Impact Factor
ISSN: 1449-1907
Int J Med Sci 2017; 14(12):1292-1300. doi:10.7150/ijms.21510 This issue Cite
Research Paper
IER5 is involved in DNA Double-Strand Breaks Repair in Association with PAPR1 in Hela Cells
1. Department of Gynecologic Oncology, Beijing Obstetrics and Gynecology Hospital, Capital Medical University, Beijing, 100006, China;
2. National Institute for Radiological Protection, Chinese Center for Disease Control and Prevention, Beijing ,100088, China;
3. Department of Radiation Toxicology and Oncology, Beijing Key Laboratory for Radiobiology, Beijing Institute of Radiation Medicine, Beijing, 100850, China.
Received 2017-6-17; Accepted 2017-9-1; Published 2017-9-30
Abstract
The immediate early response gene 5 (IER5) is a radiation response gene induced in a dose-independent manner, and has been suggested to be a molecular biomarker for biodosimetry purposes upon radiation exposure. Here, we investigated the function of IER5 in DNA damage response and repair. We found that interference on IER5 expression significantly decreased the efficiency of repair of DNA double-strand breaks induced by ionizing radiations in Hela cells. We found that IER5 participates in the non-homologous end-joining pathway of DNA breaks repair. Additionally, we identified a number of potential IER5-interacting proteins through mass spectrometry-based protein assays. The interaction of IER5 protein with poly(ADP-Ribose) polymerase 1 (PARP1) and Ku70 was further confirmed by immunoprecipitation assays. We also found that Olaparib, a PARP1 inhibitor, affected the stability of IER5. These results indicate that targeting of IER5 may be a novel DNA damage response-related strategy to use during cervical cancer radiotherapy or chemotherapy.
Keywords: cervical cancer, IER5, DNA double strand break repair, ionizing radiation, PARP1, Olaparib.
Introduction
Cervical cancer is a leading threat to women's health, especially in developing countries [1]. The treatment for cervical cancer includes surgery, radiation, and chemotherapy. Concurrent chemoradiotherapy has been regarded as the standard treatment of locally advanced cervical cancer [2-5]. Because of the widespread uptake of screening for the early detection of cervical cancer, its death rate has declined [1, 6]. However, no significant improvements have been achieved in cervical cancer survival rate over the past three decades [7]. Therefore, it is necessary to discover new and effective strategies for a favorable therapeutic outcome. Specific molecules involved in radiosensitivity have been the focus of many studies in recent years [8]. One of such molecules, immediate early response gene 5 (IER5), was found to be induced by ionizing radiation (IR) in studies using cDNA microarray technology [9, 10].
IER5 belongs to the immediate-early gene family, and encodes a protein of 327 amino acids, mostly located in the cytoplasm [11]. IER5 can be induced by growth-promoting and oncogenic signals, IR and heat shock. IER5 plays a role in cell proliferation, and it inhibits cell proliferation through the reduction of Cdc25B expression, resulting in G2/M cell cycle arrest [11, 12]. Additionally, IER5 interacts with protein phosphatase 2A (PP2A) and is involved in the generation of a hypophosphorylated active form of heat shock factor 1 (HSF1), contributing to proliferation and survival of cancer cells [13-15].
Previous studies have shown that the expression of IER5 in response to radiation response is dose-independent [16]. IER5 is involved in DNA damage repair. Therefore, it has been suggested that IER5 can be used as molecular biomarker for biodosimetry purposes [16, 17]. However, whether IER5 functions in cervical cancer cellular response to radiation is still unclear.
In the present study, we report that IER5 is involved in the DNA repair process. Specifically, IER5 participates in non-homologous end joining (NHEJ)-mediated DNA double strand breaks (DSBs) repair. Additionally, we used immunoprecipitation assays and mass spectrometry (MS) to identify the proteins that interact with IER5. Our study has established the foundation to further study the role of IER5 in cervical cancer, and hypothesized its use in cancer therapy.
Materials and methods
Patient samples and immunohistochemistry
Human cervical tissue samples were obtained from Beijing Biobank of Clinical Resources. The collection of human tissue samples was approved and supervised by the Ethics Committee of Beijing Obstetrics and Gynecology Hospital. Thirty-one cervical cancer tissues and 20 normal cervical tissues were obtained.
Immunohistochemistry was carried out using the Ultra-Sensitive S-P kit (Boster Bioscience [Hubei Province, China] Co. LTD), following the manufacturer's instructions. The staining of IER5 was scored considering both the percentage of positive cells and intensity of the staining, as described previously [18]. The number of positive cells was counted in at least ten fields for each section and analyzed with the Image Pro Plus software.
Cell culture and treatment
Hela cells were cultured in Dulbecco's modified Eagle's medium (Hyclone), supplemented with 10 % fetal calf serum (FBS, Sigma), and 100 U/ml penicillin and 100 mg/ml streptomycin (Hyclone). Cells were incubated at 37 ºC, in a 5 % CO2 incubator.
Cells were irradiated with 60Co γ-rays at a dose rate of 1.98 Gy/min at room temperature. Cells were treated with 10 μM Olaparib (Selleck), MG132 (Merck), or cycloheximide (CHX, Sigma), as indicated; stock solution were prepared in dimethyl sulfoxide (DMSO ).
Reverse transcription and quantitative polymerase chain reaction
Total RNA was extracted from cells using TRIzol (Invitrogen). Reverse transcription (RT-PCR) was carried out using the ReverTra Ace qPCR RT Master Mix with gDNA Remover kit (TOYOBO), following the manufacturer's instructions. Quantitative PCR (qPCR) was performed with a Bio-Rad iCycler&iQ Real-time PCR system (Bio-Rad) and a fluorescence-labeled SYBR Green real master Mix kit (TIANGEN Biotech [Beijing] Co. LTD). The mRNA primers were as follows: IER5 (forward: 5'CCGGGAACGTGGCTAACC3'; reverse: 5'TTCCGTAGGAGTCCCGAGAA3'); β-actin (forward: 5'GCGCGGCTACAGCTTCA3'; reverse: 5'CTTAATGTCACGCACGATTTCC3').
Plasmids, RNA interference and cell transfection
The cDNA of IER5 was amplified by RT-PCR from total RNA extracted from Hela cells and cloned into the pCMV plasmid in frame with the sequence encoding 3×FLAG epitopes (p3×FLAG-IER5-CMV). To specifically knock down gene expression in Hela cells, the following small interfering RNAs (siRNAs) were used: native control siRNA (siNC) (UUCUCCGAACGUGUCACGUTT), siIER5 (CCUCAUCAGCAUCUUCGGUUU), si53BP1 (GAAGGACGGAGUACUAAUATT), siRAD51 (GAAGCUAUGUUCGCCAUUATT). The pDR-GFP, pEGFP-Pem1-Ad2 and pCherry plasmids and I-SceI had been previously obtained by our laboratory.
Hela cells were seeded into 6-well plates or 100 mm culture plates and cultured in medium without antibiotics; siRNAs or plasmids were transfected into cells using Lipofectamine 2000 (Invitrogen), following the manufacturer's recommendations. Cells were harvested 36/48 h after transfection and subjected to the specified assays.
Western blot analysis
Cells were lysed in lysis buffer (Thermo); equal amounts of protein were separated by SDS-polyacrylamide gel electrophoresis (SDS-PAGE) and then transferred onto polyvinylidene fluoride membranes. The membranes were blocked with 5 % milk in TBST (20 mM Tris-HCl, 500 mM NaCl [pH7.5], 0.1 % [v/v] Tween 20) for 1 h at room temperature, and incubated with the primary antibodies overnight at 4 °C. Membranes were then incubated with the appropriate secondary antibody for 1 h at room temperature, and washed with TBST. Bands were visualized using the Image quant LAS500 system (GE).
Antibodies used in this study: anti-IER5 goat polyclonal antibody (Abcam), anti-IER5 rabbit polyclonal antibody (Santa Cruz), anti-GAPDH mouse monoclonal antibody (Zhong Shan Jin Qiao), anti-PARP1 rabbit monoclonal antibody (Santa Cruz), anti-Ku70 mouse monoclonal antibody (Abcam), anti-Ku80 rabbit monoclonal antibody (Santa Cruz), anti-FLAG M2 mouse monoclonal antibody (Sigma), anti-53BP1 rabbit polyclonal antibody (Santa Cruz), anti-RAD51 rabbit polyclonal antibody (Proteintech), anti-γH2AX mouse monoclonal antibody (Millipore), anti-Ubiquitinylated proteins mouse monoclonal antibody (Millipore), and anti-PADPR mouse monoclonal antibody (Abcam).
Neutral comet assay
Hela cells were collected at 0 h (mock treated), 10 min, 1 h, 4 h and 6 h after treatment with IR (4 Gy). Neutral Comet assays were performed using the Single Cell Gel Electrophoresis Assay kit (Trevigen), according to the manufacturer's instructions. Comet tail moments (TM) of 50 cells in each experiment were measured under microscope and analyzed with the CASPLab (version1.2.3) software. The results from three independent experiments were analyzed by ANOVA.
Immunoprecipitation
Hela Cells were transfected with the p3×FLAG-IER5-CMV plasmid; 36 h later, the cells were irradiated with IR (4Gy). Cells were lysed with lysis buffer (20 mM Tris-HCl [pH8.0]; 100 mM NaCl; 0.5 % Nonidet P 40; 1 mM EDTA; protease inhibitor cocktail [Roche]) and centrifuged at 12000 g for 10 min at 4 °C. Cells lysates were precleared using protein A/G agarose beads (Santa Cruz) and incubated with anti-FLAG M2 affinity gel (Sigma) for 2 h at 4 °C. The gels with bound immuno complexes were then washed with lysis buffer for three times and subjected to SDS-PAGE, Coomassie Brilliant Blue staining and western blot analysis using specific antibodies.
NHEJ and homologous recombination (HR) assays
Hela cells were transfected first with siRNAs and, after 48 h, alternatively with the pEGFP-Pem1-Ad2 (a NHEJ reporter) linearized with the restriction enzymes HindIII or I-SceI or the pDR-GPF (a HR reporter) plasmids. The pCherry plasmid was used as a control for transfection efficiency. After 12/72 h, cells were collected for fluorescence-activated cell sorter (FACS) analyses of the GFP+/pCherry+ cells, which indicated the efficiency of NHEJ or HR.
Statistical analysis
The SPSS version 18.0 software was used for all statistical analysis. The data were calculated and shown as mean ± SEM of at least three experiments. The one-way ANOVA, χ2 test and student's t-test were used to assess the statistical significance of differences. The Database for Annotation, Visualization and Integrated Discovery (DAVID) online database was used to analyze the MS report. Statistical significance was defined as p < 0.05.
Results
The expression of IER5 was higher in tumor tissues than in normal cervical tissues
We analyzed the expression of IER5 in 31 cervical cancer tissues samples and 20 normal cervical tissue samples. The expression of IER5 was largely localized in the cytoplasm and significantly increased in cancer samples (Fig. 1). Specifically, 93.5 % (29 of 31) of the cancer samples showed IER5 expression, whereas 35 % of normal cervices (7 of 20) were IER5-positive (Table 1).
Immunohistochemistry analysis of IER5 expression in cervical cancer and normal samples. × 200 magnification: the samples of cervical cancer show strong expression of IER5.
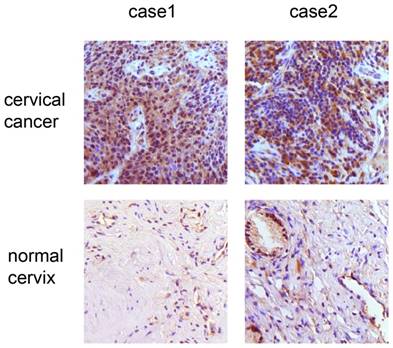
Immunohistochemical analysis of IER5 in cervical cancers and normal cervical samples
| Immunohistochemical staining | Tumor | Normal |
|---|---|---|
| Positive | 29 | 7 |
| Negative | 2 | 13 |
| Total | 31 | 20 |
| χ2 test | P <0.05 | |
Suppression of IER5 was associated with a DSB repair defect
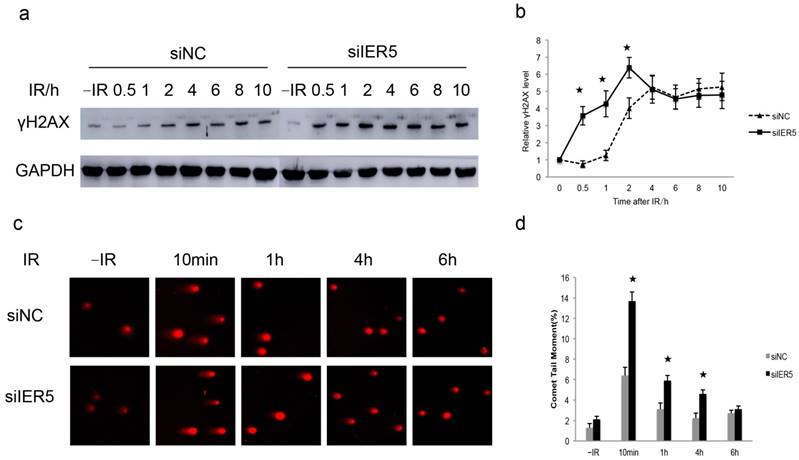
We previously reported that IER5 expression is induced in response to DSBs [19, 20]. Additionally, IER5 has been indicated as a potential biomarker to radiation exposure [16]. To address the role of IER5 in DSBs, we compared the DNA damage response in Hela cells untreated (siNC) or treated with siIER5 (to knockdown IER5 expression), upon IR exposure (4 Gy). We analyzed the phosphorylation of H2AX at Ser 139 (γH2AX), a marker of DSBs [21], in the cells at different time points after IR treatment. We found that the amount of γH2AX in IER5-depleted cells increased dramatically at 0.5 h after IR exposure compared to the control cells, and persisted up to 10 h after irradiation (Fig. 2a and b).
Next, we performed neutral comet assays to further demonstrate the suppression of IER5 was associated with a DSBs repair defect. It is known that, in a neutral comet assay, TM values are positively correlated with the degree of DNA damage [22]. The majority of cells appeared damaged 10 min after IR, showing large comet tails, while 6 h after IR, the damage was repaired in most cells and comet tails returned to background levels. In IER5-depleted cells, the TM values were significantly higher than in control cells 10 min, 1 h and 4 h after IR (Fig. 2c and d). Taken together, these results indicated that the suppression of IER5 renders the cells more sensitive to DNA damage.
IER5 regulates NHEJ-mediated DSBs repair
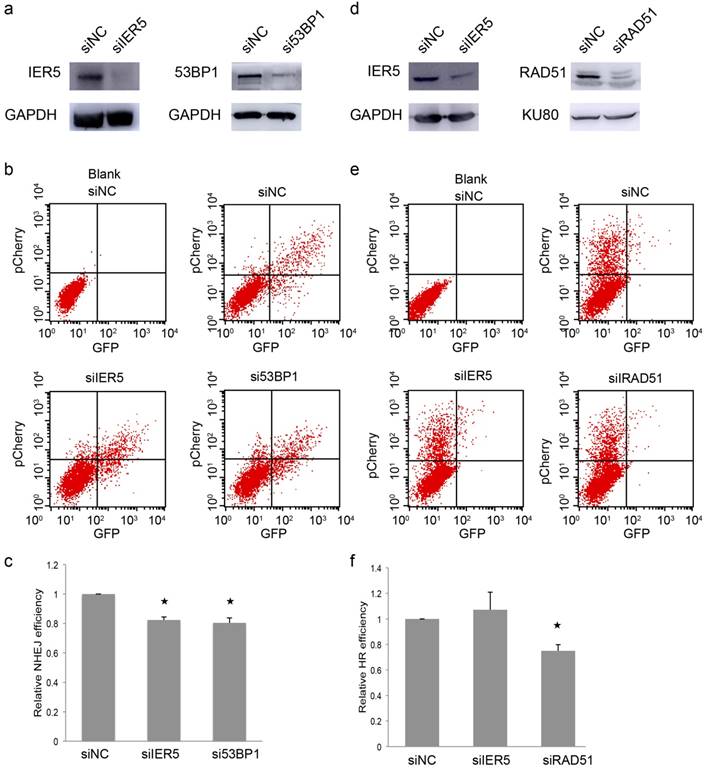
Because IER5 is a DNA damage-related protein, we decided to investigate which specific DNA repair process IER5 is involved in. NHEJ and HR are two key pathways in the repair of DSBs [23]. We first looked at the NHEJ repair pathway, by performing an NHEJ assay in which the rejoining of a previously linearized plasmid (cut between the promoter and the GFP coding sequence), allows the expression of GFP [24]. We depleted IER5 or 53BP1, a key factor in NHEJ [25], in Hela cells, through RNA interference (Fig.3a), and transfected the linearized pEGFP-Pem1-Ad2 plasmid along with the pCherry expression plasmid to monitor the transfection efficiency. The NHEJ efficiency was indicated by the percentage of GFP+/Cherry+ cells in FACS analyses. We found that the depletion of IER5, similarly to that of 53BP1, was associated with a lower efficiency of NHEJ (Fig. 3b and c).
Next, we assessed whether IER5 also plays a role in HR. The pDR-GFP plasmid contains two non-functional GFP genes and an I-SceI cleavage site in the first GFP cassette. When I-SceI produces a DSB in the GFP gene, its sequence can be repaired by HR, forming a functional GFP gene [26]. The pCherry expression plasmid was co-transfected with the HR reporter to monitor the transfection efficiency. We depleted IER5 or RAD51, a key factor in HR [27], in Hela cells using siRNAs (Fig. 3d) and measured the percentage of GFP+/Cherry+ cells in FACS analyses. Depletion of IER5 did not affect the percentage of GFP+/Cherry+ cells (Fig. 3e and f) and therefore the efficiency of HR. These results suggested that IER5 plays a role in NHEJ-mediated DSB repair, but is not involved in DSB repair through HR.
IER5-depleted cells show a DSB repair defect. a. IER5-depleted Hela cells were treated with 4 Gy of IR, and γH2AX levels were detected by western blot at different time points after irradiation. b. Quantification of γH2AX levels based on densitometric scanning of the immunoblotting signals of γH2AX shown in a. c. Detection of DSBs by a neutral comet assay in siNC- and siIER5-treated Hela cells subjected to IR (4Gy). d. Quantification of comet tail moment (averages ± SD) with the CASPLab software. Fifty cells per group were examined.
IER5 regulates NHEJ-mediated DSB repair. a. Western blot indicating IER5 and 53BP1 knockdown by specific siRNAs. b. FACS analyses of NHEJ assay in Hela cells treated with IER5 and 53BP1 siRNAs. c. Quantification of the NHEJ assay. d. Western blot indicating IER5 and RAD51 knockdown in Hela cells upon treatment with specific siRNAs. e. Efficiency of HR, as analyzed by FACS. f. Quantification of the HR assay.
IER5 interacts with poly(ADP-ribose) polymerase 1 (PARP1) and Ku70
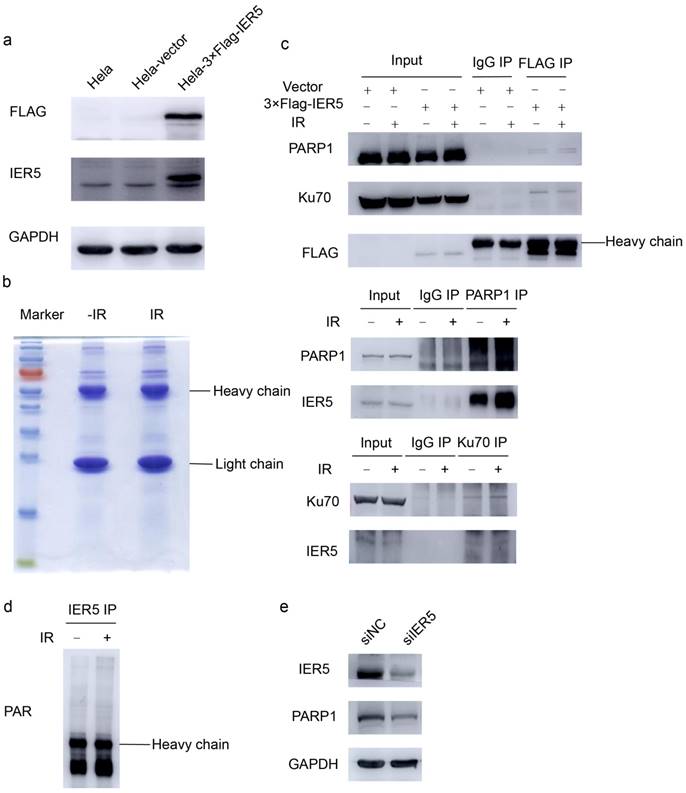
To further investigate how IER5 mediates DSB repair, we constructed the p3×FLAG-IER5-CMV plasmid to perform immunoprecipitation experiments. The expression of FLAG tagged-IER5 was verified by western blot (Fig. 4a). Next, Hela cells were transfected with the p3×FLAG-IER5-CMV plasmid and immunoprecipitation assays were performed to identify potential IER5-binding partners. The immunoprecipitates were separated by 12 % SDS-PAGE and Coomassie Brilliant Blue staining was used to visualize the proteins on the gels as bands (Fig. 4b). Chosen bands were cut from the gel and analyzed by MS. A total of 347 and 256 proteins were identified in irradiated and non-irradiated samples, respectively; these proteins were further analyzed using the DAVID online database. Ten proteins were associated with DNA metabolic processes and DNA repair (Table 2). Co-immunoprecipitation assays demonstrated that PARP1 and Ku70 interacted with IER5 in Hela cells (Fig. 4c). In western blot assays, we examined the PARylation status of IER5 under IR stress, and found it is not affected by DNA damage cells (Fig. 4d). Furthermore, PARP1 levels decreased in IER5-deficient cells compared control cells (Fig. 4e).
IER5 interacts with PARP1 and Ku70. a. the expression of the 3×FLAG tagged-IER5 fusion protein was verified by western blot. b. To identify IER5 interacting proteins, Hela cells stably expressing FLAG-tagged IER5 were irradiated with IR (4Gy). Immunoprecipitates were isolated from cell lysates using the anti-FLAG M2 affinity gel, separated on a 12% SDS-PAGE gel and subsequently subjected to Coomassie Brilliant Blue staining. c. The poly(ADP-ribose) polymerase (PARP1) protein and Ku70 were identified as IER5-interacting proteins by western blot. d. Hela cells were exposed to IR (4Gy) or mock-treated, and immunoprecipitation assays were performed using an anti- IER5 antibody. The immunoprecipitates were blotted and probed with a PAR antibody. e. Decreased level of PARP1 protein in IER5-depleted Hela cells.
MS and DAVID online database analysis of IER5 interacting protein
| protein ID | protein name | score | matches |
|---|---|---|---|
| gi|7582386 | Bcl-2-associated transcription factor short form | 435 | 17 |
| gi|1806048 | nuclear DNA helicase II | 433 | 18 |
| gi|178152 | poly(ADP-ribose) polymerase | 103 | 3 |
| gi|460771 | hnRNP-E1 | 84 | 3 |
| gi|4503841 | X-ray repair cross-complementing protein 6 | 78 | 3 |
| gi|31645 | glyceraldehyde-3-phosphate dehydrogenase | 68 | 3 |
| gi|35053 | uracil DNA glycosylase | 63 | 3 |
| gi|1575536 | regulator of nonsense transcript stability | 51 | 2 |
| gi|5881961 | Dhm1-like protein | 48 | 2 |
| gi|386772 | histone H3 | 36 | 2 |
Olaparib decreased the stability of IER5
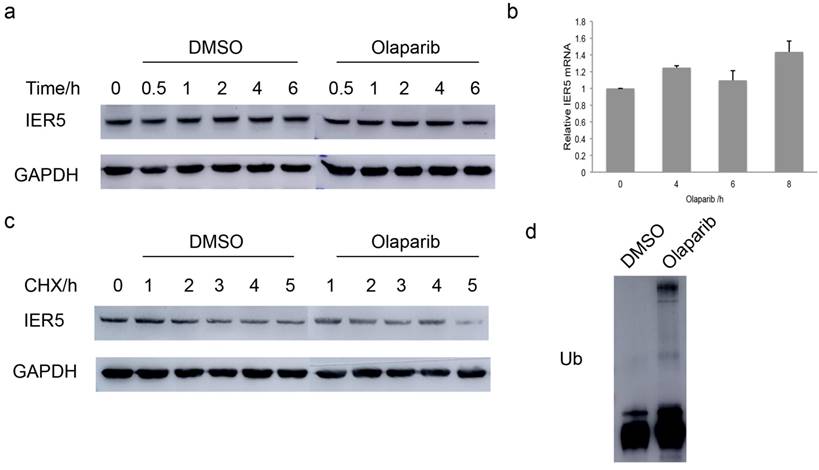
PARP1 is thought to act as a key regulator of DNA repair [28] and its inhibitor, Olaparib, has been approved for clinical cancer treatment [29]. Considering the above findings, we wondered whether the catalytic activity of PARP1 would correlate with the expression of IER5 protein in Hela cells. Western blot assays were performed to detect the amount of IER5 at different time points after treatment of the cells with Olaparib (10 μM). Olaparib was associated with the decrease in the levels of IER5 6 h after the treatment (Fig. 5a). The mRNA level of IER5 was determined by qPCR in Olaparib-treated and control cells. Surprisingly, the mRNA level of IER5 was increased in Olaparib-treated cells (Fig. 5b), suggesting that the down regulation of IER5 in these cells may be the result of increased protein degradation.
To prove this hypothesis, Hela cells were treated with Olaparib and the protein synthesis inhibitor CHX and harvested at different times after treatment. The degradation of IER5 was significantly faster in the presence of Olaparib (Fig. 5c), indicating that PARP1 activity is required for IER5 stability.
Finally, we examined whether Olaparib regulates IER5 protein stability via proteasome-mediated degradation. Hela cells were pretreated with DMSO or 10 μM Olaparib for 2 h and further incubated with the proteasome inhibitor MG132 (10 μM) for 6 h. The ubiquitination of IER5 increased in Olaparib-treated cells (Fig. 5d), suggesting that Olaparib promotes the degradation of IER5 through the ubiquitin-proteasome system.
Discussion
Previous studies have suggested that higher IER5 expression is associated with poor prognosis in various cancer patients [13]. Ahn et al. reported that IER5 is upregulated (89.3 folds) in cervical cancer specimens compared to normal ones [30]. However, the relationship between IER5 expression and function has not been fully understood, so far.
In this study, we explored the mechanisms through which IER5 is involved in DNA repair in Hela cells. After IR, DNA damage was more severe in IER5-depleted cells than in control cells. Additionally, PARP1 and Ku70, important factors in DNA repair, were identified as IER5-interacting proteins.
PARP1 is activated by DNA strand breaks and polyADP ribosylates various acceptor molecules to facilitate DNA repair: the posttranslational modification (PTM) of proteins through PARylation is one of the immediate events in DNA damage [28, 31]. DNA damage-induced PTMs, such as phosphorylation, ubiquitylation, PARylation, SUMOylation, neddylation, methylation and acetylation, play a central role in DNA damage response [32]. Interestingly, it has been reported that IER5 can change the PTMs of HSF1 under heat shock condition: specifically, IER5, forms a ternary complex with HSF1 and protein phosphatase 2 (PP2A), and dephosphorylates HSF1 at numbers of serine and threonine residues [11, 13, 14]. Our work indicates that IER5 is a possible PARP1 substrate and that the levels of PARP1 decrease in IER5-depleted Hela cells.
Olaparib decreases the stability of IER5. a. The treatment of cells with the PARP1 inhibitor Olaparib (10 μM) decreased IER5 expression. b. IER5 mRNA increased in cells treated with 10 μM Olaparib for the indicated times. c. IER5 protein stability decreased after Olaparib treatment. d. The ubiquitinylation of IER5 protein increased after Olaparib treatment.
The interactions between IER5 and Ku70 or PARP1 are valuable for further studies in the NHEJ-mediated DSB repair. Ku70, a DNA binding protein, dimerizes with Ku80, and Ku70/Ku80 complex recognizes DNA DSBs and recruits the DNA-dependent protein kinase catalytic subunit (DNA-PKcs); the Ku70/Ku80/DNA-PKcs complex mediates NHEJ and repairs DSBs [23, 33]. In addition, PARP1 also interacts with DSBs repair proteins and participates in the NHEJ repair pathway [34].
Since PARP1 inhibitors sensitize tumors to radiation and chemotherapeutic agents that induce DNA DSBs, they have been used cancer treatment [34]. Olaparib has undergone the most comprehensive evaluation in this regard: notably, it is the first inhibitor used as single antitumoral agent in the treatment of tumors associated with BRCA1 or BRCA2 mutations [29, 35] and has been approved for the clinical treatment of ovarian and breast cancer in 2014. Some studies have shown improved overall survival associated with the use of Olaparib in ovarian cancer treatment [36]. In our study, we found that the inhibition of PARP1 by Olaparib accelerates IER5 degradation in Hela cells. Our findings suggest that the interaction between IER5 and PARP1 might be important for cervical cancer. Though the mechanisms are not fully understood yet, it provides insight for possible novel therapies to treat cervical cancer.
Our observations suggest that IER5 is a novel regulator of the NHEJ pathway for DNA DSB repair, possibly through its interaction with PARP1 and Ku70. However, further studies are necessary to fully uncover mechanism of IER5 function.
Acknowledgements
This work was supported by grants from the National Natural Science Foundation of China (Grant No. 81272888), Beijing Municipal Administration of Hospitals Clinical Medicine Development of Special Funding Support (Grant No. ZYLX201705) and Beijing Municipal Science and Technology Commission (D131100005313009).
Competing Interests
The authors have declared that no competing interest exists.
References
1. Tsikouras P, Zervoudis S, Manav B, Tomara E, Iatrakis G, Romanidis C, Bothou A, Galazios G. Cervical cancer: screening, diagnosis and staging. J BUON. 2016;21:320-5
2. Keys HM, Bundy BN, Stehman FB, Muderspach LI, Chafe WE, Suggs CL 3rd, Walker JL, Gersell D. Cisplatin, radiation, and adjuvant hysterectomy compared with radiation and adjuvant hysterectomy for bulky stage IB cervical carcinoma. N Engl J Med. 1999;340:1154-61
3. Morris M, Eifel PJ, Lu J, Grigsby PW, Levenback C, Stevens RE, Rotman M, Gershenson DM, Mutch DG. Pelvic radiation with concurrent chemotherapy compared with pelvic and para-aortic radiation for high-risk cervical cancer. N Engl J Med. 1999;340:1137-43
4. Rose PG, Bundy BN, Watkins EB, Thigpen JT, Deppe G, Maiman MA, Clarke-Pearson DL, Insalaco S. Concurrent cisplatin-based radiotherapy and chemotherapy for locally advanced cervical cancer. N Engl J Med. 1999;340:1144-53
5. Whitney CW, Sause W, Bundy BN, Malfetano JH, Hannigan EV, Fowler WC Jr, Clarke-Pearson DL, Liao SY. Randomized comparison of fluorouracil plus cisplatin versus hydroxyurea as an adjunct to radiation therapy in stage IIB-IVA carcinoma of the cervix with negative para-aortic lymph nodes: a Gynecologic Oncology Group and Southwest Oncology Group study. J Clin Oncol. 1999;17:1339-48
6. Pimple S, Mishra G, Shastri S. Global strategies for cervical cancer prevention. Curr Opin Obstet Gynecol. 2016;28:4-10
7. Siegel R, Ma J, Zou Z, Jemal A. Cancer statistics, 2014. CA Cancer J Clin. 2014;64:9-29
8. Qin C, Chen X, Bai Q, Davis MR, Fang Y. Factors associated with radiosensitivity of cervical cancer. Anticancer Res. 2014;34:4649-56
9. Long XH, Zhao ZQ, He XP, Wang HP, Xu QZ, An J, Bai B, Sui JL, Zhou PK. Dose-dependent expression changes of early response genes to ionizing radiation in human lymphoblastoid cells. Int J Mol Med. 2007;19:607-15
10. Kis E, Szatmari T, Keszei M, Farkas R, Esik O, Lumniczky K, Falus A, Safrany G. Microarray analysis of radiation response genes in primary human fibroblasts. Int J Radiat Oncol Biol Phys. 2006;66:1506-14
11. Ishikawa Y, Kawabata S, Sakurai H. HSF1 transcriptional activity is modulated by IER5 and PP2A/B55. FEBS Lett. 2015;589:1150-5
12. Nakamura S, Nagata Y, Tan L, Takemura T, Shibata K, Fujie M, Fujisawa S, Tanaka Y, Toda M, Makita R, Tsunekawa K, Yamada M, Yamaoka M, Yamashita J, Ohnishi K, Yamashita M. Transcriptional repression of Cdc25B by IER5 inhibits the proliferation of leukemic progenitor cells through NF-YB and p300 in acute myeloid leukemia. PLoS One. 2011;6:e28011
13. Asano Y, Kawase T, Okabe A, Tsutsumi S, Ichikawa H, Tatebe S, Kitabayashi I, Tashiro F, Namiki H, Kondo T, Semba K, Aburatani H, Taya Y, Nakagama H, Ohki R. IER5 generates a novel hypo-phosphorylated active form of HSF1 and contributes to tumorigenesis. Sci Rep. 2016;6:19174
14. Kawabata S, Ishita Y, Ishikawa Y, Sakurai H. Immediate-early response 5 (IER5) interacts with protein phosphatase 2A and regulates the phosphorylation of ribosomal protein S6 kinase and heat shock factor 1. FEBS Lett. 2015;589:3679-85
15. Ishikawa Y, Sakurai H. Heat-induced expression of the immediate-early gene IER5 and its involvement in the proliferation of heat-shocked cells. FEBS J. 2015;282:332-40
16. Tavakoli H, Manoochehri M, Modarres Mosalla SM, Ghafori M, Karimi AA. Dose-dependent and gender-related radiation-induced transcription alterations of Gadd45a and Ier5 inhuman lymphocytes exposed to gamma ray emitted by (60)Co. Radiat Prot Dosimetry. 2013;154:37-44
17. Li MJ, Wang WW, Chen SW, Shen Q, Min R. Radiation dose effect of DNA repair-related gene expression in mouse white blood cells. Med Sci Monit. 2011;17:BR290-7
18. Bachtiary B, Schindl M, Pötter R, Dreier B, Knocke TH, Hainfellner JA, Horvat R, Birner P. Overexpression of hypoxia-inducible factor 1alpha indicates diminished response to radiotherapy and unfavorable prognosis in patients receiving radical radiotherapy for cervical cancer. Clin Cancer Res. 2003;9:2234-40
19. Ding KK, Shang ZF, Hao C, Xu QZ, Shen JJ, Yang CJ, Xie YH, Qiao C, Wang Y, Xu LL, Zhou PK. Induced expression of the IER5 gene by gamma-ray irradiation and its involvement in cell cycle checkpoint control and survival. Radiat Environ Biophys. 2009;48:205-13
20. Li Xiu-Nan, Ji Chen, Zhou Ping-Kun, Wu Yu-Mei. Establishments of IER5 silence and overexpression cervical cancer SiHa cell lines and analysis of radiosensitivity. Int J Clin Exp Pathol. 2016;9:6672-82
21. Chowdhury D, Keogh MC, Ishii H, Peterson CL, Buratowski S, Lieberman J. gamma-H2AX dephosphorylation by protein phosphatase 2A facilitates DNA double-strand break repair. Mol Cell. 2005;20:801-9
22. Speit G, Rothfuss A. The comet assay: a sensitive genotoxicity test for the detection of DNA damage and repair. Methods Mol Biol. 2012;920:79-90
23. Shrivastav M, De Haro LP, Nickoloff JA. Regulation of DNA double-strand break repair pathway choice. Cell Res. 2008;18:134-47
24. Lin H, Ha K, Lu G, Fang X, Cheng R, Zuo Q, Zhang P. Cdc14A and Cdc14B Redundantly Regulate DNA Double-Strand Break Repair. Mol Cell Biol. 2015;35:3657-68
25. Gupta A, Hunt CR, Chakraborty S, Pandita RK, Yordy J, Ramnarain DB, Horikoshi N, Pandita TK. Role of 53BP1 in the Regulation of DNA Double-Strand Break Repair Pathway Choice. Radiation Research. 2014;181:1-8
26. Mamouni K, Cristini A, Guirouilh-Barbat J, Monferran S, Lemarie A, Faye JC, Lopez BS, Favre G, Sordet O. RhoB promotes gammaH2AX dephosphorylation and DNA double-strand break repair. Mol Cell Biol. 2014;34:3144-55
27. Baumann P, West SC. Role of the human RAD51 protein in homologous recombination and double-stranded-break repair. Trends Biochem Sci. 1998;23(7):247-51
28. Khodyreva SN, Lavrik OI. Poly(ADP-Ribose) polymerase 1 as a key regulator of DNA repair. Molecular Biology. 2016;50:580-595
29. Drew Y. The development of PARP inhibitors in ovarian cancer: from bench to bedside. Br J Cancer. 2015;113(Suppl 1):S3-9
30. Ahn WS, Bae SM, Lee JM, Namkoong SE, Han SJ, Cho YL, Nam GH, Seo JS, Kim CK, Kim YW. Searching for pathogenic gene functions to cervical cancer. Gynecol Oncol. 2004;93:41-8
31. Wei H, Yu X. Functions of PARylation in DNA Damage Repair Pathways. Genomics Proteomics Bioinformatics. 2016;14:131-9
32. Dantuma NP, van Attikum H. Spatiotemporal regulation of posttranslational modifications in the DNA damage response. Embo j. 2016;35:6-23
33. Hu H, Gatti RA. MicroRNAs: new players in the DNA damage response. J Mol Cell Biol. 2011;3:151-8
34. Li N, Chen J. ADP-ribosylation: activation, recognition, and removal. MolCells. 2014;37:9-16
35. Cerrato A, Morra F, Celetti A. Use of poly ADP-ribose polymerase [PARP] inhibitors in cancer cells bearing DDR defects: the rationale for their inclusion in the clinic. J Exp Clin Cancer Res. 2016;35:179
36. Kehoe S. Olaparib and ovarian cancer-overall survival outcomes. Lancet Oncol. 2016;17:1474-5
Author contact
![]() Corresponding author: Yu-Mei Wu, Beijing Obstetrics and Gynecology Hospital, Capital Medical University, 17 Qi-he-lou St, Dongcheng District, Beijing 100006, China. E-mail: wuyumeiedu.cn
Corresponding author: Yu-Mei Wu, Beijing Obstetrics and Gynecology Hospital, Capital Medical University, 17 Qi-he-lou St, Dongcheng District, Beijing 100006, China. E-mail: wuyumeiedu.cn

 Global reach, higher impact
Global reach, higher impact