Impact Factor
ISSN: 1449-1907
Int J Med Sci 2018; 15(1):86-94. doi:10.7150/ijms.21164 This issue Cite
Research Paper
In vitro and in vivo effects of miRNA-19b/20a/92a on gastric cancer stem cells and the related mechanism
1. Department of Oncology, The First Affiliated Hospital with Nanjing Medical University, Jiangsu Province Hospital, Guangzhou Road 300, Nanjing 210029, Jiangsu Province, China;
2. Department of General Surgery, Cancer Hospital of Nanjing Medical University, Baiziting 42, Nanjing 210009, Jiangsu Province, China;
3. Department of Pathology, Cancer Hospital of Nanjing Medical University, Baiziting 42, Nanjing 210009, Jiangsu Province, China;
4. Department of General Surgery, Cancer Hospital of Nanjing Medical University, Baiziting 42, Nanjing 210009, Jiangsu Province, China.
Received 2017-5-24; Accepted 2017-10-11; Published 2018-1-1
Abstract
We aimed to analyze the in vitro and in vivo effects of miRNA-19b/20a/92a on gastric cancer stem cells (GCSCs) and the related mechanism. GCSCs were cultured until adherence and differentiation, and subjected to miRNA microarray analysis to find and to verify miRNA deletion. Cells stably expressing lentivirus carrying miRNA-19b/20a/92a were constructed by transfection. The relationship between miRNA-19b/20a/92a and renewal of GCSCs was studied by the tumor sphere assay, and that between miRNA-19b/20a/92a and their proliferation was explored with MTT and colony formation assays. Target genes of miRNA for promoting the proliferation and self-renewal of GCSCs were found by using bioinformatics database, and verified by the reporter gene assay and Western blot. The expressions of miRNA-19b/20a/92a gradually decreased during the adherence and differentiation of GCSCs. The expressions of lentivirus carrying miRNA-17-19 gene in MKN28 and CD44-/EpCAM- cells were increased significantly. Transient transfection with pre-miRNA-19b/20a/92a elevated miRNA expressions in CD44-/EpCAM- and MKN28 cells, whereas transfection with pre-miRNA-19b/20a/92a antagonists reduced the expressions in SGC7901 and CD44+/EpCAM+ cells. Overexpression of lenti-miRNA-19b/20a/92a significantly enhanced the capability of GCSCs to form tumor spheres. In the presence of chemotherapeutic agent, the survival of lenti-miRNA-19b/20a/92a-infected cells was prolonged. Transient transfection with pre-miRNA-19b/20a/92a significantly increased the number of CD44+/EpCAM+ cells, but transfection with antagonists had the opposite outcomes. The stable miRNA-19b/20a/92a expression groups proliferated faster than the control group did. The proliferation of cells transfected with pre-miRNA-19b/20a/92a was accelerated, whereas that of cells transfected with the antagonists was decelerated. Compared with the control group, the number of colonies in the former group was higher, but that in the latter group was lower. miRNA-19b and miRNA-92a could bind the 3' untranslated region of HIPK1, while miRNA-20a was able to bind that of E2F1. Expressions of miRNA-20a and miRNA-92a in gastric cancer samples were negatively correlated with the prognosis of patients. miRNA-19b/20a/92a facilitated the self-renewal of GCSCs by targeting E2F1 and HIPK1 on the post-transcriptional level and activating the β-catenin signal transduction pathway. miRNA-92a was an independent factor and index predicting the prognosis of gastric cancer.
Keywords: gastric cancer, miRNA-19b/20a/92a, molecular mechanism.
Introduction
Currently, tumors have been widely accepted to originate from cancer stem cells (CSCs). Like normal stem cells, CSCs are capable of self-renewal and multipotential differentiation to maintain cancer onset, progression, metastasis and recurrence [1]. Gastric cancer is the second most common malignant tumor worldwide, so it is of great significance to perform studies on gastric cancer stem cells (GCSCs) [2, 3]. As a class of non-coding single-stranded small-molecule RNAs with about 19-22 nucleotides discovered in recent years, miRNAs usually regulate the expressions of target gene proteins on the post-transcriptional level, dominantly participating in the onset and progression of tumors [4]. miRNAs are also involved in regulating the self-renewal and multipotential differentiation of stem cells and CSCs [5-7]. For example, miRNAs can regulate the self-renewal and differentiation of embryonic stem cells through targeted self-renewal of related genes such as nanog, SOX2 and OCT4 [8,9]. Let-7 can regulate the self-renewal and proliferation of breast CSCs.
It is now well-established that malignant tumors are generated and maintained by a small group of cancer cells capable of self-renewal and multipotential differentiation. Such cells are referred to as CSCs or tumor-initiating cells, which are closely related to tumor onset, progression, metastasis, as well as resistances to chemotherapy and radiotherapy. Self-renewal is not only one of the most important characteristics differentiating CSCs from common cancer cells, but also the root cause for CSCs to maintain their “stemness” and for inducing metastasis and recurrence eventually. However, the regulatory mechanism for CSC self-renewal has not been fully clarified hitherto. Thereby motivated, we herein studied the molecular mechanism by which miRNA-19b/20a/92a promoted the self-renewal and proliferation of GCSCs.
Materials and Methods
Ethics
This study has been approved by the ethics committee of our hospital. All experimental animals were given humane care to minimize their suffering.
Cell lines
Gastric cancer cell lines SGC7901 and MKN28 were purchased from the PLA Academy of Military Medical Sciences (China). CD44+/EpCAM+ GCSCs and CD44-/EpCAM- non-GCSCs were isolated from SGC7901 cells by flow cytometry.
Gastric cancer tissue samples
Paraffin samples of gastric cancer and paracancerous tissues were preserved by Department of Pathology of our hospital. The patients with gastric cancer were followed up at regular intervals.
Main reagents
High-glucose DMEM, epidermal growth factor (EGF), basic fibroblast growth factor (bFGF), low-adhesion culture flasks, Trizol and LipofectaminTM transfection reagent were purchased from Invitrogen (USA). Fetal bovine serum (FBS) was bought from Gibco (USA). 0.05% Trypsin and phosphate saline solution (PBS) were obtained from Shanghai Jingtian Biotechnology Co., Ltd. (China). Anti-Ep CAM and anti-CD44 antibodies were provided by BD (USA). CFSE was purchased from Beijing Zhongshan Golden Bridge Biological Technology Co., Ltd. (China). RNA enzyme-free water, real-time fluorescent quantitative PCR probe, real-time fluorescent quantitative PCR kit, siPORTTM Neo FXTM transfection reagent, miRNA mimic, miRNA inhibitors, miRNA probe and RecoverAllTM total RNA extraction kit were bought from Ambion (USA). Dual luciferase reporter assay kit was obtained from Promega (USA). ECL Western blot detection kit and nitrocellulose (NC) membrane were provided by Amersham (USA).
Main apparatus
CO2 incubator was purchased from Forma Scientific (USA). Ultra-clean bench was bought from Suzhou Cleanroom Equipment Factory (China). Microscope was obtained from Olympus (Japan). Real-time quantitative PCR system was provided by Roche (Shanghai, China). Air-bath shaker was purchased from Wuhan Medical Apparatus and Instrument Factory (China). Water purification system was bought from Milipore (USA). Micro-vertical electrophoresis system and microplate reader were obtained from Bio-Rad (USA).
miRNA microarray analysis and verification
Culture of GCSCs
Gastric cancer cell lines SGC7901 and MKN28 were cultured in RPMI 1640 medium containing 10% FBS (v/v) in an incubator with 5% CO2 at 37°C. The above cells were digested with trypsin, and washed with PBS and serum-free DMEM. Then 1000 cells were collected, placed into a low-adhesion flask and cultured in serum-free high-glucose DMEM (including EGF and bFGF). The culture was terminated on Day 7, and then the cells were observed under an inverted microscope.
Cell preparation for miRNA microarray analysis
The tumor sphere cells that had been cultured for seven days were centrifuged, washed, and cultured in ordinary medium and medium containing FBS respectively. Afterwards, the adherent cells cultured for 8 h, 24 h and 72 h were digested and collected.
Extraction of total RNA
The cells were digested, centrifuged, washed and centrifuged again after repeated pipetting and shaking. The upper layer of aqueous sample was collected, transferred into a clean test tube, added with isopropanol, shaken and centrifuged for 10 min to remove the upper layer of suspension. The remaining was precipitated with ethanol and centrifuged again.
miRNA microarray analysis
Small RNA was isolated from total RNA by microcentrifuge spin column. A poly (A) tail was added to its 3'-end and then ligated with an oligonucleotide tag. Hybridization reaction was conducted in 6× SSPE buffer containing formamide, after which monitoring was conducted using Cy5 fluorescent dye. Images were collected by a laser scanner to perform digitalized conversion using the Array-Pro image analysis software.
PCR detection
Total RNA was extracted by trypsin digestion and reverse-transcribed in RNA enzyme-free EP tubes. The product was thereafter subjected to real-time fluorescent quantitative PCR.
miRNA-19b/20a/92a promoted self-renewal of GCSCs
Construction of cells stably expressing lentivirus
Lentiviral vector Pgcsil-008 (kl1496) was subjected to NheI digestion. Primers were synthesized. Target genes were amplified by PCR and competent cells were prepared. The PCR product was inserted into linearized lentiviral vectors for transformation, cloning and sequencing. Transient transfection was made for the prepared cell suspension.
Drug sensitivity test
The cells with stable expressions of miRNA were cultured and centrifuged to prepare a single cell suspension which was cultured in serum-free DMEM (including EGF and bFGF). The cells were added with 5-fluorouracil on the second day of culture and dimethyl sulfoxide on the second day of treatment, and detected after culture by a microplate reader.
Flow cytometry
The transiently transfected cells were collected by centrifugation, washed with PBS, incubated with 20 μL of antibody, and washed again with PBS before detection.
Tumor formation in NOD-SCID mice
SGC7901-Luc cells stably expressing miRNA were digested, centrifuged, washed twice with PBS, once with serum-free culture medium and once with serum-free DMEM containing 20 ng/ml EGF and 10 ng/ml bFGF. Then 1000 cells were counted, added in low-adhesion culture plates, and cultured in serum-free DMEM containing 20 ng/ml EGF and 10 ng/ml bFGF for one week. Afterwards, the cells were collected by centrifugation, of which 2000 were injected into the back of NOD-SCID mice to observe tumor growth. Every three days, the tumor fluorescent intensity was observed by using IVIS 100 Imaging System 2 min after 100 mg/kg D-luciferin was injected.
miRNA-19b/20a/92a promoted proliferation of GCSCs
MTT assay
The cells in logarithmic growth phase were cultured in culture plates (1000 cells per well), and transient transfection was conducted 24 h later. After 24 h of transfection, dimethyl sulfoxide was added to the wells every day for 30 min-4 h of culture, and then they were detected by a microplate reader.
Colony formation assay
One hundred cells were seeded in 6-well plates and cultured until typical colonies formed. The cell colonies were counted under an inverted microscope after fixing and staining (a colony contained over 50 cells).
Construction of reporter gene vector
Primers for the 3' untranslated regions of HIPK1 and E2F1 were designed. The PCR-screened primers were ligated to the p GL3 luciferase reporter gene vector.
Cell transient transfection
The cells were digested with trypsin, spread evenly into 6-well culture plates by using siPORT transfection reagent (Ambion, USA), miRNA precursor and miRNA inhibitors according to the technical manual.
Reporter gene transfection and luciferase assay
Cells in the logarithmic growth phase were inoculated into 24-well plates at the density of 5×105 and cultured to 80% confluence. Subsequently, each well was added successively with 0.2 μg, 0.4 μg, 0.8 μg plasmids, 100 ng PM, 5 ng PRL-TK internal reference vector, 2 μL of LipofectaminTM and 200 μL of serum-free culture medium. Forty-eight hours after transfection, the supernatant was aspirated and each well was washed with PBS. Afterwards, 200 μL of lysis buffer was added into each well and centrifuged at 12,000 rpm for 10 min, and the supernatant was collected into a clean centrifuge tube. Then 20 μL of supernatant and 100 μL of luciferase assay reagent II (LARII) were mixed to detect the firefly luciferase activity on TD-20/20 Luminometer. The Renilla luciferase activity was detected after addition of 100 μL Stop & Glo™ reagent. All activities were normalized based on the Renilla luciferase activity. Average of the activities of three samples was used, and each experiment was repeated twice.
Western blot
Protein samples were mixed with a quarter of volume of 4× SDS loading buffer, and denatured at 95°C for 10 min. Then 20 μL of protein sample per lane was loaded for SDS-PAGE.
After the electrophoresis, the gel was equilibrated in transferring buffer for 10 min and thereafter transferred onto the NC membrane with the semi-dry method at 0.8 mA/cm2 for 20-30 min. Subsequently, the membrane was stained with Ponceau staining solution. After the positions of target protein bands were marked with a marker pen, the staining solution was rinsed with deionized water. The membrane was then blocked in TBS containing 10% skimmed milk (10 mmol/L Tri-base, 150 mmol/L NaCl) for 1 h at room temperature, and incubated with rabbit anti-human E2F1 polyclonal antibody (1:1000 diluted by TBS containing 10% skimmed milk), mouse anti-human β-actin monoclonal antibody (1:10000), mouse anti-human HIPK1 monoclonal antibody (1:500) and mouse anti-human β-catenin monoclonal antibody (1:2000) overnight at 4°C. Then the membrane was washed with TBST (10 mmol/L Tri-base, 150 mmol/L NaCl, 0.1% Tween20, pH 8.0) 5 times by shaking at room temperature (5 min each time), incubated with HRP-labeled secondary antibody that had been diluted with TBS containing 10% skimmed milk for 2 h at room temperature, washed with TBST 5 times by shaking (5 min each time), color-developed using an ECL system and developed by developing device. The gray values of protein bands were detected by Quantity One (BioRad, USA), with the ratio of the gray value of a target band to that of β-actin as the index to compare the target protein expressions.
Extraction of total RNA from paraffin sections of gastric cancer tissues
Paraffin section with the thickness of 5-20 μm was added into an RNase-free EP tube that was then added 1 mL of dimethylbenzene, mixed by vortexing, heated at 50°C for 3 min and centrifuged at 12,000 rpm for 2 min. After dimethylbenzene was removed, the residue was washed twice by 1 mL of 100% ethanol and centrifuged at 12,000 rpm for 2 min. After vacuum suction or drying of the precipitate, the solution was heated at 40-45°C for 15 min to remove ethanol as much as possible. Then 200 μL of digestion buffer was added, and heated at 50°C for 15 min and at 80°C for 15 min. Subsequently, 240 μL of isolation additive and 550 μL of 100% ethanol were added in each tube and mixed. The mixture was washed once by 700 μL of Wash 1 solution and centrifuged at 12,000 rpm for 1 min, and then once by 500 of Wash 2/3 solution each and centrifuged at 12,000 rpm for 1 min. Afterwards, the residue was added 60 μL of DNase, mixed and incubated at room temperature for 30 min. Then the washing with Wash 1/2/3 solutions and centrifugation were repeated. After liquid was removed by centrifugation at 12,000 rpm for 1 min, the residue was finally eluted by 60 μL of eluent or RNase-free water.
Statistical analysis
All data were analyzed by SPSS 20.0. The continuous variables were compared by analysis of variance. The inter-group differences with significant variance homogeneity were detected by Mann-Whitney U and Kruskal-Wallis H tests.
Results
miRNA microarray analysis results
As listed in Table 1, the expressions of miRNA-19b, miRNA-92a and miRNA-20a, the members of miRNA-17-92 gene cluster, gradually decrease along with the adherence and differentiation of tumor spheres.
Microarray detection results of miRNA-17-92 gene cluster members
| miRNA-17-92 gene cluster member | Adherence for 8 h | Adherence for 24 h | Adherence for 72 h |
|---|---|---|---|
| miR-19b expression | 20330 | 16935a | 14565ab |
| miR-92a expression | 7345 | 4280a | 2850ab |
| miR-20a expression | 11565 | 9545a | 7540ab |
Compared with adherence for 8 h, aP<0.05; compared with adherence for 24 h, bP<0.05.
Effects of miRNA-19b/20a/92a on renewal of GCSCs
Lentiviral transfection and expressions
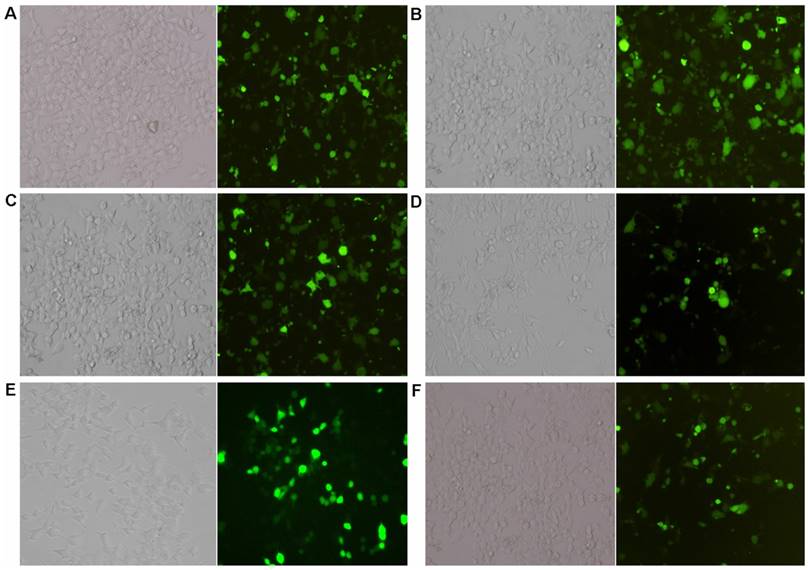
The expressions of lentivirus carrying miRNA-17-19 gene cluster members in MKN28 and CD44-/EpCAM- cells significantly increased over 10-fold (Figure 1).
Transient transfection expressions
Transient transfection with pre-miRNA-19b/ 20a/92a increased miRNA expressions in CD44-/EpCAM- and MKN28 cells, whereas transfection with pre-miRNA-19b/20a/92a antagonists decreased their expressions in SGC7901 and CD44-/EpCAM- cells (Figure S1).
Tumor sphere assay results
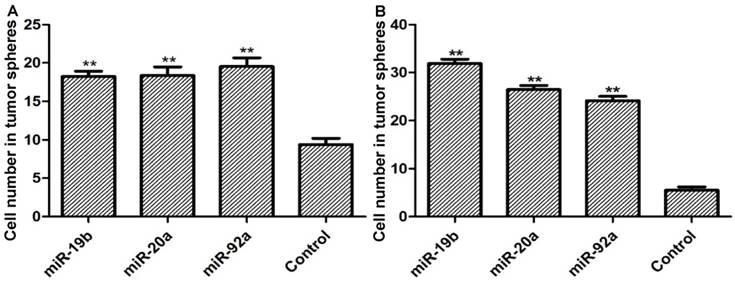
Overexpression of lenti-miRNA-19b/20a/92a significantly boosted the ability of GCSCs to form tumor spheres in which the number of cells evidently increased (Figure 2).
Drug sensitivity test results
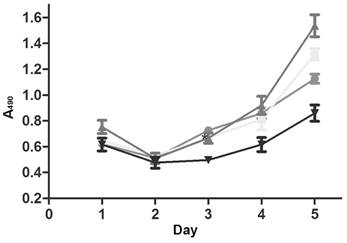
CSCs can resist chemotherapeutic agents, leading to multi-drug resistance and secondary recurrence. After treatment with anti-gastric cancer drug 5-fluorouracil, the survival of lenti-miRNA-19b/20a/92a-infected cells was prolonged compared with that of control (Figure 3).
Lentiviral transfection and expressions (×200). A: Lenti-miRNA-19b expression in CD44-/EpCAM- cells; B: lenti-miRNA-20a expression in CD44-/EpCAM- cells; C: lenti-miRNA-92a expression in CD44-/EpCAM- cells; D: lenti-miRNA-19b expression in MKN28 cells; E: lenti-miRNA-20a expression in MKN28 cells; F: lenti-miRNA-92a expression in MKN28 cells. Left: Expressions of specific miRNAs; right: expressions of green fluorescent protein.
Cell numbers in tumor spheres formed by (A) MKN28 and (B) CD44-/EpCAM- cells. Compared with control group, *P<0.05, **P<0.01.
Growth curves of lenti-miRNA-19b/20a/92a-infected cells. ●: Lenti-miRNA-19b; ■: lenti-miRNA-20a; ▲: lenti-miRNA-92a; ▼: lenti-NC.
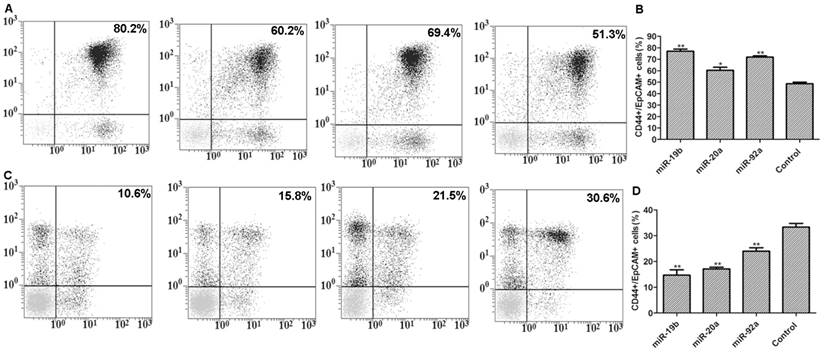
Flow cytometry results
Transient transfection with pre-miRNA-19b/20a/92a significantly increased the number of CD44+/EpCAM+ cells, but transfection with antagonists had the opposite results (Figure 4).
In vivo results
Twenty-eight days after injection of lenti-miRNA-19b/20a/92a-infected cells, each mouse formed tumor in the back, as evidenced by the fluorescence signals (Figure S2). In contrast, only one mouse in the lenti-NC group did so (P<0.05).
Promotive effects of miRNA-19b/20a/92a on proliferation of GCSCs
MTT assay results
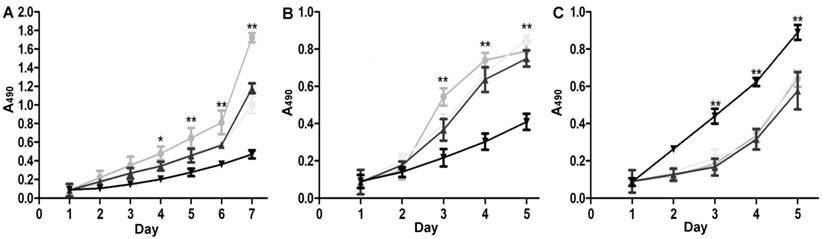
The stable miRNA-19b/20a/92a expression groups proliferated more quickly than the control group did. The proliferation of cells transfected with pre-miRNA-19b/20a/92a was speeded up, whereas that of cells transfected with antagonists was slowed down (Figure 5).
Flow cytometry results. A: Flow cytometry results of pre-miRNA-19b/20a/92a-transfected cells with positive expressions (from left to right: miRNA-19b, miRNA-20a, miRNA-92a and control); B: corresponding histogram; C: flow cytometry results of antagonist-transfected cells with positive expressions (from left to right: miRNA-19b, miRNA-20a, miRNA-92a and control); D: corresponding histogram. Compared with control group, *P<0.05, **P<0.01.
MTT assay results for SGC7901 cells. A: Stable miRNA-19b/20a/92a expression groups, ●: lenti-miRNA-19b; ■: lenti-miRNA-20a; ▲: lenti-miRNA-92a; ▼: lenti-NC; B: cells transfected with pre-miRNA-19b/20a/92a, ●: lenti-miRNA-19b; ■: lenti-miRNA-20a; ▲: lenti-miRNA-92a; ▼: pre-NC; C: cells transfected with antagonists, ●: miRNA-19b-inh; ■: miRNA-20a-inh; ▲: miRNA-92a-inh; ▼: pre-NC. Compared with control group, *P<0.05, **P<0.01.
Colony formation assay results. A: Lenti-miRNAs SGC7901 cells; B: lenti-miRNAs MKN28 cells; C: pre-miRNA SGC7901 cells; D: miRNA-inh SGC7901 cells. Compared with control group, **P<0.01.
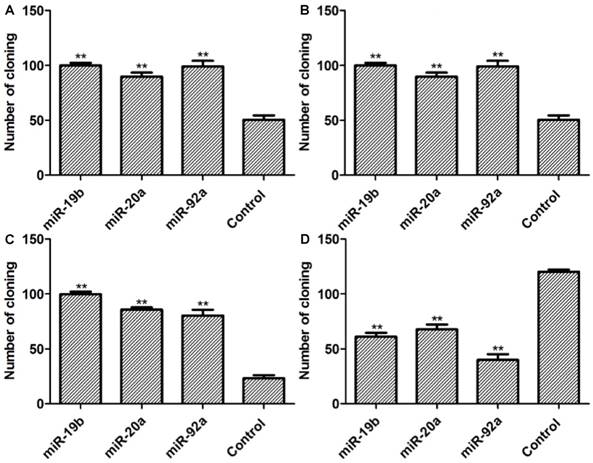
Colony formation assay results
As presented in Figure 6, the numbers of colonies in stable miRNA-19b/20a/92a expression groups significantly exceed that of the control group. Compared with the control group, the numbers of colonies in groups transfected with pre-miRNA-19b/20a/92a were higher, whereas those of groups transfected with antagonists were lower.
In vivo results
We also evaluated the effects of miRNA-17-92 on the proliferation of GCSCs in vivo. The mice injected with miRNA-19b/20a/92a had significantly higher tumor formation capacities than those of NC mice (Figure S3).
Bioinformatics searching results
The target genes of miRNA-17-92 were searched in bioinformatics database MiRanda. There were two miRNA-20a-binding conserved domains in human E2F1, and there were one miRNA-19b- and one miRNA-92a-binding conserved domains in human HIPK1.
Reporter gene assay results
It has previously been reported that miRNA-20a can target E2F1 and then induce miRNA-17-92 gene cluster expression. To further validate these targets, we inserted the 3' untranslated regions of E2F1 and HIPK1 into pGL3 vector and performed the reporter gene assay. miRNA-19b and miRNA-92a bound the 3' untranslated region of HIPK1, and miRNA-20a bound that of E2F1.
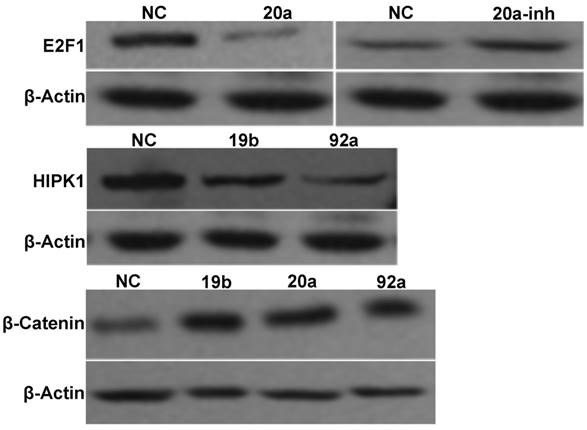
Western blot results
The Western blot results are displayed in Figure 7. Compared with NC, transient transfection with pre-miRNA-20a inhibited endogenous E2F1 expression, but transfection with the antagonist promoted its expression. Since transient transfection with pre-miRNA-19b/92a suppressed HIPK1 expression, E2F1 and HIPK1 were the target genes of miRNA-20a and miRNA-19b/92a respectively. Besides, β-catenin expressions of the cells transfected with pre-miRNA-19b/20a/92a increased compared with that of NC, indicating that β-catenin was activated in them.
Expressions and clinical significance of miRNA-19b/20a/92a in gastric cancer tissue samples
Survival analysis was performed (Figure S4) based on real-time PCR results and clinical pathological data (Table 2). Clearly, the expressions of miRNA-20a and miRNA-92a in gastric cancer samples were negatively correlated with the prognosis of patients. miRNA-92a was an independent factor predicting the prognosis of gastric cancer.
Western blot results of miRNA-17-92 gene cluster and target genes.
Univariate and multivariate analysis results of clinical pathological data and overall survival
| Clinical feature | Overall survival in univariate analysis | Overall survival in multivariate analysis | ||
|---|---|---|---|---|
| P value | HR (95%CI) | P value | HR (95%CI) | |
| Age | 0.312 | 1.012(0.998-1.037) | ||
| Gender | 0.619 | 1.207(0.575-2.537) | ||
| Tumor differentiation | 0.339 | 1.250(0.791-1.977) | ||
| Tumor stage | <0.001 | 2.685(1.744-4.136) | 0.016 | 1.811(1.115-2.943) |
| MiR-17 expression | 0.264 | 1.005(0.996-1.054) | ||
| MiR-20a expression | <0.001 | 1.016(1.007-1.026) | 0.260 | 1.006(0.995-1.017) |
| MiR-19a expression | 0.012 | 1.000(1.000-1.000) | 0.033 | 1.000(1.000-1.000) |
| MiR-19b expression | 0.356 | 1.017(0.981-1.054) | ||
| MiR-18a expression | 0.100 | 1.002(1.000-1.005) | ||
| MiR-92a expression | <0.001 | 1.001(1.000-1.001) | <0.001 | 1.001(1.000-1.001) |
Discussion
Malignant tumor tissue, as a heteroplasmon, consists of cells at different stages of differentiation, of which there are a small number of stem cell-like cells with renewal and differentiation potentials, referred to as CSCs. CSCs are typified by specific markers within tumors, which can form xenografts in immunodeficient mice [10]. Han et al. [11] cultured gastric cancer cells and isolated those with specific markers, which were subcutaneously implanted into rats to form tumors, suggesting the existence of GCSCs.
Until now, gastric cancer still cannot be well treated mainly because some GCSCs escape chemotherapy drugs, which has become one of the main reasons for recurrence and metastasis [12]. Targeted therapy provides new hope for gastric cancer patients, and eligible drugs should be able to inhibit the damage to GCSCs without affecting normal cells. Whether GCSCs markers can become suitable targets needs further studies [13, 14]. Yashiro et al. [15] found that inhibition of c-met gene increased the sensitivity of GCSCs to chemotherapy. GCSCs are also closely related to the prognosis of gastric cancer, and high expression of CD44+ stem cell-like cells can predict biological invasion behaviors, also as an independent predictor for treatment outcomes [16]. Golestaneh et al. [17] reported that GCSCs had different mRNA expression levels in the tumorigenic process, and that these mRNAs were involved in the biological regulation of cancer cells [18-20].
In this study, the expressions of lentivirus carrying miRNA-17-19 gene in MKN28 and CD44-/EpCAM- cells increased significantly. Transient transfection with pre-miRNA-19b/20a/92a elevated the expressions of miRNA in CD44-/EpCAM- and MKN28 cells, whereas transfection with the antagonists reduced their expressions in SGC7901 and CD44+/EpCAM+ cells. Overexpression of lenti-miRNA-19b/20a/92a significantly enhanced the capability of GCCs to form tumor spheres. Under the action of chemotherapeutic agent, the survival of lenti-miRNA-19b/20a/92a-infected cells was prolonged. Transient transfection with pre-miRNA-19b/20a/92a significantly increased the number of CD44+/EpCAM+ cells, but transfection with the antagonists reduced this number. MTT assay showed that the proliferation rates of stable miRNA-19b/20a/92a expression groups surpassed that of the control group. Transient transfection with pre-miRNA-19b/20a/92a accelerated the proliferation rate of gastric cancer cells, but transfection with the antagonists slowed down the proliferation. The colony formation assay showed that the number of colonies formed by the cells with stable miRNA-17-92 expression was significantly higher than that of the control group. Compared with the control group, the numbers of colonies in the precursor-transfected groups were higher, whereas those of the antagonist-transfected groups were lower.
In addition, bioinformatics analysis revealed another inhibitory molecule of the Wnt/β-catenin pathway, HIPK1, which was also a potential target gene of miRNA-17-92 gene cluster. HIPK1 can suppress the activation of Wnt/β-catenin in embryonic kidney cells. Moreover, many Wnt/β-catenin-related genes need the activation of HIPK1 in the development of the stomach. In this study, we not only proved by the reporter gene assay and Western blot that HIPK1 was a target gene of miRNA-17-92, but also found that transfection with precursors elevated the expression of β-catenin. Based on this, we hypothesized that miRNA-17-92 gene cluster may indirectly activate the Wnt/β-catenin pathway through directly targeting E2F1 and HIPK1, increasing the number of EpCAM+ GCSCs simultaneously. Indirectly activating Wnt/β-catenin and increasing the number of EpCAM+ GCSCs may be one of the mechanisms by which miRNA-17-92 promotes the self-renewal of GCSCs, so in-depth studies are still in need.
Conclusion
In summary, miRNA-19b/20a/92a genes were continuously deleted during the differentiation of GCSCs, and miRNA-17-92 gene facilitated their renewal and proliferation. Meanwhile, miRNA-19b/20a/92a promoted GCSCs self-renewal by targeting E2F1 and HIPK1 at the post-transcriptional level and activating the β-catenin signaling pathway. The expressions of miRNA-20a and miRNA-92a in gastric cancer samples were negatively correlated with the prognosis of patients. miRNA-92a was an independent factor and index predicting the prognosis of gastric cancer.
Supplementary Material
Supplementary figures.
Competing Interests
The authors have declared that no competing interest exists.
References
1. Ohtsu K, Yao K, Matsunaga K. et al. Lipid is absorbed in the stomach by epithelial neoplasms (adenomas and early cancers): a novel functional endoscopy technique. Endosc Int Open. 2015;3:E318-22
2. Hsu SD, Tseng YT, Shrestha S. et al. miRTarBase update 2014: an information resource for experimentally validated miRNA-target interactions. Nucleic Acids Res. 2014;42:78-85
3. Li JH, Liu S, Zhou H, Qu LH, Yang JH. starBase v2.0: decoding miRNA-ceRNA, miRNA-ncRNA and protein-RNA interaction networks from large-scale CLIP-Seq data. Nucleic Acids Res. 2014;42:92-7
4. Gajos-Michniewicz A, Duechler M, Czyz M. MiRNA in melanoma-derived exosomes. Cancer Lett. 2014;347:29-37
5. Shuang L, Feng Y, Zhang J. et al. Regulatory roles of miRNA in the human neural stem cell transformation to glioma stem cells. J Cell Biochem. 2014;115:1368-80
6. Jones MF, Hara T, Francis P. et al. The CDX1-microRNA-215 axis regulates colorectal cancer stem cell differentiation. Proc Natl Acad Sci U S A. 2015;112:E1550-8
7. Takahashi RU, Miyazaki H, Takeshita F. et al. Loss of microRNA-27b contributes to breast cancer stem cell generation by activating ENPP1. Nat Commun. 2015;6:7318
8. Tulsyan S, Agarwal G, Lal P, Mittal B. Significant association of combination of OCT4, NANOG, and SOX2, gene polymorphisms in susceptibility and response to treatment in North Indian breast cancer patients. Cancer Chemother Pharmacol. 2014;74:1065-78
9. Huang G, Ye S, Zhou X, Liu D, Ying QL. Molecular basis of embryonic stem cell self-renewal: from signaling pathways to pluripotency network. Cell Mol Life Sci. 2015;72:1741-57
10. Shiozawa Y, Nie B, Pienta KJ, Morgan TM, Taichman RS. Cancer stem cells and their role in metastasis. Pharmacol Ther. 2013;138:285-93
11. Han ME, Jeon TY, Hwang SH. et al. Cancer spheres from gastric cancer patients provide an ideal model system for cancer stem cell research. Cell Mol Life Sci. 2011;68:3589-605
12. Zhang X, Hua R, Wang X. et al. Identification of stem-like cells and clinical significance of candidate stem cell markers in gastric cancer. Oncotarget. 2016;7:9815-31
13. Nishikawa S, Konno M, Hamabe A. et al. Surgically resected human tumors reveal the biological significance of the gastric cancer stem cell markers CD44 and CD26. Oncol Lett. 2015;9:2361-7
14. Wang B, Chen Q, Cao Y. et al. LGR5 Is a Gastric Cancer Stem Cell Marker Associated with Stemness and the EMT Signature Genes NANOG, NANOGP8, PRRX1, TWIST1, and BMI1. PLoS One. 2016;11:e0168904
15. Yashiro M, Nishii T, Hasegawa T. et al. A c-Met inhibitor increases the chemosensitivity of cancer stem cells to the irinotecan in gastric carcinoma. Br J Cancer. 2013;109:2619-28
16. Ryu HS, Park do J, Kim HH, Kim WH, Lee HS. Combination of epithelial-mesenchymal transition and cancer stem cell-like phenotypes has independent prognostic value in gastric cancer. Hum Pathol. 2012;43:520-8
17. Golestaneh AF, Atashi A, Langroudi L, Shafiee A, Ghaemi N, Soleimani M. miRNAs expressed differently in cancer stem cells and cancer cells of human gastric cancer cell line MKN-45. Cell Biochem Funct. 2012;30:411-8
18. Zabala M, Lobo NA, Qian D, van Weele LJ, Heiser D, Clarke MF. Overview: Cancer Stem Cell Self-Renewal. Cambridge, USA: Elsevier. 2016:25-58
19. Raza U, Zhang JD, Sahin O. MicroRNAs: master regulators of drug resistance, stemness, and metastasis. J Mol Med (Berl). 2014;92:321-36
20. Takahashi RU, Miyazaki H, Ochiya T. The role of microRNAs in the regulation of cancer stem cells. Front Genet. 2014;4:295
Author contact
![]() Corresponding author: Haitao Zhu Email: zhuhtchnmunet
Corresponding author: Haitao Zhu Email: zhuhtchnmunet

 Global reach, higher impact
Global reach, higher impact