially expressed in cervical cancer tissues and was highly associated with cervical cancer cell growth regulation. In addition, bioinformatic predictions and experimental validation were used to identify whether eIF4E-binding protein 3 (eIF4EBP3) was a direct target of miR-22-3p; eIF4EBP3 protein levels were generally low in the cervical cancer tissues. Furthermore, functional studies revealed that either a miR-22-3p inhibitor or eIF4EBP3 overexpression could induce apoptosis in cervical cancer cells in vitro. Importantly, we found that eIF4EBP3 accumulation could significantly attenuate cervical cancer cell proliferation triggered by a miR-22-3p mimic as well as enhance apoptosis in cervical cancer cells. Conclusion: Taken together, our data provide primary proof that miR-22-3p can induce cervical cancer cell growth at least in part by up-regulating its expression to decrease eIF4EBP3 expression levels; miR-22-3p thus holds promise as a prognostic biomarker and potential therapeutic target for treating cervical cancer.
Keywords: MicroRNA-22-3p (miR-22-3p), eIF4E-binding protein 3 (eIF4EBP3), Apoptosis, Human cervical squamous carcinoma cells.
Introduction
Cervical carcinoma is the fourth most common female cancer and is responsible for high rates of incidence and mortality worldwide [1]. In China, cervical cancer is one of the most serious diseases affecting the physical and mental health of women; there were 87,982 new patients diagnosed with cervical cancer and 23,375 deaths in 2011 [2]. Cervical cancer develops when the normal cervical epithelium transforms into preneoplastic cervical intraepithelial neoplasia (CIN), which ultimately progresses to invasive cervical cancer cells; this is a multi-step process that involves multiple genes [3-4]. Clearly, understanding the molecular mechanisms of cervical cancer would have significant clinical impacts.
The eIF4E-binding proteins (eIF4EBPs) play essential roles in preventing translation initiation and inhibiting protein synthesis at a global or message-specific level [5]. During translation initiation, eIF4EBPs negatively regulate cell proliferation and act as tumour suppressors, and these actions are frequently dysregulated in cancer [6]. In addition, using eIF4EBPs for genetic or pharmacologic strategies can induce apoptosis and inhibit proliferation, leading to suppressive effects on various cancers [7-8]. As we know, cervical cancer is an epigenetic disease that is subject to typical epigenetic alterations, such as changes in DNA methylation, histone modification and microRNA expression levels [9]. MicroRNAs (miRNAs), a family of single-stranded non-coding RNAs that are19-24 nucleotides in length, regulate the transcription and translation of target mRNAs via binding to their 3'-untranslated regions (3'-UTRs); this action ultimately results in mRNA deregulation and transcriptional repression [10-11]. There is evidence that functional miRNAs mediate many biological tumour processes, including cell proliferation, invasion, apoptosis and cellular metabolism [12]. Currently, multiple studies have confirmed that altering miRNA expression facilitates the development of cervical cancer. Potential miRNAs involved in the regulation of eIF4EBPs have yet to be discovered.
This study aimed to determine whether a particular subtype of eIF4EBP is directly down-regulated by miR-22-3p; these findings have implications for the therapeutic treatment of cervical cancer development and progression.
Materials and Methods
Reagents
Cervical squamous carcinoma cell lines C33a (HPV-16 negative) and SiHa (HPV-16 positive) were obtained from Hangzhou Hibio Bio-tech Co., Ltd. (Hangzhou, Zhejiang, China). An Annexin V-FITC/ Propidium Iodide (PI) Flow Cytometry Assay Kit was purchased from Invitrogen (Carlsbad, CA, USA). A Phototope-HRP Western Blot Detection System, including anti-mouse IgGs, a biotinylated protein ladder, HRP-linked antibodies, 20X LumiGLO Reagent and 20X peroxide, was purchased from Cell Signaling Technology (Beverly, MA, USA). Antibodies directed against eIF4EBP3 and actin were purchased from Santa Cruz (Santa Cruz, CA, USA). pcDNA-eIF4EBP3, pcDNA-eIF4EBP2, pcDNA-eIF4EBP3 mutant (mut) and pcDNA-eIF4EBP2 mutant (mut) plasmids were kindly supplied by Nanjing Dongji Bio-tech Co., Ltd. Cell culture supplies were purchased from Life Technologies (Gaithersburg, MD, USA). Unless otherwise specified, all of the other reagents were of analytical grade.
Tissue procurement and preparation
This study was approved by the Ethics Committee of the Chinese Academy of Sciences and the Nanjing Maternity and Child Health Care Hospital in Nanjing. The need for written informed consent was specially waived by the Ethics Committee Board because all clinical materials were deidentified.
For the tissue and blood sample collections, we recruited women who underwent cervical lesions from January 2009 to January 2015 at Nanjing Maternity and Child Health Care Hospital. Human cervical cancer specimens were obtained from 30 HPV-16/18-positive patients (median age of 43 years, age range of 25-56 years). Thirty patients (median age of 45 years, age range of 26-59 years) whose pathological diagnoses were mild cervicitis or who had no obvious pathological changes were chosen as the non-cancerous cervical tissue counterparts (Controls). Regarding the source of cervical tissues in control group, some cervical tissues were collected from non-cervical cancer patients who had a hysterectomy for hysteromyoma or adenomyosis; other tissues came from patients who had a tissue biopsy for non-cancer diagnoses. Those tissues (positive for HPV-16 or HPV-18; HPV typing was performed using a gene chip technique) were all examined by pathologists. Before HPV analyses, the presence or absence of CT, NG, GV, MG, TV, MH, and HSV-2 sexually transmitted pathogens was determined by routine clinical microbiology methods.
C33a and SiHa Cell Culture and DNA Transfection Conditions
C33a and SiHa cells were grown in Dulbecco's modified Eagle's medium (Gibco BRL, Grand Island, NY, USA) supplemented with 1% nonessential amino acids, 2 mM glutamine and 10% foetal bovine serum in a 37°C incubator with 5% CO2. Wild-type eIF4EBP3 and eIF4EBP2 cDNA was cloned using an RNA PCR Core Kit (Applied Biosystems). The primers used for PCR were as follows: eIF4EBP3, 5'-AAG TTC CTG CTG GAG TGC AAG A-3' (sense) and 5'-TCT CCT GCT CCT TCA GCT CCT C-3' (antisense); eIF4EBP2, 5'-TTT GCA TTC ACC CTC CTT CCC A-3' (sense) and 5'-AGG GCA CCA AAT CCA ACC AGA A-3' (antisense); the PCR cycling parameters used were as follows: 94°C for 45 s; 60°C for 30 s; and 68°C for 80 s for 30 cycles, followed by 68°C for 20 s. Complementary DNA (cDNA) for eIF4EBP3 and eIF4EBP2 was cloned in frame using BamHI/EcoRI sites into the pcDNA 3.1 expression plasmid (Invitrogen, Carlsbad, CA). The resulting pcDNA-eIF4EBP3 and pcDNA-eIF4EBP2 vectors were then transfected into C33a and SiHa cells. Following serum starvation for an additional 24 h, the cells were transfected using LipofectamineTM reagent (Life Technologies, Gaithersburg, MD, USA) according to the manufacturer's protocol. Briefly, 0.05-1.5 μg/ml plasmid DNA and 12 μg/ml LipofectamineTM reagent were diluted in serum-free DMEM. Subsequently, the Lipofectamine 2000/eIF4EBP3 vector or the Lipofectamine 2000/eIF4EBP2 vector mixture was added the cells and incubated at 37 °C in a 5% CO2 at mosphere for 12 h. Finally, 1 ml of growth medium (20% FCS) per well was added. Reporter gene activities were normalised to total protein levels, and all of the results are an average from triplicate experiments.
miR-22-3p Lentivirus Construction
The lentivirus gene transfer vectors carrying the precursor of hsa-miR-22-3p and encoding mCherry as a markerwere constructed by Genechem Co., Ltd., Shanghai, China, and confirmed by DNA sequencing. The RNA primers were 5'-GGG AAG CTG CCA GTT GAA G-3' (sense) and 5'-GTG CGT GTC GTG GAG TCG-3' (antisense). Mutant 3'-UTRs were generated by the overlap-extension PCR method. Both wild-type and mutant 3'-UTR fragments were subcloned into the pGL3-control vector (Promega, Madison, WI) downstream of the stop codon for the luciferase gene.
Electron Microscopy
Cervical tissue was obtained by cutting longitudinal sections with a 3-5 mm maximum thickness. Next, the blocks were immersed immediately for 2 h in 2.5% glutaraldehyde. After an overnight wash in sodium phosphate buffer, the tissue blocks were postfixed in 1% OsO4 for 1 h and stained with 1% uranyl acetate. Next, the tissue blocks were dehydrated, flat-embedded in Durcupan (Fluka Chemic AG, Sweden) and sectioned ata 60-70nm thickness onto 300 mesh copper slot grids. Finally, the ultrathin sections were examined at 3700X and 12500X magnification, and photographs were taken using a Zeiss 109 electron microscope.
Real-time Quantitative Polymerase Chain Reaction (Real-time qPCR)
According to the manufacturer's instructions, total RNA was extracted from cervical tissue using Trizol reagent (Invitrogen, Carlsbad, CA, USA). RNA was quantified according to its absorption at 260nm. The isolated RNA was then DNase-treated and reverse-transcribed according to manufacturer's protocol. Briefly, miRNAs were reverse transcribed using a PrimeScript reverse transcription kit, miScript SYBRGreen PCR kit and miScript primer assays according to the manufacturer's instructions (Qiagen, Valencia, CA, USA). Quantitative real-time PCR was performed using an ABI PRISM 7300 sequence detection system. Cycling parameters were 2 min at 50 °C and 10 min at 95 °C, followed by a total of 40 cycles of 15 s at 95 °C and 1 min at 60 °C. All of the reactions were performed in triplicate. The gene expression △△CT values of the miRNA were calculated by normalizingto the internal control β-actin. The relative amounts of mRNA were calculated using the following Eqn1: 2-△△CT=2-(CT.gC1qR- CT.actin)Time x + (CT.gC1qR- CT.actin)Time 0.
Luciferase assays
The mutant construct of the eIF4EBP3 or eIF4EBP2 3'-UTR were obtained by introducing the mutation into 8 nucleotides (GGCAGCUA) of the seed region for miR-22-3p. The miR-22-3p target sequences in the coding region of eIF4EBP3 or eIF4EBP2 were amplified by PCR and cloned into the GV143 vector containing a firefly luciferase reporter gene. The wild-type eIF4EBP3/eIF4EBP2 3'-UTR or mutant eIF4EBP3/eIF4EBP2 3'-UTR and the empty 3'-UTR vector were cotransfected into HEK293 cells; after incubation for 48 h, the cells were harvested and assayed for Renilla and firefly luciferase activity using adual-luciferase reporter assay system (Promega, Madison, WI, USA). The relative luciferase activities were calculated by normalizing to the Renilla luciferase activities (Renilla luciferase vector transfection was used as reference); empty 3'-UTR vector-transfected cells were used as a negative control (NC).
Western Blot Analysis
C33a and SiHa cells were collected in sample buffer and then incubated in lysis buffer and protease inhibitors for 30 min on ice. Next, the supernatants were collected after centrifugation at 13,000 × g and 4 °C for 15 min. Proteins were electrophoresed on a 10-15% denaturing polyacrylamide gel and subsequently transferred to PVDF membranes. The membranes were then blocked for 1 h in 5% non-fat milk in PBST (PBS containing 0.05% Tween 20), and the membranes were incubated at 4 °C overnight with primary antibodies. Following incubation with horseradish peroxidase-conjugated secondary antibodies for 1 h at RT, reactive protein bands were detected using an enhanced chemiluminescence (ECL) western detection system.
Cell Proliferation Assay
Methyl thiazolyl tetrazolium (MTT) assays were used to measure proliferation in C33a and SiHa cells. Cells were seeded in 96-well plates at a density of (5-7) × 103 cells per well. Cells from the different treatment groups were cultured for another 24 h, 48 h or 72 h, and 10 μL of MTT (5 mg/mL) was added to each well and incubate for 4 h at 37 °C. Then, the reaction was stopped with 100 mL of dimethyl sulfoxide (DMSO), and the absorbance was measured at 490 nm on a microplate reader.
Detection of Apoptotic Cells
Apoptosis was analysed by flow cytometry analyses using Annexin V-FITC/propidium iodide staining. After receiving different treatments for different times, C33a and SiHa cells were harvested, washed and resuspended in a binding buffer comprising10 mM HEPES, 140 mM NaCl and 2.5 mM CaCl2 (pH 7.4). Then, the cells were incubated with Annexin V-FITC and propidium iodide in the dark for 15 min. Finally, binding buffer was added, and the stained cells were analysed using a Beckman Coulter Epic XL flow cytometer. Q1_LL represents normal cells, and the early and late apoptotic cells arelocated in the Q1_LR and Q1_UR regions. The necrotic cells are distributed in the Q1_UL region. The relative ratio of early and lately apoptotic cells was chosen for further comparison.
Statistical analysis
All experiments were repeated at least three times and performed in triplicate. Data areshown as the means ± standard deviation (SD). P-values less than 0.05 were considered to be statistically significant (*p < 0.05; **p < 0.01; ***p < 0.001; #p > 0.05). Student's t-tests were used to determine differences between the experimental groups.
Results
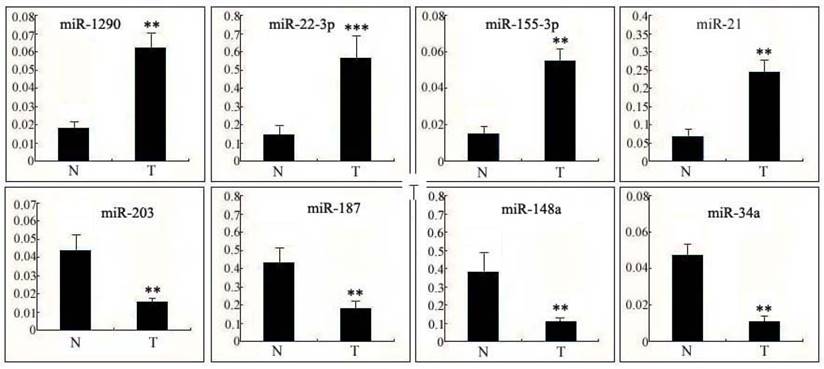
Expression of miRNAs in human cervical tissue
To further verify the results of the microarray analysis, miRNAs with more than a 5-fold difference compared with non-cancerous cervix tissues were our first choices for this experiment. The 4 most up-regulated miRNAs (miR-1290, miR-22-3p, miR-155-3p and miR-21) and 4 down-regulated miRNAs (miR-203, miR-187, miR-148a and miR-34a) in human cervical squamous cell carcinoma tissues (T) relative to non-cancerous cervix tissues (N) were confirmed using qPCR (Fig. 1). The data collected were in good agreement with the results of the preliminary screening, suggesting that the results from the microarray analysis were precise and repeatable. From the data obtained, we chose the miRNA with the greatest fold change- miR-22-3p, which is hypothesized to be closely correlated with cell growth regulation in cervical carcinoma cells.
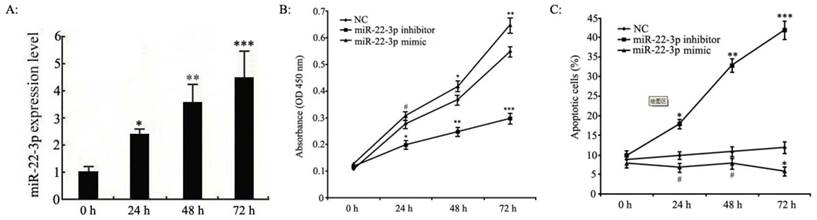
miR-22-3p overexpression induces growth in C33a and SiHa cells in vitro
In previous experiments, microarray analyses have shown that miR-22-3p expression is up-regulated in human cervical squamous cell carcinoma tissues; thus, miR-22-3p is likely to play an important role in cervical carcinoma cell growth. However, the effects of miR-22-3p on cervical carcinoma cell function have not yet been determined. Consequently, we transfected miR-22-3p mimics into C33a and SiHa cells to assess the function of miR-22-3p. As shown in Fig.2A, we used qPCR analyses to confirm that miR-22-3p expression levels were increased in a time-dependent manner in cervical carcinoma cells at 0 h, 24 h, 48 h and 72 h post-transfection.
Next, proliferation in C33a and SiHa cells treated with miR-22-3p mimic or inhibitor was determined by MTT assays; the results indicated that miR-22-3p overexpression led to an obvious increase in cell viability compared to that in the NC group, whereasproliferationwas blunted by the additionof a miR-22-3p inhibitor (Fig. 2B). Next, apoptosis was assessed by flow cytometry; the results indicated that miR-22-3p overexpression in C33a and SiHa cells inhibited apoptosis compared with the NC group, whereas a miR-22-3p inhibitor significantly enhanced miR-22-3p-induced apoptosis (Fig. 2C). Those observations suggest that miR-22-3p could play a key role in promoting cell growth.
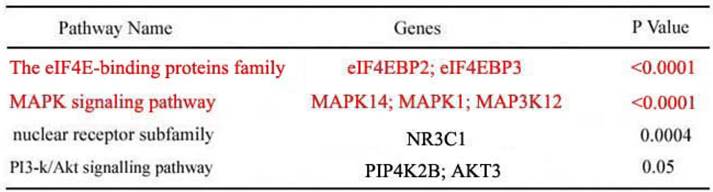
Identification of the target genes and pathways of miR-22-3p
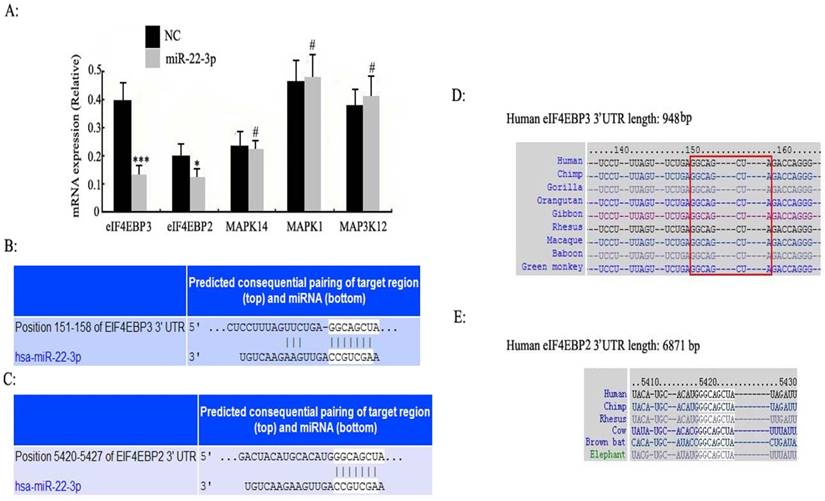
According to the above results of the bioinformatic analysis (www.targetscan.org), the eIF4E-binding protein family and MAPK signalling pathway had the highest correlation with miR-22-3p (P <0.0001); we thus reasoned that the eIF4E-binding protein family and MAPK were more likely to be potential targets of miR-22-3p than either PI3-k/Akt or the nuclear receptor subfamily (Table 1). As a result, the eIF4E-binding protein family and MAPK were selected to determine whether miR-22-3p had effects on them using qPCR. The most prominent change observed was a decrease in eIF4EBP3, which was decreased to a greater extent than eIF4EBP2 when miR-22-3p was overexpressed; no changes in MAPK14, MAPK1 and MAP3K12 expression in the MAPK signalling pathway were observed (Fig. 3A). These data revealed a significant relationship between miR-22-3p expression and the most common targets in the eIF4E-binding protein family. Additionally, the Target Scan results showed that both eIF4EBP3 (Position 151-158) and eIF4EBP2 (Position 5420-5427) had one perfect site in their 3'-UTR that could interact with the miR-22-3p seed sequence GGCAGCUA (Fig. 3B, C); importantly, both of these siteare highly evolutionarily conserved in multiple species, and its homology reaches up to 100% (Fig. 3D, E).
Effect of the eIF4EBP3 gene on apoptosis in C33a and SiHa cells in vitro
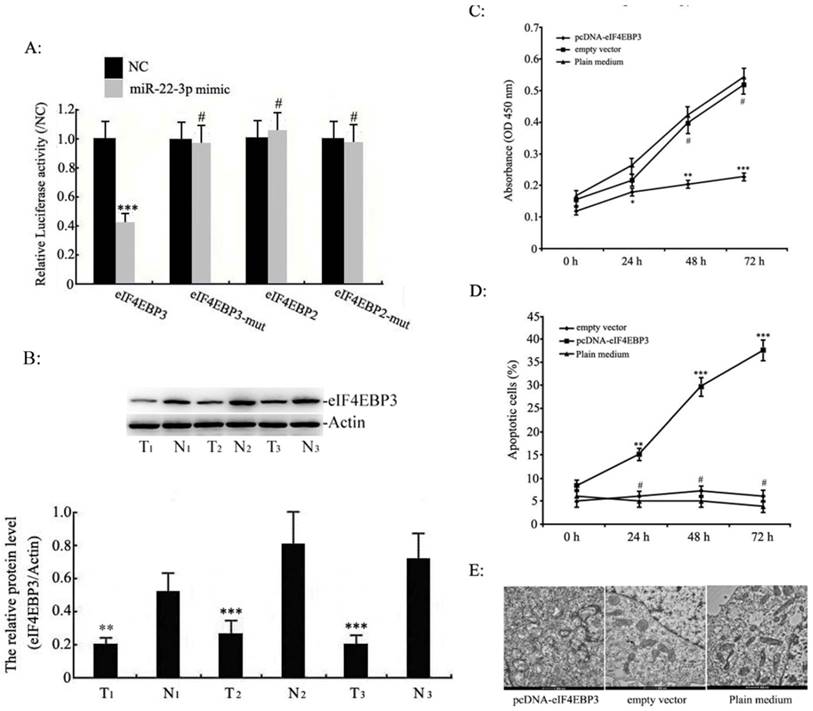
Because miRNAs suppress gene expression primarily through sequence-specific interactions with the 3'-UTR of their target genes, we constructed luciferase reporter plasmids containing the eIF4EBP3 or eIF4EBP2 3'-UTR sequence or their mutants to evaluate the potential role of the putative miR-22-3p seed sites. The luciferase activity of eIF4EBP3 cotransfected with miR-22-3p was reduced, where as the mutant eIF4EBP3 3'-UTR abolished this miR-22-3p-dependent suppression. However, the luciferase reporter that was fused with the 3'-UTR of eIF4EBP2 or its mutant failed to respond to miR-22-3p expression. These results suggest that miR-22-3p directly targeted the 3'-UTR sequence of eIF4EBP3 rather than eIF4EBP2 (Fig. 4A). Based on these data, we further studied the role of eIF4EBP3 and miR-22-3p in cervical cancer cell apoptosis and in the progression of cervical cancer. The protein levels of eIF4EBP3 were measured in human cervical tissues using Western blot analyses. In addition, the results revealed that compared with the levels in non-cancerous cervical tissues (N), eIF4EBP3 expression levels were significantly decreased in human cervical squamous cell carcinoma tissues (T) (Fig. 4B). These data further provide evidence that eIF4EBP3 acts as a direct target of miR-22-3p.
Furthermore, proliferation in C33a and SiHa cells treated with pcDNA-eIF4EBP3 or empty vector was assessed by MTT assay; the results indicated that eIF4EBP3 overexpression led to an obvious defect in cell viability compared to the plain medium group, whereas cell proliferation was not different between the empty vector group and the plain medium group (Fig. 4C). Next, apoptosis was assessed by flow cytometry; the results indicated that compared with the plain medium group, the eIF4EBP3 overexpression upregulated cell apoptosis in a time-dependent manner the C33a and SiHa cells, whereas apoptosis was unchanged in the empty vector group (Fig. 4D). The cellular ultrastructure changes in C33a and SiHa cells were detected by TEM; these results agreed with the flow cytometry results. There were no distinct changes in the empty vector group or the plain medium control group. However, obvious ultrastructure changes, such as organelles loss and apoptotic bodies, were present in cells treated with the pcDNA-eIF4EBP3 vector (Fig. 4E). These observations suggest that the eIF4EBP3 gene plays a key role in promoting cell apoptosis.
Expression of miRNAs in human cervical tissue. Differential expression levels of the 4 most up-regulated miRNAs (miR-1290, miR-22-3p, miR-155-3p andmiR-21) and the 4 most down-regulated miRNAs (miR-203, miR-187, miR-148a andmiR-34a) in human cervical squamous cell carcinoma tissues (T) relative to non-cancerous cervical tissues (N) were confirmed using qPCR. The data are presented as the means ± S.D. of three independent experiments. Student's t-test: T versus N. ***, p<0.001; **, p<0.01.
Effects of miR-22-3p overexpression on cervical squamous carcinoma cell proliferation and apoptosis. A: C33a and SiHa cells treated with miR-22-3p mimic were assessed at 0 h, 24 h, 48 h and 72 h post-transfection as follows. The levels of miR-22-3p in transfected C33a and SiHa cells were measured at various time points using qPCR. The data represent the means ± S.D. of three independent experiments (***p<0.001; **p<0.01; *p<0.05 vs 0 h). B: Proliferation analyses of C33a and SiHa cells treated with miR-22-3p mimic or inhibitor using MTT assays. Untreated cells were chosen as a negative control (NC). The results are expressed as the mean ± SD from 3 independent experiments. (***p<0.001; **p<0.01; *p<0.05; #p>0.05 vs NC). C: C33a and SiHa cells were stained and trypsinized with annexin V and propidium iodide (PI) and then analysed using flow cytometry. Apoptosis was calculated as a percentage of the total colonies counted. The data represent the means ± S.D. of three independent experiments; untreated cells were chosen as a negative control (NC) (***p<0.001; **p<0.01; *p<0.05; #p>0.05 vs NC).
Enriched target genes and pathways related to miR-22-3p. Top four enriched signalling pathways regulated by miR-22-3p according to p value or gene count. GO terms are grouped into four categories. The candidate pathways included the eIF4E-binding proteins, MAPK signalling pathway, nuclear receptor subfamily and PI3-Akt signalling pathway; of these, the eIF4E-binding proteins and MAPK signalling pathway were most likely to be correlated with miR-22-3p with p<0.0001 (nuclear receptor subfamily, p<0.0004; PI3-Akt, p<0.05).
Role of the eIF4EBP3 gene in miR-22-3p-mediated cervical cancer cell growth
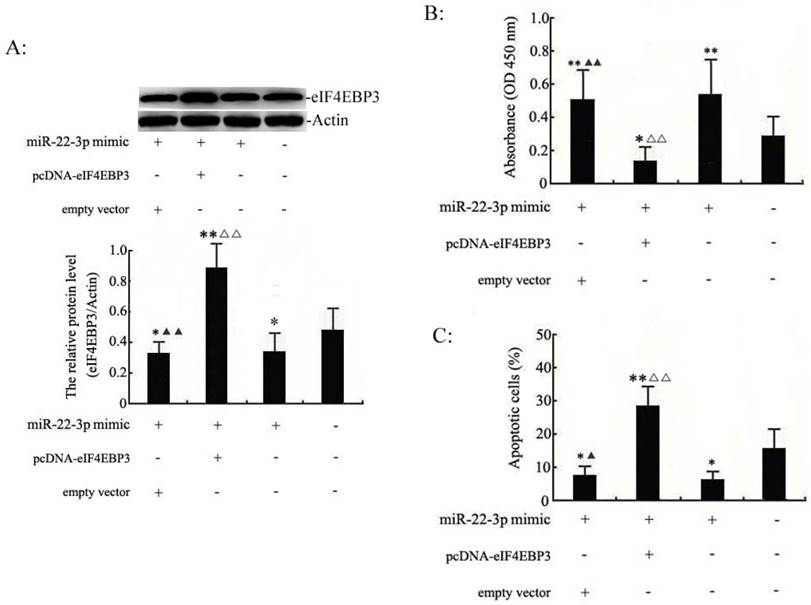
We set out to identify the interaction between miR-22-3p and eIF4EBP3 to shed light on the mechanism of the pro-growth effects of miR-22-3p on cervical cancer cells. Our previous experiments demonstrated that miR-22-3p overexpression could promote cellular growth. Therefore, miR-22-3p + empty vector, miR-22-3p + pcDNA-eIF4EBP3 or miR-22-3p mimic alone were transfected into C33a and SiHa cells to determine the role of the eIF4EBP3 gene in miR-22-3p-mediated growth. First of all, western blot analysis results demonstrated that the eIF4EBP3 expression levels was significantly increased in the miR-22-3p mimic + pcDNA-eIF4EBP3 group compared with the plain medium. Moreover, eIF4EBP3 expression level in the miR-22-3p mimic + empty vector group and miR-22-3p mimic group was slight increased when compared with the plain medium groups (Fig. 5A). Then, MTT assays were performed to measure cell proliferation. In C33a and SiHa cells, we found that cell viability was obviously higher inthe miR-22-3p + empty vector group and the miR-22-3p mimic alone group than the plain medium group, whereas proliferation was partially decreased in the miR-22-3p + pcDNA-eIF4EBP3 group, and there was an apparent difference compared with the plain medium group (Fig. 5B). Flow cytometry apoptosis assays revealed that cotransfection of the empty vector with miR-22-3p mimic led to decreased apoptosis compared with the plain medium control, and there were apparent changes in cells treated with miR-22-3p + pcDNA-eIF4EBP3 vector compared with the plain medium control; however, eIF4EBP3 overexpression significantly attenuated the miR-22-3p-induced growth by overcoming the promoting effects of miR-22-3p (Fig. 5C). In summary, these data suggest that miR-22-3p-induced cervical cancer cell growth mediated by the eIF4EBP3 gene was likely to be the molecular mechanism underlying cervical cancer development.
Enriched target genes and pathways related to miR-22-3p. A: C33a and SiHa cells were treated with miR-22-3p mimic, and untreated cells were chosen as a negative control (NC) (mCherry was used as a marker). At 48 h post-transfection, the mRNA expression levels of eIF4EBP3, eIF4EBP2, MAPK14, MAPK1 and MAP3K12 were analysed using qPCR. The data are represented as the means ± S.D. of three independent experiments (***p<0.001; *p<0.05; #p>0.05 vs NC). (B, C) 3'-UTRs of the predicted target genes eIF4EBP3 (Position 151-158) and eIF4EBP2 (Position 5420-5427) are complementary to the miR-22-3p sequence according to TargetScan. (D, E) The complementary GGCAGCUA sequence of miR-22-3p and eIF4EBP3 or eIF4EBP2 is highly evolutionarily conserved, and its homology reaches up to 100%.
Discussion
The regulation of protein synthesis is a ubiquitous and reversible progress in eukaryotes [13]. During translation initiation, one important regulatory step is that the eukaryotic initiation factor (eIF) 4E protein recognizes the mRNA m7 GpppN (where N is any nucleotide) cap structure and recruits the eIF4F complex, which promotes the initiation of mRNA translation [14]. Phosphorylation and association with eIF4E-binding proteins (eIF4EBPs) are important for eIF4E binding to the cap structure [15]. Unphosphorylated or hypophosphorylated eIF4EBPs exhibit a high affinity for eIF4E and thus prevent translation initiation, whereas hyperphosphorylated eIF4EBPs lose their affinity foreIF4E [16].
Three related eIF4EBPs have been studied thus far; of these, eIF4EBP1 and eIF4EBP2 are the best studied. eIF4E is partially activated by two translational repressors, eIF4EBP1 and eIF4EBP2, which bind to eIF4E and prevent its assembly with eIF4F. eIF4EBP1 and eIF4EBP2 phosphorylation leads to their separation from eIF4E; then, eIF4E can bind to form eIF4F complexes [17-18]. In the present study, a new subtype of the 4E-BP family is reported, eIF4EBP3, which is homologous to eIF4EBP1 and eIF4EBP2 and exhibits 57% and 59% identity, respectively. In the middle region of the eIF4EBP3 protein, the homology is highly consistent with the residues of eIF4EBP1 and eIF4EBP2, which contain the eIF4E binding motif [19]. Moreover, eIF4EBP3 overexpression specifically decreases eIF4E-dependent translation. Our observations demonstrate that the protein expression levels of eIF4EBP3 are significantly decreased in human cervical squamous cell carcinoma tissues and that when constitutively expressed in cervical cancer cell lines, eIF4EBP3 causes growth inhibition, morphological abnormalities and apoptosis.
eIF4EBP3 has been shown to associate with eIF4E and inhibit eIF4E-dependent translation [20]. Recent studies have also shown that eIF4EBP3 is present in the nucleus as well as in the cytoplasm. Therefore, assessing the function of nuclear eIF4EBP3 and cytoplasmic eIF4EBP3 is very important for future studies. In recent years, along with the rapid development of life science and technology, miRNA, a small non-coding RNA molecule that is highly evolutionarily conserved, has attracted attention in every field and greatly expanded our knowledge of biological and disease development [21-22]. In mammals, miRNA can regulate up to 30% or more of human gene expression at the post-transcriptional level [23]. In particular, its unique one-to-multi and multi-to-one interactions with its target genes are undoubtedly a strategic target that should be used in the treatment of refractory and complex diseases. At present, correlations between miRNAs and tumours have been reported by many studies. However, little is knownabout the relationship between miRNAs and eIF4EBP3 in cervical cancer; thus, correlation studies are urgently needed. In our study, both cervical cancer and mild cervicitis tissues were collected to compare differentially expressed miRNA profiles using a miRNA microarray. In addition, we obtained multiple differentially expressed miRNAs; four miRNAs were highly up-regulated (miR-1290, miR-22-3p, miR-155-3p and miR-21), and an additional four miRNAs were highly down-regulated (miR-203, miR-187, miR-148a and miR-34a).
Effects of eIF4EBP3 on C33a and SiHa cell growth. A: The eIF4EBP3, eIF4EBP3 mut, eIF4EBP2 and eIF4EBP2 mut 3'-UTR luciferase reporters were co-transfected with miR-22-3p mimic or the empty virus vector (NC) in HEK293 cells (mCherry was used as a marker). At 48 h post-transfection, luciferase activity was assessed using the dual-luciferase reporter assay system. The relative luciferase activities were calculated by normalizing to renilla luciferase activities, and the data are presented as the means ± S.D. of three independent experiments (***p<0.001; #, p>0.05 vs NC). B: Relative expression levels of eIF4EBP3 protein in human cervical squamous cell carcinoma tissues (T) relative to non-cancerous cervix tissues (N) were evaluated using Western blots, and actin was used as the loading control. The data are presented as the means ± S.D. of three independent experiments (***p<0.001 vsN). C: Proliferation in C33a and SiHa cells treated with pcDNA-eIF4EBP3 or the empty vector was measured by MTT assay at 0 h, 24 h, 48 h and 72 h post-transfection. The results are expressed as the mean ± SD from 3 independent experiments. ***p<0.001; **p<0.01; *p<0.05; #p>0.05 vs plain medium group (Control). D: C33a and SiHa cells were stained and trypsinized with annexin V and propidium iodide (PI) and then analysed by flow cytometry. Apoptosis was calculated as a percentage of the total colonies counted. The data are presented as the means ± S.D. of three independent experiments. ***p<0.001; **p<0.01; #p>0.05 vs plain medium group (Control). E: At 48 h post-transfection, no abnormal ultrastructural changes were found in the empty vector or plain medium groups, whereas typical characteristics of apoptosis, such as chromatin condensation and edge accumulation at the nuclear membrane and apoptotic bodies, were evident in the pcDNA-eIF4EBP3-treated group. The scale bar is 500 nm.
eIF4EBP3 regulates miR-22-3p-mediated apoptosis in C33a and SiHa cells. C33a and SiHa cells treated with miR-22-3p + empty vector, miR-22-3p + pcDNA-eIF4EBP3 or miR-22-3p mimic alone at 48 h post-transfection were analysed using the following methods. A: Relative expression levels of eIF4EBP3 protein in C33a and SiHa cells were evaluated using Western blots, and actin was used as the loading control. The data are presented as the means ± S.D. of three independent experiments (**p<0.01; *p<0.05 vs NC; ▲▲p< 0.01 vs miR-22-3p + pcDNA-eIF4EBP3 group; △△p< 0.01 vs miR-22-3p mimic group). B: Proliferation in C33a and SiHa cells was measured by MTT assay. Untreated cells were chosen as a negative control (NC). The results are expressed as the mean ± SD from 3 independent experiments. (**p<0.01; *p<0.05 vs NC; ▲▲p< 0.01 vs miR-22-3p + pcDNA-eIF4EBP3 group; △△p< 0.01 vs miR-22-3p mimic group). C: At 48 h post-transfection, cells were stained and trypsinized with annexin V and then analysed by flow cytometry. Apoptosis was calculated as a percentage of the total colonies counted, and the data are presented as the means ± S.D. of three independent experiments (**p< 0.01; *p< 0.05; #p>0.05 vs NC; ▲p< 0.05 vs miR-22-3p + pcDNA-eIF4EBP3 group; △△p< 0.05 vs miR-22-3p mimic group).
Of these miRNAs, we found that miRNA-22-3p had the greatest fold change, which indicates that may regulate cervical carcinoma cell growth. Based on these results, we speculated that miR-22-3p would be closely associated with the occurrence of cervical cancer. Previous studies have shown that miR-22-3p is frequently dysregulated to varying degrees in human diseases; for example, it is up-regulatedin colorectal cancer [24], prostatic cancer [25] and endometriosis [26] and down-regulated in oesophageal cancer [27]. Furthermore, miR-22-3p can exert its tumour-suppressing or tumour-promoting functions by targeting genes. However, there are few reports about the relationship between cervical cancer cell growth and the functional role of miR-22-3p; hence, researching the role of cervical cancer-related miRNAs in cervical cancer cell growth is necessary. In the present study, we found that miR-22-3p overexpression in C33a and SiHa cell lines induced significant proliferation, which revealed that miR-22-3p may act as a key regulator in cervical cancer development. In summary, we acquired primary evidence that miR-22-3p might play an important role in cervical cancer growth. It is necessary for us to determine the downstream regulatory mechanisms of miR-22-3p in cervical cancer.
Currently, no evidence-based mechanism underlying miR-22-3p-induced cervical cancer cell growth has been reported in previous studies. To look for clues about this mechanism, a bioinformatics analysis was applied to predict the downstream target genes of miR-22-3p. We predicted four molecular pathways (the eIF4E-binding protein family, MAPK signalling pathway, PI3-k/Akt signalling pathway and nuclear receptor subfamily) that appeared to be regulated by miR-22-3p (www.targetscan.org). Among them, the eIF4E-binding protein family and MAPK signalling pathways had the highest correlation with miR-22-3p (P <0.001); we thus reasoned that the eIF4E-binding protein family and MAPK signalling pathway were more likely to be potential targets of miR-22-3p than the other two. Previous studies have reported that increased hepatic miR-22-3p expression levels impair gluconeogenesis by silencing the Wnt-responsive transcription factor Tcf7 [28]. In our study, the potential roles of the eukaryotic initiation factor subfamily and MAPK in targeting miR-22-3p were further evaluated using qPCR. The results revealed that the miR-22-3p overexpression in C33a and SiHa cells suppressed both eIF4EBP3 and eIF4EBP2 expression, but there were no effects on MAP14, MAPK1 and MAPK3K12 expression in the MAPK signalling pathway. Therefore, the eIF4E-binding protein family was identified as the candidate target genes of miR-22-3p. In the next experiments, we found that the activity of a reporter gene bearing the 3'-UTR of eIF4EBP2 could not be suppressed by miR-22-3p, whereas the reporter gene containing eIF4EBP3 3'-UTR was successfully inhibited by miR-22-3p due to sequence-specific interactions with the 3'-UTR of eIF4EBP3. Based on these data, we ultimately identified eIF4EBP3 as the direct target gene of miR-22-3p.
Conclusion
Collectively, our findings revealed that miR-22-3p is up-regulated in tissues from patients with cervical cancer and identified eIF4EBP3 as a direct target of miR-22-3p. In particular, functional experiments implied that overexpressing miR-22-3p could induce growth by suppressing its target gene eIF4EBP3. In summary, our results show that the interaction between miR-22-3p and its target gene eIF4EBP3 could be utilized as a therapeutic target to improve clinical cervical cancer treatment.
Abbreviations
miRNAs: microRNAs; eIF4EBPs: eIF4E-binding proteins; eIF4EBP3: eIF4E-binding protein 3; eIF4EBP2: eIF4E-binding protein2; p38 MAPK: p38 mitogen-activated protein kinase; PI: propidium iodide; cDNA: complementary DNA; SD: standard deviation; real-time PCR: real-time quantitative polymerase chain reaction; mt: mutant; MTT: methyl thiazolyl tetrazolium.
Acknowledgements
We are grateful to Wenqu Li, Rong Huang and Rongbing Shi for their critical reading of the manuscript. We thank all the staff at the Clinical Laboratory Department of Nanjing Maternity and Child Health Care Hospital affiliated to Nanjing Medical University for their guidance. We declare no conflict of interest.
Funding
This study was supported by grants from the National Natural Science Foundation of China (Contract grant number: 81571437), the National Natural Science Foundation of Jiangsu Province (Contract grant number: BK20151078), the Jiangsu Maternal and Child Health Fund (Contract grant number: F201621), and the Science and Technology Commission Foundation of Huangdao District of Qingdao (Contract grant number: 2014-1-97).
Ethics approval and consent to participate
See ethics approval.
Consent for publication
All persons have given their approval for submission and publication and declare that the work described here has not been published before.
Availability of data and material
The data sets analysed in the current study are available from the corresponding author upon reasonable request.
Authors' contributions
LJG and JPC conceived the study and drafted the manuscript. KTL and ZL participated in its design and helped draft the manuscript. TH performed the molecular biological studies and the statistical analyses. WYD collected patient information. JF and WZ helped revise the manuscript and performed statistical analyses. All of the authors read and approved the final manuscript.
Competing Interests
The authors have declared that no competing interest exists.
References
1. Deng Z, Hasegawa M, Yamashita Y, Matayoshi S, Kiyuna A, Agena S. et al. Prognostic value of human papillomavirus and squamous cell carcinoma antigen in head and neck squamous cell carcinoma. Cancer Sci. 2012;103:2127-34
2. Jin T, Wu X, Yang H, Liu M, He Y, He X. et al. Association of the miR-17-5p variants with susceptibility to cervical cancer in a Chinese population. Oncotarget. 2016;7(47):76647-55
3. Origoni M, Salvatore S, Perino A, Cucinella G, Candiani M. Cervical Intraepithelial Neoplasia (CIN) in pregnancy: the state of the art. Eur Rev Med Pharmacol Sci. 2014;18(6):851-60
4. Sharma A, Rajappa M, Saxena A, Sharma M. Cytokine profile in Indian women with cervical intraepithelial neoplasia and cancer cervix. Int J Gynecol Cancer. 2007;17(4):879-85
5. Yoqev O, Williams VC, Hinits Y, Huqhes SM. IF4EBP3L acts as a gatekeeper of TORC1 in activity-dependent muscle growth by specifically regulating Mef2ca translational initiation. PloS Biol. 2013;11(10):e1001679
6. Lee M, Kim EJ, Jeon MJ. MicroRNAs 125a and 125b inhibit ovarian cancer cells through post-transcriptional inactivation of EIF4EBP1. Oncotarget. 2016;7(8):8726-42
7. Cha YL, Li PD, Yuan LJ, Zhang MY, Zhang YJ, Rao HL. et al. EIF4EBP1 overexpression is associated with poor survival and disease progression in patients with hepatocellular carcinoma. PLoS One. 2015;10(2):e0117493
8. Fang Z, Lu L, Tian Z, Luo K. Overexpression of phosphorylated 4E-binding protein 1 predicts lymph node metastasis and poor prognosis of Chinese patients with hilar cholangiocarcinoma. Med Oncol. 2014;31(5):940
9. Zhou Q, Han LR, Zhou YX, Li Y. MiR-195 Suppresses Cervical Cancer Migration and Invasion Through Targeting Smad3. Int J Gynecol Cancer. 2016;26(5):817-24
10. Bartel DP. MicroRNAs: target recognition and regulatory functions. Cell. 2009;136:215-33
11. Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116(2):281-97
12. Fesler A, Liu H, Wu N, Liu F, Ling P, Ju J. Autophagy regulated by miRNAs in colorectal cancer progression and resistance. Cancer Transl Med. 2017;3(3):96-100
13. Haraguchi K, Yada N, Sato S, Habu M, Hayakawa M, Takahashi O. et al. The methylation status and expression of human telomerase reverse transcriptase is significantly high in oral carcinogenesis. APMIS. 2017;125(9):797-807
14. Morino S, Ueda H, Doi M, Ishida T. Gene expression of human eukaryotic initiation factor-4E for protein synthesis and study of its recognition mechanism of mRNA cap structure. Yakugaku Zasshi. 1995;115(6):401-419
15. Ramírez-Carvajal L, Singh N, de los Santos T, Rodríguez LL, Long CR. Depletion of elongation initiation factor 4E binding proteins by CRISPR/Cas9 enhances the antiviral response in porcine cells. Antiviral Res. 2016;125:8-13
16. Singh M, Shin YK, Yang X, Zehr B, Chakrabarti P, Kandror KV. eIF4EBPs Control Fat Storage by Regulating the Expression of Egr1 and ATGL. J Biol Chem. 2015;290(28):17331-8
17. Kamenska A, Simpson C, Standart N. eIF4E-binding proteins: new factors, new locations, new roles. Biochem Soc Trans. 2014;42(4):1238-45
18. Steiner JL, Pruznak AM, Deiter G, Navaratnarajah M, Kutzler L, Kimball SR. et al. Disruption of genes encoding eIF4E binding proteins-1 and -2 does not alter basal or sepsis-induced changes in skeletal muscle protein synthesis in male or female mice. PLoS One. 2014;9(6):e99582
19. Coleman LJ, Peter MB, Teall TJ, Brannan RA, Hanby AM, Honarpisheh H. et al. Combined analysis of eIF4E and 4E-binding protein expression predicts breast cancer survival and estimates eIF4E activity. Br J Cancer. 2009;100(9):1393-9
20. Yogev O, Williams VC, Hinits Y, Hughes SM. eIF4EBP3L acts as a gatekeeper of TORC1 in activity-dependent muscle growth by specifically regulating Mef2ca translational initiation. PLoS Biol. 2013;11(10):e1001679
21. Abiko F, Tomoo K, Mizuno A, Morino S, Imataka H, Ishida T. Binding preference of eIF4E for 4E-binding protein isoform and function of eIF4E N-terminal flexible region for interaction, studied by SPR analysis. Biochem Biophys Res Commun. 2007;355(3):667-72
22. Tsukumo Y, Alain T, Fonseca BD, Nadon R, Sonenberg N. Translation control during prolonged mTORC1 inhibition mediated by 4E-BP3. Nat Commun. 2016;7:11776
23. Xu Z, Xiao SB, Xu P, Xie Q, Cao L, Wang D. et al. miR-365, a novel negative regulator of interleukin-6 gene expression, is cooperatively regulated by Sp1 and NF-kappaB. J Biol Chem. 2011;286(24):21401-12
24. Schee K, Lorenz S, Worren MM, Gunther CC, Holden M, Hoviq E. et al. Deep Sequencing the MicroRNA Transcriptome in Colorectal Cancer. PloS One. 2013;8(6):e66165
25. Knyazev EN, Samatov TR, Fomicheva KA, Nyushko KM, Alekseev BY, Shkurnikov MY. MicroRNA hsa-miR-4674 in Hemolysis-Free Blood Plasma Is Associated with Distant Metastases of Prostatic Cancer. Bull Exp Biol Med. 2016;161(1):112-5
26. Lv Y, Gao S, Zhang Y, Wang L, Chen X, Wang Y. miRNA and target gene expression in menstrual endometria and early pregnancy decidua. Eur J Obstet Gynecol Reprod Biol. 2016;197:27-30
27. Warnecke-Eberz U, Chon SH, Hölscher AH, Drebber U, Bollschweiler E. Exosomal onco-miRs from serum of patients with adenocarcinoma of the esophagus: comparison of miRNA profiles of exosomes and matching tumor. Tumour Biol. 2015;36(6):4643-53
28. Kaur K, Vig S, Srivastava R, Mishra A, Singh VP, Srivastava AK. et al. Elevated Hepatic miR-22-3p Expression Impairs Gluconeogenesis by Silencing the Wnt-Responsive Transcription Factor Tcf7. Diabetes. 2015;64(11):3659-69
Author contact
![]() Corresponding authors: Ling-juan Gao and Jing-ping Chu, Clinical Laboratory, Nanjing Maternity and Child Health Care Hospital, Tianfei Alley, Nanjing Mochou Road, 210004, Nanjing, P.R. China Department of Gynaecology and Obstetrics, Huangdao District of Traditional Chinese Medicine, Hainan Island Road, 266500, Qingdao, P.R. China Tel.: 86 25 83362160, Fax: 86 25 84460507 E-mail: gaolingjuanedu.cn
Corresponding authors: Ling-juan Gao and Jing-ping Chu, Clinical Laboratory, Nanjing Maternity and Child Health Care Hospital, Tianfei Alley, Nanjing Mochou Road, 210004, Nanjing, P.R. China Department of Gynaecology and Obstetrics, Huangdao District of Traditional Chinese Medicine, Hainan Island Road, 266500, Qingdao, P.R. China Tel.: 86 25 83362160, Fax: 86 25 84460507 E-mail: gaolingjuanedu.cn

 Global reach, higher impact
Global reach, higher impact