Impact Factor
ISSN: 1449-1907
Int J Med Sci 2018; 15(1):77-85. doi:10.7150/ijms.22345 This issue Cite
Research Paper
Gene and Protein Expression Profiles in a Mouse Model of Collagen-Induced Arthritis
1. Department of Biomedical Laboratory Science, College of Health Science, Eulji University, Seongnam-si, Gyeonggi-do, 13135, Republic of Korea;
2. Department of Senior Healthcare, BK21 plus Program, Graduated School, Eulji University, Daejeon, 34824, Republic of Korea;
3. Department of Biomedical Laboratory Science, College of Health Sciences, Yonsei University at Wonju, Wonju, Gangwon-do 26493, Republic of Korea.
Received 2017-8-12; Accepted 2017-10-12; Published 2018-1-1
Abstract
The risk of rheumatoid arthritis (RA), an autoimmune disease, in the elderly population increases along with that of atherosclerosis, cardiovascular disease, type 2 diabetes, and Alzheimer's disease. Identifying specific biomarkers for RA can clarify the underlying molecular mechanisms and can aid diagnosis and patient care. To this end, the present study investigated the genes and proteins that are differentially expressed in RA using a mouse collagen-induced arthritis (CIA) model. We performed gene microarray and proteome array analyses using blood samples from the mice and found that 50 genes and 24 proteins were upregulated and 48 genes were downregulated by more than 2-fold in the CIA model relative to the control. The gene microarray and proteome array results were validated by evaluating the expression levels of select genes and proteins by real-time PCR and western blotting, respectively. We found that the level of integrin α2, which has not been previously reported as a biomarker of RA, was significantly increased in CIA mice as compared to controls. These findings provide a set of novel biomarkers that can be useful for diagnosing and evaluating the progression of RA.
Keywords: collagen-induced arthritis, microarray, proteome analysis, biomarker, integrin α2.
Introduction
The incidence of rheumatoid arthritis (RA) is rising in the elderly population; according to a report by the National Institutes of Health, approximately 1.3 million adults are afflicted with RA [1]. It is estimated that up to 1% of the global population has been diagnosed with RA. The symptoms include swelling, pain, and joint stiffness from the knuckles to the knees. RA can also affect other organs such as lungs and heart, and is a progressively debilitating disease that can dramatically reduce the quality of life. The exact cause of RA is unknown, although it is assumed that both genetic and environmental factors are involved [2, 3]. It has been reported that RA is related to the binding of autoantibodies to the host synovium [4], qualifying RA as an autoimmune disease. The incidence of RA is higher in women, suggesting that sex hormones influence disease etiology [5]. Cigarette smoking and dust are also proposed risk factors for RA [6, 7]. Similar to atherosclerosis, cardiovascular disease, and non-insulin-dependent diabetes, RA is an age-associated disease [8, 9]. Rheumatoid factor (RF) and circulating anti-cyclic citrullinated peptide levels are biomarkers for RA diagnosis; however, only a subset of patients expresses both factors [4, 10]. Patients are also diagnosed based on symptoms and family history [11].
A DBA1/J mouse model of collagen-induced arthritis (CIA) is widely used for the study of RA [12, 13]. These mice exhibit the pathological features of RA, including synovial hyperplasia, inflammatory cell infiltration, and cartilage erosion [14]. Transferring CIA mouse serum to healthy mice induces arthritis via passive immunity [15, 16].
Tumor necrosis factor (TNF)-α is a key cytokine involved in RA. Transgenic mice overexpressin human TNF-α develop RA, and treatment of arthritic mice with anti-TNF-α antibody prevents disease development [17, 18]. Interleukin (IL)-1, a component of TNF-α signaling, plays an important role in cartilage erosion [18, 19]. Several genes have been linked to RA susceptibility [20, 21], including signal transducer and activator of transcription (STAT)4, which is a risk factor for systemic lupus erythematosus [22] and is associated with IL-12/23 and interferon (IFN)-α/β in T cell signaling [23]. Despite these findings, there are few specific biomarkers that are useful for diagnosing and monitoring the progression of RA.
To address this issue, we analyzed the gene and protein expression profiles of RA using the CIA model. A previous gene expression profiling study using CIA mice reported that major histocompatibility complex class I, II, basigin, fibroblast activation protein, cathepsin K, cluster of differentiation (CD)53, RAF-1, glucagon, and retinal taurine transporter contribute to CIA susceptibility or severity [24]. In the present study, we identified the integrin α2 gene (Itga2) as an additional and novel biomarker for RA.
Materials and Methods
Materials
Antibodies for western blotting were purchased from Bio-Rad (Hercules, CA, USA). The ProteomeProfiler Mouse Cytokine Array Panel A (ARY006) was from R&D Systems (Minneapolis, MN, USA). Collagen (Chondrex, 20022) and complete (Chondrex, 7001) and incomplete (Chondrex, 7002) Freund's adjuvant were purchased from Central Lab. Animal Inc. (Seoul, Korea).
Animals
Male DBA1/J mice (6-8 weeks old) were purchased from Central Lab. Animal Inc. and Orient Bio (Seongnam, Korea). Animal maintenance and experiments were in accordance with the guidelines of the Eulji University Institutional Animal Care and Use Committee (approval No. EUIACUC16-17, approval date 10 August 2016).
In vivo experiments
Bovine type II collagen was used to induce arthritis in mice as previously described [16]. Briefly, bovine type II collagen (2 mg/ml) was mixed at a 1:1 volume ratio with complete Freund's adjuvant. Each mouse was injected with 100 mg of bovine type II collagen in 0.1 ml of emulsion. A booster injection of 100 mg of bovine type II collagen was administered subcutaneously as a solution in 0.1 ml of incomplete Freund's adjuvant 14 days later. Mice were continuously observed for swelling of the distal joints after the primary immunization. Arthritis developed between 34 and 40 days after the primary immunization based on the arthritis score [16] (data not shown). At the end of the experiment, blood and paws were collected from each mouse. Whole blood was stored in a PAXgene tube (Qiagen, Valencia, CA, USA) at -80°C until RNA and protein extraction. Paws were fixed in 10% buffered formalin, decalcified in 10% formic acid, and then embedded in paraffin. Sagittal serial sections of the whole paws were cut and stained with hematoxylin and eosin for light microscopy examination.
RNA extraction, cDNA synthesis, and quantitative real-time (qRT-)PCR
Total RNA was extracted using the QIAamp RNA Blood Mini kit (Qiagen) according to the manufacturer's protocols, and 1 µg was used for cDNA synthesis with the SensiFAST cDNA Synthesis kit (Bioline, Taunton, MA, USA), with a primer annealing step at 25°C for 10 min, followed by reverse transcription at 42°C for 15 min, inactivation at 85°C for 10 min, and storage at 4°C. qRT-PCR was performed on an ABI StepOnePlus system (Applied Biosystems, Foster City, CA, USA). Forward and reverse primer sequences were as follows: IL-1β, 5'-GCTCATCTGGGATCCTCTCC-3' and 5'-CCTGCCTGAAGCTCTTGTTG-3' [54]; IL-6, 5'-ACGGCCTTCCCTACTTCACA-3' and 5'-CATTTCCACGATTTCCCAGA-3' [55]; TNF-α, 5'-GCCTCTTCTCATTCCTGCTTG-3' and 5'-CTGATGAGAGGGAGGCCATT-3' [55]; integrin α2, 5'-CGCTCCTTCTGTCATCAAGAGTGTC-3' and 5'-GGAATGTGGATAGTCACCAATGCC-3' [56]; and β-actin, 5'- CGTGCGTGACATCAAAGAGAA-3' and 5'- TGGATGCCACAGGATTCCAT-3' [55]. β-Actin was used as an internal control to normalize target gene expression levels, which were determined with the 2-ΔΔCT method [57].
Protein extraction and western blotting
Blood from control and CIA mice was mixed with radioimmunoprecipitation assay buffer (Thermo Fisher Scientific, Waltham, MA, USA) containing protease inhibitor (GE Healthcare, Little Chalfont, UK). After incubation on ice for 20 min, samples were centrifuged at 15,000 × g and 4°C for 15 min. The supernatant was used to determine the protein concentration with the Quick start Bradford reagent (Bio-Rad). A total of 100 µg of extracted protein was used for immunoblotting. Samples were separated by sodium dodecyl sulfate polyacrylamide gel electrophoresis and transferred to a polyvinylidene difluoride membrane. After incubation with 5% skim milk in Tris-buffered saline with Tween 20 (TBST) composed of 10 mM Tris (pH 8.0), 150 mM NaCl, and 0.05% Tween 20 for 1 h, the membrane was incubated overnight at 4°C with antibodies against the following proteins: IL-1β (#12242) and glyceraldehyde 3-phosphate dehydrogenase (GAPDH; #5174) (both from Cell Signaling Technology, Danvers, MA, USA); TNF-α (ab66579) and integrin α2 (ab133557) (both from Abcam, Cambridge, MA, USA); IL-6 (sc-1265-R) (Santa Cruz Biotechnology, Santa Cruz, CA, USA). The membrane was washed four times for 5 min and incubated for 2 h with a 1:10,000 dilution of horseradish peroxidase-conjugated anti-mouse or -rabbit antibody. The membrane was washed six times with TBST for 10 min and developed with the enhanced chemiluminescence system (GE Healthcare) and blue X-ray film (Agfa HealthCare NV, Mortsel, Belgium) according to the manufacturer's protocols. After the transfer, the gel was stained with Coomassie Blue reagent (Bio-Rad). GAPDH was used as the loading control. The membrane was stained with Ponceau S (Sigma-Aldrich, St. Louis, MO, USA) after immunoblotting.
Microarray
Blood was collected from mice in a PAXgene blood RNA tube (PreAnalytiX, Hombrechtikon, Switzerland) and RNA was isolated using the PAXgene Blood RNA kit (PreAnalytiX) according to the manufacturer's protocol. RNA purity and integrity were determined based on the optical density 260/280 ratio on an Agilent 2100 Bioanalyzer (Agilent Technologies, Palo Alto, CA, USA). Microarray analysis with a GeneChip Mouse Gene 2.0 ST Array was performed Macrogen Co. (Seoul, Korea). Raw data were extracted using Affymetrix Expression Console software and were filtered when P < 0.05. The statistical significance of expression data was determined with the independent Student's t test based on fold change, where the null hypothesis was that no difference existed between the two groups. Gene enrichment analysis and functional annotation were performed based on Kyoto Encyclopedia of Genes and Genomes (KEGG) pathways.
Proteome array
The ProteomeProfiler Mouse Cytokine Array Panel A (R&D Systems) was used according to the manufacturer's protocols to obtain protein expression profiles using 50-μl blood samples. Spot density was determined using HLImage software (Western Vision Software, Salt Lake City, UT, USA).
Statistical analysis
Differences between groups were evaluated with the Student's t test using Excel software (Microsoft, Redmond, WA, USA). P < 0.05 was considered statistically significant.
Results
Murine model of arthritis
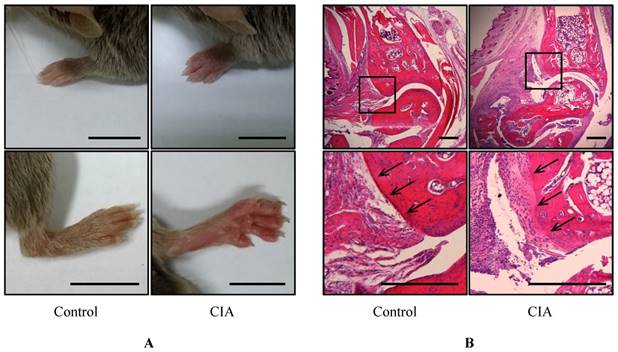
To identify potential biomarkers of RA, we established a mouse CIA model by injecting male DBA1/J mice with bovine type II collagen. Pathological changes were observed after 49 days; the mice had swollen paws and ankles typical of arthritis (Figure 1A). Histological examination of the mouse foot revealed increased inflammation and immune cell infiltration (Figure 1B), and the cartilage boundaries appeared crushed. These results confirm that RA was induced in the CIA mice after 49 days.
Gross morphological and histological examination of CIA. (A) Gross observation of mouse paws. Shown are the fore paws (top) and hind paws (bottom) of control (n = 12) and CIA (n = 14) mice at 14 weeks of age. Scale bars = 10 mm. (B) H&E staining of sagittal sections of control and CIA mouse joints. Lower panels show enlarged views of the areas delineated by a box in the upper panels. Arrows indicate cartilage boundaries. Scale bars = 1 mm.
Genes differentially expressed in the blood of control and CIA mice
| Increased genes | Decreased genes | |||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| No. | Gene symbol | Fold change | No. | Gene symbol | Fold change | No. | Gene symbol | Fold change | No. | Gene symbol | Fold change | |||
| 1 | Irf7 | 24.7 | 26 | Olfr1386 | 2.2 | 1 | Sec24a | -3.9 | 25 | Rps2 | -2.2 | |||
| 2 | Isg15 | 6.6 | 27 | Olfr1502 | 2.2 | 2 | Cd27 | -3.3 | 26 | Il7r | -2.2 | |||
| 3 | Ifit1 | 6.1 | 28 | Cyp2d26 | 2.2 | 3 | Cd8b1 | -3.3 | 27 | Ccnd2 | -2.2 | |||
| 4 | Oas3 | 4.9 | 29 | Mir423 | 2.2 | 4 | Ccl5 | -3.2 | 28 | Rgs14 | -2.2 | |||
| 5 | Mir107 | 4.2 | 30 | Ppp1r15a | 2.2 | 5 | Cd3g | -3.2 | 29 | Pck2 | -2.2 | |||
| 6 | H2-Q8 | 3.4 | 31 | Spta1 | 2.2 | 6 | Cd3e | -2.9 | 30 | Cd59a | -2.2 | |||
| 7 | Fn1 | 3.2 | 32 | Rps15 | 2.2 | 7 | Lat | -2.7 | 31 | Tcf7 | -2.2 | |||
| 8 | C1ra | 3.0 | 33 | Vwf | 2.2 | 8 | Hist1h2ba | -2.7 | 32 | Rps11 | -2.2 | |||
| 9 | Ifih1 | 2.9 | 34 | Pla2g2a | 2.2 | 9 | Ncr1 | -2.7 | 33 | Ugcg | -2.1 | |||
| 10 | Fpr2 | 2.9 | 35 | Cmpk2 | 2.1 | 10 | Eif4a1 | -2.6 | 34 | Rpl6 | -2.1 | |||
| 11 | Clca3a1 | 2.7 | 36 | F5 | 2.1 | 11 | Rps8 | -2.6 | 35 | Olfr373 | -2.1 | |||
| 12 | Olfr774 | 2.7 | 37 | Fos | 2.1 | 12 | Cd4 | -2.6 | 36 | Olfr705 | -2.1 | |||
| 13 | Hist1h2bj | 2.7 | 38 | Olfr917 | 2.1 | 13 | Il2rb | -2.5 | 37 | Cd209a | -2.1 | |||
| 14 | Ifi204 | 2.6 | 39 | Ptpn11 | 2.0 | 14 | Ddx5 | -2.5 | 38 | Lck | -2.1 | |||
| 15 | Fads2 | 2.6 | 40 | C3 | 2.0 | 15 | Rpl30 | -2.5 | 39 | Klrd1 | -2.1 | |||
| 16 | Plxna4 | 2.5 | 41 | Hist2h4 | 2.0 | 16 | Mirlet7f-2 | -2.5 | 40 | Olfr1501 | -2.1 | |||
| 17 | Sp100 | 2.5 | 42 | Flna | 2.0 | 17 | Ppp2r2d | -2.5 | 41 | Rpl37a | -2.1 | |||
| 18 | Vcl | 2.3 | 43 | Cks1b | 2.0 | 18 | Atp1b3 | -2.4 | 42 | Mir103-2 | -2.0 | |||
| 19 | Tuba3b | 2.3 | 44 | Olfr1057 | 2.0 | 19 | Ube3c | -2.4 | 43 | Ppil2 | -2.0 | |||
| 20 | Dusp3 | 2.3 | 45 | Olfr726 | 2.0 | 20 | Olfr875 | -2.4 | 44 | Ddx46 | -2.0 | |||
| 21 | Hist1h2aa | 2.3 | 46 | Olfr1298 | 2.0 | 21 | Rmrp | -2.3 | 45 | Stat4 | -2.0 | |||
| 22 | H2-T24 | 2.3 | 47 | Igtp | 2.0 | 22 | Ctsw | -2.3 | 46 | Itgb7 | -2.0 | |||
| 23 | Rps6ka2 | 2.3 | 48 | Mir7-1 | 2.0 | 23 | Slc40a1 | -2.3 | 47 | Bcl2a1c | -2.0 | |||
| 24 | Itga2 | 2.3 | 49 | Thbs1 | 2.0 | 24 | Sec61b | -2.2 | 48 | Olfr1299 | -2.0 | |||
| 25 | Sort1 | 2.2 | 50 | Olfr38 | 2.0 | |||||||||
Gene expression profiling
Gene expression analysis was performed using RNA from blood samples from control and CIA mice. In total, 98 genes showed ≥ 2-fold change in blood mRNA expression in CIA as compared to control mice (Table 1 and Table S1). Of these genes, 50 were upregulated and 48 were downregulated. The expression levels of eight genes [interferon regulatory factor (Irf)7; interferon-stimulated gene 15; interferon induced protein with tetratricopeptide repeats 1; 2'-5'-oligoadenylate synthetase 3; microRNA 107; histocompatibility 2, Q region locus 8; fibronectin 1; and complement component 1, r subcomponent A] were > 3-fold higher in CIA than in control mice, with the level of Irf7 showing a > 24-fold difference. The expression of five genes [Sec24a, Cd27, Cd8b1, chemokine (C-C motif) ligand (Ccl)5, and Cd3g] was decreased by > 3 fold in CIA as compared to control mice. A KEGG pathway analysis of 98 genes whose expression differed by ≥ 2-fold between the two groups revealed that Cd4 was associated with seven different KEGG pathways (Tables 1 and 2). Genes in five of nine analyzed pathways were downregulated in CIA. For example, Th1 and Th2 cell differentiation in the immune system category showed downregulation of seven genes. Of the 26 analyzed pathways, five contained only genes that were upregulated in CIA, such as those related to mitogen-activated protein kinase signaling, extracellular matrix (ECM), and focal adhesion. All of the analyzed pathways had P < 0.05.
Protein expression profiling
Changes in protein levels in CIA mice were evaluated with a proteome array using whole blood. Because most genes identified by the KEGG pathway analysis were related to the immune system and immune-related diseases, we used the ProteomeProfiler Mouse Cytokine Array Panel A for protein expression profiling. We found that the levels of all 40 cytokines were slightly increased in the blood of CIA as compared to control mice (Table S2), with 24 showing a > 2-fold increase (Table 3B). These genes were grouped into 18 categories based on the KEGG classification scheme (Table 3A). Most of the upregulated cytokines were associated with cytokine-cytokine receptor interaction, Janus kinase-STAT signaling, and helper T cell (Th)17 differentiation pathways, whereas seven were associated with RA and the hematopoietic cell lineage. The cytokines associated with the RA pathway included IL-17; IL-23; IL-1β; monocyte chemoattractant protein (MCP)-5; TNF-α; regulated upon activation, normally T-expressed, and presumably secreted (RANTES); IL-6; and IL-16. IL-2 expression showed the greatest difference between CIA and control mice. The levels of IL-1β, TNF-α, and IL-6, which are the major pro-inflammatory cytokines [25], were 6.41, 3.35, and 2.41-fold higher, respectively; IL-17 and -23, which are involved in Th17 cell differentiation [26], were upregulated by 13.09- and 7.21-fold, respectively; and RANTES, also known as Ccl5 [27, 28], was upregulated 2.5-fold in the RA model.
Validation of differentially expressed genes and proteins
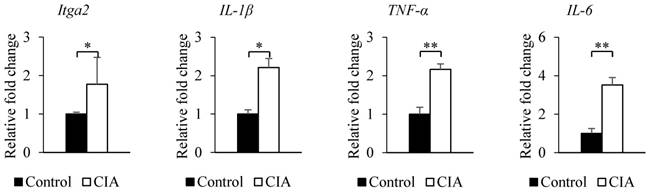
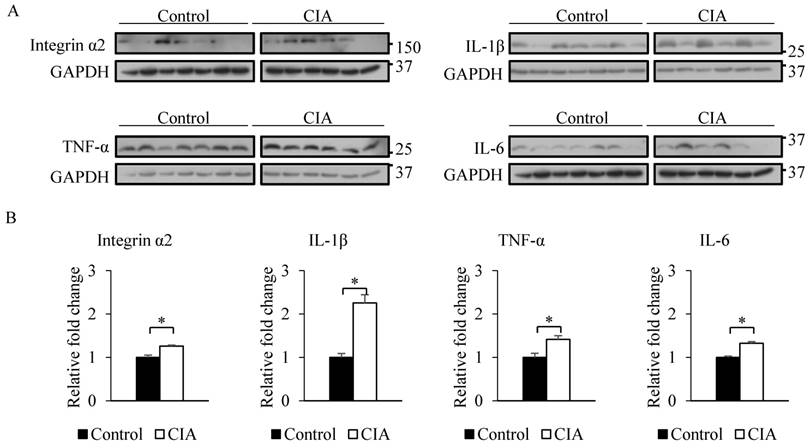
The results from the gene expression microarray and proteome array were validated by qRT-PCR and western blot analysis of selected genes and proteins, including integrin α2, IL-1β, TNF-α, and IL-6, that showed ≥ 2-fold difference in expression relative to the control. Consistent with the gene microarray results, Itga2 expression was significantly higher in CIA than in control mice by qRT-PCR (Figure 2). A similar result was obtained for the genes encoding IL-1β, TNF-α, and IL-6. Western blot analysis revealed that blood protein levels of integrin α2, IL-1β, TNF-α, and IL-6 were increased in CIA as compared to control mice (Figure 3). Thus, the qRT-PCR and western blotting results support the validity of the microarray and protein array data.
KEGG pathway analysis of differentially expressed genes identified by microarray analysis
| No. | KEGG classification | Pathway | Number of significant genes | Increased genes | Decreased genes | P value |
|---|---|---|---|---|---|---|
| 1 | Immune system | Th1 and Th2 cell differentiation | 7 | Stat4, Lck, Il2rb, Cd4, Lat, Cd3e, Cd3g | < 0.001 | |
| 2 | Th17 cell differentiation | 6 | Lck, Il2rb, Cd4, Lat, Cd3e, Cd3g | < 0.001 | ||
| 3 | T cell receptor signaling pathway | 6 | Lck, Cd4, Lat, Cd3e, Cd3g, Cd8b1 | < 0.001 | ||
| 4 | Antigen processing and presentation | 5 | H2-Q8, H2-T24 | Klrd1, Cd4, Cd8b1 | < 0.001 | |
| 5 | Hematopoietic cell lineage | 5 | Il7r, Cd4, Cd3e, Cd3g, Cd8b1 | < 0.001 | ||
| 6 | Complement and coagulation cascades | 5 | C1ra, Vwf, F5, C3 | Cd59a | < 0.001 | |
| 7 | Natural killer cell mediated cytotoxicity | 4 | Klrd1, Lck, Ncr1, Lat | < 0.001 | ||
| 8 | NOD-like receptor signaling pathway | 4 | Irf7, Oas3, Ifi204 | Ccl5 | < 0.01 | |
| 9 | RIG-I-like receptor signaling pathway | 3 | Irf7, Isg15, Ifih1 | < 0.01 | ||
| 10 | Immune diseases | Primary immunodeficiency | 5 | Lck, Il7r, Cd4, Cd3e, Cd8b1 | < 0.001 | |
| 11 | Systemic lupus erythematosus | 5 | Hist1h2bj, Hist1h2aa, C3, Hist2h4 | Hist1h2ba | < 0.001 | |
| 12 | Graft-versus-host disease | 3 | H2-Q8, H2-T24 | Klrd1 | < 0.01 | |
| 13 | Signal transduction | PI3K-Akt signaling pathway | 10 | Fn1, Itga2, Vwf, Thbs1 | Pck2, Ppp2r2d, Itgb7, Ccnd2, Il7r, Il2rb | < 0.001 |
| 14 | Jak-STAT signaling pathway | 4 | Stat4, Ccnd2, Il7r, Il2rb | < 0.01 | ||
| 15 | MAPK signaling pathway | 4 | Dusp3, Rps6ka2, Fos, Flna | < 0.01 | ||
| 16 | NF-kappa B signaling pathway | 3 | Bcl2a1c, Lck, Lat | < 0.05 | ||
| 17 | Signaling molecules and interaction | Cell adhesion molecules (CAMs) | 5 | H2-Q8, H2-T24 | Itgb7, Cd4, Cd8b1 | < 0.001 |
| 18 | ECM-receptor interaction | 4 | Fn1, Itga2, Vwf, Thbs1 | < 0.001 | ||
| 19 | Cytokine-cytokine receptor interaction | 4 | Il7r, Il2rb, Ccl5, Cd27 | < 0.01 | ||
| 20 | Cellular community | Focal adhesion | 6 | Fn1, Vcl, Itga2, Vwf, Flna, Thbs1 | < 0.001 | |
| 21 | Cell growth and death | Apoptosis | 4 | Tuba3b, Spta1 | Bcl2a1c, Ctsw | < 0.01 |
| 22 | Transport and catabolism | Phagosome | 9 | H2-Q8, C1ra, Tuba3b, H2-T24, Itga2, C3, Thbs1 | Sec61b, Cd209a | < 0.01 |
| 23 | Lipid metabolism | alpha-Linolenic acid metabolism | 2 | Fads2, Pla2g2a | < 0.05 | |
| 24 | Translation | Ribosome | 7 | Rps15 | Rps2, Rpl30,Rpl37a, Rpl6, Rps11, Rps8 | < 0.01 |
| 25 | Nervous system | Neurotrophin signaling pathway | 3 | Rps6ka2, Sort1, Ptpn11 | < 0.05 | |
| 26 | Sensory system | Olfactory transduction | 12 | Clca3a1, Olfr774, Olfr1386, Olfr1502, Olfr726, Olfr1298, Olfr38 | Olfr1299,Olfr1501, Olfr705, Olfr373, Olfr875 | < 0.001 |
Validation of microarray and proteome array results by qRT-PCR. The expression of each gene shown in the figure was confirmed by qRT-PCR using specific primers. β-Actin served as an internal control. Data represent the mean ± SEM. *P < 0.05, **P < 0.01
Proteins differentially expressed in the blood of control and CIA mice. (A) Classification of upregulated proteins
| No. | Classification | Identified proteins | Number of proteins |
|---|---|---|---|
| 1 | Cytokine-cytokine receptor interaction | IL-2, IL-27, IL-17, MIP-1beta, IL-23, IL-1beta, IL-1ra, MIP-2, MCP-5, TARC, IL-3, TNF-alpha, IP-10, MIG, BLC, I-TAC, RANTES, IL-4, IL-6, IL-5, IL-10 | 21 |
| 2 | Jak-STAT signaling pathway | IL-2, IL-27, IL-23, IL-3, IL-4, IL-6, IL-5, IL-10 | 8 |
| 3 | Th17 cell differentiation | IL-2, IL-27, IL-17, IL-23, IL-1beta, IL-1ra, IL-4, IL-6 | 8 |
| 4 | Rheumatoid arthritis | IL-17, IL-23, IL-1beta, MCP-5, TNF-alpha, RANTES, IL-6, IL-16 | 7 |
| 5 | Hematopoietic cell lineage | IL-1beta, IL-1ra, IL-3, TNF-alpha, IL-4, IL-6, IL-5 | 7 |
| 6 | NOD-like receptor signaling pathway | IL-1beta, MIP-2, MCP-5, TNF-alpha, RANTES, IL-6 | 6 |
| 7 | Graft-versus-host disease | IL-2, IL-1beta, TNF-alpha, IL-6, IL-10 | 5 |
| 8 | NF-kappa B signaling pathway | MIP-1beta, IL-1beta, IL-1ra, TNF-alpha, BLC | 5 |
| 9 | T cell receptor signaling pathway | IL-2, TNF-alpha, IL-4, IL-5, IL-10 | 5 |
| 10 | PI3K-Akt signaling pathway | IL-2, IL-3, IL-4, IL-6 | 4 |
| 11 | Th1 and Th2 cell differentiation | IL-2, IL-4, IL-5 | 3 |
| 12 | MAPK signaling pathway | IL-1beta, IL-1ra, TNF-alpha | 3 |
| 13 | Systemic lupus erythematosus | TNF-alpha, IL-10 | 2 |
| 14 | HIF-1 signaling pathway | TIMP-1, IL-6 | 2 |
| 15 | RIG-I-like receptor signaling pathway | TNF-alpha, IP-10 | 2 |
| 16 | Apoptosis | IL-3, TNF-alpha | 2 |
| 17 | Natural killer cell mediated cytotoxicity | TNF-alpha | 1 |
| 18 | Antigen processing and presentation | TNF-alpha | 1 |
Densitometry analysis of upregulated proteins (n = 24)
| No. | Protein name | Relative fold change |
|---|---|---|
| 1 | IL-2 | 27.4 |
| 2 | IL-27 | 19.3 |
| 3 | IL-17 | 13.1 |
| 4 | MIP-1beta | 10.2 |
| 5 | IL-23 | 7.2 |
| 6 | IL-1beta | 6.4 |
| 7 | IL-1ra | 4.6 |
| 8 | MIP-2 | 4.2 |
| 9 | MCP-5 | 4.1 |
| 10 | TIMP-1 | 4.0 |
| 11 | TARC | 3.9 |
| 12 | IL-3 | 3.9 |
| 13 | TREM-1 | 3.8 |
| 14 | TNF-alpha | 3.4 |
| 15 | IP-10 | 2.9 |
| 16 | MIG | 2.9 |
| 17 | BLC | 2.7 |
| 18 | I-TAC | 2.5 |
| 19 | RANTES | 2.5 |
| 20 | IL-4 | 2.4 |
| 21 | IL-6 | 2.4 |
| 22 | IL-16 | 2.2 |
| 23 | IL-5 | 2.2 |
| 24 | IL-10 | 2.0 |
Discussion
RA initially occurs as non-specific inflammation in the joints; however, other organs are also affected in 15%-25% of individuals [29]. Following T cell activation, chronic inflammation occurs accompanied by tissue injury due to activation of the pro-inflammatory cytokines IL-1 and -6 and TNF-α. Our microarray results showed that Itga2 and Irf7 were upregulated, whereas cd4 was downregulated in CIA mice. A KEGG pathway analysis indicated that pathways related to the immune system were highly represented among the differentially expressed genes. Accordingly, IL-2, -27, -17, IL-1β, -6, and TNF-α levels were > 2-fold higher in the RA model relative to control mice, which was confirmed by qRT-PCR and western blotting.
Integrin α2 is a component of the very late-activation antigen 2 complex (integrin α2β1) [30] and binds to collagen via the I-domain [31, 32]. Integrin α2β1 is expressed only by effector Th1 and Th17 cells and attaches to collagen I/II-expressing cells of the synovial matrix [32, 33], resulting in the stimulation of T cell receptor-dependent IL-17 production [34]. IL-17 secreted by Th17 cells induces the production of pro-inflammatory cytokines such as IL-1 and -6 and TNF-α by macrophages, chondrocytes, and fibroblast-like synoviocytes, and was found to cause bone erosion via expression of receptor activator of nuclear factor-κB ligand (RANKL) in fibroblast-like synoviocytes and osteoclasts [35]. Blockade of integrin α2β1 reduced synovial inflammation, cartilage destruction, and bone loss in the joints of CIA mice [36]. According to the RNA microarray results and KEGG pathway analysis, integrin α2 is involved in phosphoinositide 3-kinase (PI3K)-Akt signaling, ECM receptor interaction, focal adhesion, and phagosome formation. PI3K-Akt signaling maintains basics cellular functions such as proliferation and differentiation [37], and inhibition of this pathway is a therapeutic strategy for RA treatment. Although the exact role of integrin α2 in the pathways identified by KEGG analysis is unclear, our results suggest that integrin α2 plays an important role in RA etiology, and is thus a candidate biomarker for RA diagnosis.
Validation of microarray and proteome array results by western blotting. (A) Western blotting was performed using blood samples from control and CIA mice. GAPDH served as an internal control. (B) Relative fold change in band intensity of target proteins normalized to GAPDH level. Data represent the mean ± SEM. *P < 0.05
Irf7 was another gene that was identified by microarray analysis as being upregulated in RA. IRF7 regulates the transcription of IFN-stimulated genes such as IFN-β, RANTES, and IFN-γ-inducible protein 10 that are expressed in the joints of RA patients [38-40]. Irf7 knockdown was found to decrease IFN-stimulated response element promoter activity [41], resulting in a decrease in the expression of genes associated with Th17 cell differentiation; however, this was accompanied by an increase in IL-17 secretion by Th17 cells. Further research is needed to resolve this discrepancy.
IL-1 and TNF-α are involved in joint inflammation and erosion in RA [42]. TNF-α-induced upregulation by TNF-α in synovial T cells was shown to increase RANKL expression and stimulate osteoclastogenesis in RA [43]. IL-27 is produced by antigen-presenting cells and regulates T cell differentiation and function [44]; it has pro- or anti-inflammatory functions depending on the disease stage [45, 46]. IL-27 levels were found to be higher in RA patients than in healthy individuals [47]. IL-27Ra-/- mice showed reduced severity of proteoglycan-induced arthritis [46], whereas injection of exogenous IL-27 improved RA symptoms in the CIA model [45]. Consistent with these earlier studies, we found here that IL-27 was upregulated in CIA as compared to control mice. In contrast, we observed that IL-2 expression was also increased in the RA model, although previous reports suggest that the IL-2 level is lower in rheumatoid synovial fluid, synovial tissue, and peripheral blood of RA patients than in those of control subjects [48, 49]. This discrepancy may be due to differences between species.
Our proteome array results showed that RANTES (or CCL5) was upregulated in CIA as compared to control mice. In contrast, the Ccl5 transcript (encoding RANTES) showed the opposite trend. RANTES is a chemotactic factor that recruits monocytes, memory T cells, and natural killer cells [50-52]. Others have reported higher RANTES levels in CIA mice relative to controls [53]. Therefore, additional research is necessary to clarify the exact role of RANTES in RA.
In summary, we found that integrin α2, IL-1β and -6, and TNF-α were upregulated in a mouse model of RA. In particular, integrin α2 was identified for the first time as a potential biomarker that can expedite RA diagnosis and be used to monitor disease progression.
Abbreviations
RA, rheumatoid arthritis; CIA, collagen-induced arthritis; RF, rheumatoid factor; TNF, tumor necrosis factor; IL, interleukin; STAT, signal transducer and activator of transcription; IFN, interferon; CD, cluster of differentiation; Itga2, integrin α2 gene; KEGG, Kyoto encyclopedia of genes and genomes; GAPDH, glyceraldehyde 3-phosphate dehydrogenase; ECM, extracellular matrix; CAMs, cell adhesion molecules; Th cell, helper T cell; MCP, monocyte chemoattractant protein; RANTES, regulated upon activation, normally T-expressed, and presumably secreted; integrin α2β1, very late-activation antigen 2 complex; RANKL, receptor activator of nuclear factor-κB ligand; PI3K, phosphoinositide 3-kinase.
Supplementary Material
Supplementary tables.
Acknowledgements
This research was supported by the Bio & Medical Technology Development Program of the National Research Foundation (NRF) & funded by the Korean government (MSIP&MOHW) (No. 2016M3A9B6904244).
Authors' Contributions
Sun-Yeong Gwon and Ho Joong Sung conceived and designed the experiments; Sun-Yeong Gwon performed the experiments; Sun-Yeong Gwon and Ho Joong Sung analyzed the data; Ho Joong Sung contributed reagents/materials/analysis tools; Sun-Yeong Gwon, Ki-Jong Rhee and Ho Joong Sung wrote the paper.
Competing Interests
The authors have declared that no competing interest exists.
References
1. Helmick CG, Felson DT, Lawrence RC, Gabriel S, Hirsch R, Kwoh CK. et al. Estimates of the prevalence of arthritis and other rheumatic conditions in the United States. Part I. Arthritis and rheumatism. 2008;58:15-25
2. Raychaudhuri S. Recent advances in the genetics of rheumatoid arthritis. Current opinion in rheumatology. 2010;22:109-18
3. Edwards CJ, Cooper C. Early environmental factors and rheumatoid arthritis. Clinical and experimental immunology. 2006;143:1-5
4. van Gaalen F, Ioan-Facsinay A, Huizinga TW, Toes RE. The devil in the details: the emerging role of anticitrulline autoimmunity in rheumatoid arthritis. J Immunol. 2005;175:5575-80
5. Valentino R, Savastano S, Tommaselli AP, Riccio A, Mariniello P, Pronesti G. et al. Hormonal pattern in women affected by rheumatoid arthritis. Journal of endocrinological investigation. 1993;16:619-24
6. Källberg H, Ding B, Padyukov L, Bengtsson C, Rönnelid J, Klareskog L. et al. Smoking is a major preventable risk factor for rheumatoid arthritis: estimations of risks after various exposures to cigarette smoke. Annals of the rheumatic diseases. 2011;70:508-11
7. Lundström E, Källberg H, Alfredsson L, Klareskog L, Padyukov L. Gene-environment interaction between the DRB1 shared epitope and smoking in the risk of anti-citrullinated protein antibody-positive rheumatoid arthritis: All alleles are important. Arthritis & Rheumatism. 2009;60:1597-603
8. Chung HY, Cesari M, Anton S, Marzetti E, Giovannini S, Seo AY. et al. Molecular Inflammation: Underpinnings of Aging and Age-related Diseases. Ageing research reviews. 2009;8:18-30
9. Tamil Iniyan G, Takbum O. MicroRNAs as Novel Biomarkers for the Diagnosis of Alzheimer`s Disease and Modern Advancements in the Treatment. Biomedical Science Letters. 2015;21:1-8
10. Park CE. Comparison of the Usefulness of Diagnostic Tests for Rheumatoid Arthritis. Korean Journal of Clinical Laboratory Science. 2015;47:168-74
11. Aletaha D, Neogi T, Silman AJ, Funovits J, Felson DT, Bingham CO. et al. 2010 Rheumatoid arthritis classification criteria: an American College of Rheumatology/European League Against Rheumatism collaborative initiative. Annals of the rheumatic diseases. 2010;69:1580-8
12. Asquith DL, Miller AM, McInnes IB, Liew FY. Animal models of rheumatoid arthritis. European journal of immunology. 2009;39:2040-4
13. Caplazi P, Baca M, Barck K, Carano RAD, DeVoss J, Lee WP. et al. Mouse Models of Rheumatoid Arthritis. Veterinary pathology. 2015
14. Williams RO. Collagen-induced arthritis in mice. Methods in molecular medicine. 2007;136:191-9
15. Holmdahl R, Rubin K, Klareskog L, Larsson E, Wigzell H. Characterization of the antibody response in mice with type II collagen-induced arthritis, using monoclonal anti-type II collagen antibodies. Arthritis and rheumatism. 1986;29:400-10
16. Brand DD, Latham KA, Rosloniec EF. Collagen-induced arthritis. Nat Protocols. 2007;2:1269-75
17. Keffer J, Probert L, Cazlaris H, Georgopoulos S, Kaslaris E, Kioussis D. et al. Transgenic mice expressing human tumour necrosis factor: a predictive genetic model of arthritis. The EMBO journal. 1991;10:4025-31
18. Berg WBvd. Lessons from animal models of arthritis over the past decade. Arthritis Research & Therapy. 2009;11:250 -
19. Niki Y, Yamada H, Seki S, Kikuchi T, Takaishi H, Toyama Y. et al. Macrophage- and neutrophil-dominant arthritis in human IL-1 alpha transgenic mice. The Journal of clinical investigation. 2001;107:1127-35
20. Plenge RM, Cotsapas C, Davies L, Price AL, de Bakker PI, Maller J. et al. Two independent alleles at 6q23 associated with risk of rheumatoid arthritis. Nature genetics. 2007;39:1477-82
21. Lee S-Y, Yu J-I, Chae S-C. Associations of IFITM3 haplotypes with rheumatoid arthritis in a Korean population. Genes & Genomics. 2012;34:493-8
22. Remmers EF, Plenge RM, Lee AT, Graham RR, Hom G, Behrens TW. et al. STAT4 and the risk of rheumatoid arthritis and systemic lupus erythematosus. The New England journal of medicine. 2007;357:977-86
23. Kaplan MH. STAT4: a critical regulator of inflammation in vivo. Immunol Res. 2005;31:231-42
24. Ibrahim SM, Koczan D, Thiesen HJ. Gene-expression profile of collagen-induced arthritis. Journal of autoimmunity. 2002;18:159-67
25. Zhang J-M, An J. Cytokines, Inflammation and Pain. International anesthesiology clinics. 2007;45:27-37
26. Fossiez F, Djossou O, Chomarat P, Flores-Romo L, Ait-Yahia S, Maat C. et al. T cell interleukin-17 induces stromal cells to produce proinflammatory and hematopoietic cytokines. The Journal of experimental medicine. 1996;183:2593-603
27. Adler EP, Lemken CA, Katchen NS, Kurt RA. A dual role for tumor-derived chemokine RANTES (CCL5). Immunology Letters. 2003;90:187-94
28. Schall TJ, Jongstra J, Dyer BJ, Jorgensen J, Clayberger C, Davis MM. et al. A human T cell-specific molecule is a member of a new gene family. J Immunol. 1988;141:1018-25
29. Turesson C, O'Fallon WM, Crowson CS, Gabriel SE, Matteson EL. Extra-articular disease manifestations in rheumatoid arthritis: incidence trends and risk factors over 46 years. Annals of the rheumatic diseases. 2003;62:722-7
30. Takada Y, Hemler ME. The primary structure of the VLA-2/collagen receptor alpha 2 subunit (platelet GPIa): homology to other integrins and the presence of a possible collagen-binding domain. The Journal of cell biology. 1989;109:397-407
31. Tuckwell D, Calderwood DA, Green LJ, Humphries MJ. Integrin alpha 2 I-domain is a binding site for collagens. Journal of cell science. 1995;108:1629-37
32. Heino J. The collagen receptor integrins have distinct ligand recognition and signaling functions. Matrix biology: journal of the International Society for Matrix Biology. 2000;19:319-23
33. Chan BM, Wong JG, Rao A, Hemler ME. T cell receptor-dependent, antigen-specific stimulation of a murine T cell clone induces a transient, VLA protein-mediated binding to extracellular matrix. J Immunol. 1991;147:398-404
34. Boisvert M, Chetoui N, Gendron S, Aoudjit F. Alpha2beta1 integrin is the major collagen-binding integrin expressed on human Th17 cells. European journal of immunology. 2010;40:2710-9
35. Komatsu N, Takayanagi H. Autoimmune arthritis: the interface between the immune system and joints. Advances in immunology. 2012;115:45-71
36. El Azreq M-A, Boisvert M, Cesaro A, Pagé N, Loubaki L, Allaeys I. et al. α2β1 Integrin Regulates Th17 Cell Activity and Its Neutralization Decreases the Severity of Collagen-Induced Arthritis. The Journal of Immunology. 2013;191:5941-50
37. Willems L, Tamburini J, Chapuis N, Lacombe C, Mayeux P, Bouscary D. PI3K and mTOR signaling pathways in cancer: new data on targeted therapies. Current oncology reports. 2012;14:129-38
38. Haringman JJ, Smeets TJ, Reinders-Blankert P, Tak PP. Chemokine and chemokine receptor expression in paired peripheral blood mononuclear cells and synovial tissue of patients with rheumatoid arthritis, osteoarthritis, and reactive arthritis. Annals of the rheumatic diseases. 2006;65:294-300
39. Garcia-Vicuna R, Gomez-Gaviro MV, Dominguez-Luis MJ, Pec MK, Gonzalez-Alvaro I, Alvaro-Gracia JM. et al. CC and CXC chemokine receptors mediate migration, proliferation, and matrix metalloproteinase production by fibroblast-like synoviocytes from rheumatoid arthritis patients. Arthritis and rheumatism. 2004;50:3866-77
40. van Holten J, Smeets TJ, Blankert P, Tak PP. Expression of interferon beta in synovial tissue from patients with rheumatoid arthritis: comparison with patients with osteoarthritis and reactive arthritis. Annals of the rheumatic diseases. 2005;64:1780-2
41. Sweeney SE, Kimbler TB, Firestein GS. Synoviocyte innate responses: II. Pivotal role of interferon regulatory factor 3. Journal of immunology (Baltimore, Md: 1950). 2010;184:7162-8
42. Mosmann TR, Coffman RL. Heterogeneity of cytokine secretion patterns and functions of helper T cells. Advances in immunology. 1989;46:111-47
43. Wei S, Kitaura H, Zhou P, Ross FP, Teitelbaum SL. IL-1 mediates TNF-induced osteoclastogenesis. The Journal of clinical investigation. 2005;115:282-90
44. Kastelein RA, Hunter CA, Cua DJ. Discovery and biology of IL-23 and IL-27: related but functionally distinct regulators of inflammation. Annu Rev Immunol. 2007:25
45. Niedbala W, Cai B, Wei X, Patakas A, Leung BP, McInnes IB. et al. Interleukin 27 attenuates collagen-induced arthritis. Annals of the rheumatic diseases. 2008;67:1474-9
46. Cao Y, Doodes PD, Glant TT, Finnegan A. IL-27 induces a Th1 immune response and susceptibility to experimental arthritis. J Immunol. 2008;180:922-30
47. Lai X, Wang H, Cao J, Li Y, Dai Y, Xiang Y. et al. Circulating IL-27 Is Elevated in Rheumatoid Arthritis Patients. Molecules (Basel, Switzerland). 2016:21
48. Combe B, Pope RM, Fischbach M, Darnell B, Baron S, Talal N. Interleukin-2 in rheumatoid arthritis: production of and response to interleukin-2 in rheumatoid synovial fluid, synovial tissue and peripheral blood. Clinical and experimental immunology. 1985;59:520-8
49. Kitas GD, Salmon M, Farr M, Gaston JS, Bacon PA. Deficient interleukin 2 production in rheumatoid arthritis: association with active disease and systemic complications. Clinical and experimental immunology. 1988;73:242-9
50. Wogensen L, Lee MS, Sarvetnick N. Production of interleukin 10 by islet cells accelerates immune-mediated destruction of beta cells in nonobese diabetic mice. The Journal of experimental medicine. 1994;179:1379-84
51. Grewal IS, Grewal KD, Wong FS, Picarella DE, Janeway CA Jr, Flavell RA. Local expression of transgene encoded TNF alpha in islets prevents autoimmune diabetes in nonobese diabetic (NOD) mice by preventing the development of auto-reactive islet-specific T cells. The Journal of experimental medicine. 1996;184:1963-74
52. Moritani M, Yoshimoto K, Wong SF, Tanaka C, Yamaoka T, Sano T. et al. Abrogation of autoimmune diabetes in nonobese diabetic mice and protection against effector lymphocytes by transgenic paracrine TGF-beta1. The Journal of clinical investigation. 1998;102:499-506
53. Thornton S, Duwel LE, Boivin GP, Ma Y, Hirsch R. Association of the course of collagen-induced arthritis with distinct patterns of cytokine and chemokine messenger RNA expression. Arthritis and rheumatism. 1999;42:1109-18
54. Meador BM, Krzyszton CP, Johnson RW, Huey KA. Effects of IL-10 and age on IL-6, IL-1β, and TNF-α responses in mouse skeletal and cardiac muscle to an acute inflammatory insult. Journal of Applied Physiology. 2008;104:991-7
55. Yamakawa I, Kojima H, Terashima T, Katagi M, Oi J, Urabe H. et al. Inactivation of TNF-α ameliorates diabetic neuropathy in mice. American Journal of Physiology - Endocrinology and Metabolism. 2011;301:E844-E52
56. Jafri M, Donnelly B, Allen S, Bondoc A, McNeal M, Rennert PD. et al. Cholangiocyte expression of α<sub>2</sub>β<sub>1</sub>-integrin confers susceptibility to rotavirus-induced experimental biliary atresia. American Journal of Physiology - Gastrointestinal and Liver Physiology. 2008;295:G16-G26
57. Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001;25:402-8
Author contact
![]() Corresponding author: Tel.: +82-31-740-7108; Fax: +82-31-740-7425; E-mail: hjsungac.kr
Corresponding author: Tel.: +82-31-740-7108; Fax: +82-31-740-7425; E-mail: hjsungac.kr

 Global reach, higher impact
Global reach, higher impact