Impact Factor
ISSN: 1449-1907
Int J Med Sci 2017; 14(12):1276-1283. doi:10.7150/ijms.20479 This issue Cite
Research Paper
Cell Recognition Molecule L1 Regulates Cell Surface Glycosylation to Modulate Cell Survival and Migration
1. Department of Colorectal Surgery, Cancer Hospital of China Medical University, Liaoning Cancer Hospital and Institute, Shenyang, Liaoning 110042, China;
2. Dalian Medical University, Dalian, Liaoning 116044, China;
3. National University Hospital, Singapore 119074, Singapore;
4. Department of Clinical Laboratory, the Second Affiliated Hospital of Dalian Medical University, Dalian, Liaoning 116027, China;
5. Department of Neurology, Forth Affiliated Hospital of China Medical University, Shenyang, Liaoning 110000, China.
Received 2017-4-9; Accepted 2017-9-12; Published 2017-9-30
Abstract
Background: Cell recognition molecule L1 (L1) plays an important role in cancer cell differentiation, proliferation, migration and survival, but its mechanism remains unclear.
Methodology/Principal: Our previous study has demonstrated that L1 enhanced cell survival and migration in neural cells by regulating cell surface glycosylation. In the present study, we show that L1 affected cell migration and survival in CHO (Chinese hamster ovary) cell line by modulation of sialylation and fucosylation at the cell surface via the PI3K (phosphoinositide 3-kinase) and Erk (extracellularsignal-regulated kinase) signaling pathways. Flow cytometry analysis indicated that L1 modulated cell surface sialylation and fucosylation in CHO cells. Activated L1 upregulated the protein expressions of ST6Gal1 (β-galactoside α-2,6-sialyltransferase 1) and FUT9 (Fucosyltransferase 9) in CHO cells. Furthermore, activated L1 promoted CHO cells migration and survival as shown by transwell assay and MTT assay. Inhibitors of sialylation and fucosylation blocked L1-induced cell migration and survival, while decreasing FUT9 and ST6Gal1 expressions via the PI3K-dependent and Erk-dependent signaling pathways.
Conclusion: L1 modulated cell migration and survival by regulation of cell surface sialylation and fucosylation via the PI3K-dependent and Erk-dependent signaling pathways.
Keywords: Cell adhesion molecule L1, Glycosylation, Sialylation, Fucosylation, CHO cells.
Introduction
Metastatic cancer cells usually express high density of sialic acid-rich glycoproteins on cell surfaces and help cancer cells enter the circulatory system [1]. Glycosylation is a post- or co-translational modification for most proteins and play important roles in cancer development [2]. In a previous study, we have demonstrated that the upregulation of cell adhesion molecule L1 (L1) in neural cells increased the expressions of sialic acid and fucose on the cell surface, which subsequently, enhanced cell survival [3]. Fucosylation is a common modification involving oligosaccharides and many synthesis pathways are involved in the regulation of fucosylation [4, 5]. Fucosylation of glycoproteins modulates the biological functions of adhesion molecules and plays an important role in cell survival and metastasis [6].
L1 is a type of transmembrane cell adhesion glycoprotein which belongs to a large immunoglobulin superfamily of cell adhesion molecules and mediates interactions between cells [7]. L1 promotes cell survival, migration and axon guidance in the nervous system [8]. The overexpression of L1 has been shown to indicate poor prognosis in a variety of human carcinomas including ovarian, lung, gastric, colorectal and pancreatic cancers [9-13]. Recently, we have demonstrated that L1 upregulated the protein expressions of ST3Gal4 and FUT9 via activation of the PLCɣ (Phospholipase Cγ) pathway, which increased cell surface sialylation and fucosylation [14]. CHO cell line was derived from the Chinese hamster ovary and can provide a high expression of recombinant glycoproteins which are equipped with a glycosylation mechanism very similar to that found in humans [15]. Sialic acid occupies the terminal end on oligosaccharide chains in these glycoproteins and influences the biological behavior of cells [16]. Previous studies have demonstrated that L1 regulated the Erk signaling pathway [17]. Cells expressing L1 activated the phosphoinositide 3-kinase/ Protein kinase B (PI3K/Akt) pathway to stimulate motility in gastric cancer and induce proliferation in renal cell carcinoma [18]. However, the precise mechanism of L1 in cell migration and survival is still unclear.
In this study, we investigated the effects of L1 on CHO cell survival and migration by regulation of cell surface glycosylation. We demonstrate that L1 regulated cell surface sialylation and fucosylation via the PI3K and Erk signaling pathways.
Results
L1 modulated the expression of specific carbohydrates on the cell surface of CHO cell line
Given that L1 is one of many carbohydrate-carrying molecules at the cell surface and mediates interactions between other adhesion molecules in the nervous system, we hypothesized that L1 might modulate specific glycosylation patterns at cell surfaces. To test this hypothesis, we compared cell surface glycosylation patterns between CHO cells and L1-transfected CHO (L1-CHO) cells by flow cytometry. The expression of carbohydrates recognized by SNA (Sambucus nigra lectin) and L5 antibodies were significantly upregulated in L1-transfected versus non-transfected CHO cells (Fig. 1). SNA recognized terminal sialic acids while L5 antibodies recognized terminal fucose (Fig.2A). These results demonstrated that L1 plays a role in modulation of the sialylation and fucosylation at cell surfaces.
Glycosylation patterns on cell surface of CHO cells and L1-transfected CHO cells. CHO cells and L1-CHO cells were subjected to flow cytometry analysis using a panel of carbohydrate surface markers, including lectins and antibodies against carbohydrates. A. In the flow cytometry histograms, the areas in green show the number of unstained cells and the areas outlined in red represent cells binding to carbohydrates antibodies L5 and various lectins including SNA (Sambucus nigra lectin), MAA (Maackia amurensis lectin), UEAI (Ulex europaeus agglutinin I), DSL (Datura stramonium lectin) and JAC (Jacalin). B. The quantitative results showed that the expression of carbohydrates recognized by SNA as well as L5 antibodies were significantly increased in L1-CHO cells versus CHO cells. *: p<0.05, by Student's test.
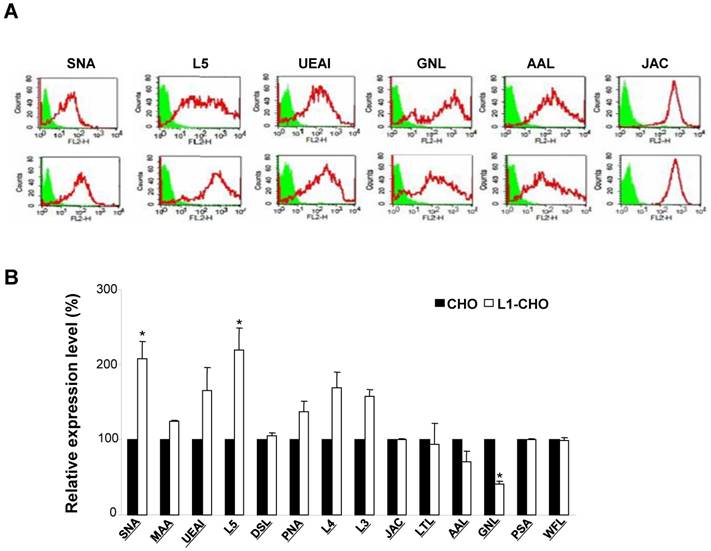
The protein expressions of ST6Gal1 and FUT9 were modulated by L1. A. The carbohydrate structures for terminal sialylation (a and b) and fucosylation (b) with related transferases were recognized by SNA and L5 antibodies on cell surfaces. B. Western blot was used to detect the expression of transferases. The protein expressions of ST6Gal1 and FUT9 were significantly upregulated in the L1-CHO cells versus CHO cells.
L1 regulated the expression of sialyltransferases, ST6Gal1 and fucosyltransferase, FUT9
Since L1 is involved in the regulation of sialylation and fucosylation at cell surfaces, we hypothesized that activated L1 may regulate the expression of specific sialyltransferases and fucosyltransferases. Western blot was used to assess this hypothesis. The results showed that the expressions of FUT9 and ST6Gal1 were significantly upregulated in CHO cells transfected with L1 versus non-transfected CHO cells (Fig.2B). Hence, the protein expressions of ST6Gal1 and FUT9 in CHO cells were upregulated upon L1 activation, indicating changes in sialylation and fucosylation activities.
Activated L1 promoted cell migration of CHO cells
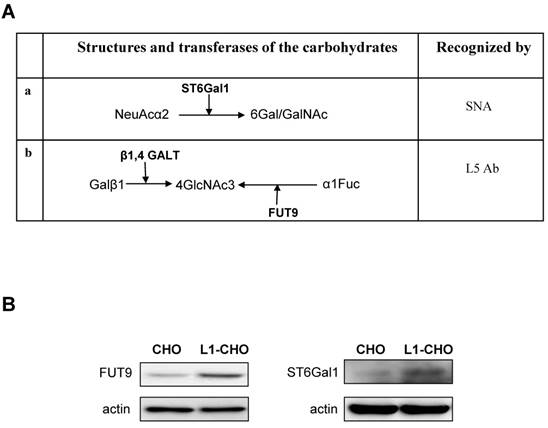
To investigate the role of activated L1 in cell migration, transwell membranes were coated with L1 antibodies (L1Ab). Thus, only cells that express L1 at the cell surface will be stimulated. As expected, under such conditions, cell migration was significantly increased in L1-CHO cells treated with L1Ab, compared to L1Ab-treated non-transfected CHO cells (Fig. 3A, 3B).
Activated L1 promoted cell survival of CHO cells
To investigate whether L1 plays a role in cell survival, MTT analysis was performed. In agreement with our previous study, cell survival was significantly enhanced in L1-CHO cells versus CHO cells (Fig. 3C). Together, these observations demonstrated that changes in glycosylation patterns induced by L1 may also regulate cell migration and cell survival.
Inhibitors of sialylation and fucosylation blocked L1-induced cell migration and cell survival
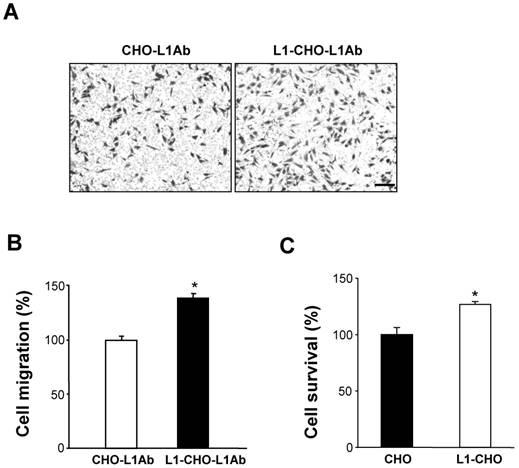
We investigated whether sialylation and fucosylation could be involved in L1-inducedcell migration and survival by using Soyasaponin I, a potent and specific sialyltransferase inhibitor, and Tunicamycin, which prevents N-glycosylation of fucosyltransferase leading to inactivation of the enzyme. Both Tunicamycin and Soyasaponin I could significantly decreased the cell migration of L1-CHO cells after L1 antibody stimulation in a dose-dependent manner (Fig 4A). Additionally, cell survival of L1-CHO cells stimulated with L1 antibody were significantly reduced after treatment with Soyasaponin I and Tunicamycin in a dose-dependent manner (Fig 4B). The strongest inhibition effects were produced after the sialyltransferase inhibitor and fucosyltransferase inhibitor were used together (Fig 4C and 4D). The results demonstrated that sialylation and fucosylation may also contribute to L1-induced cell migration and cell survival.
Activation of L1 promoted cell migration and cell survival. To verify the role of L1 in cell migration, the undersurface of transwell membranes was coated with L1Ab. Relative number of cells transmigrating through the membrane were determined by staining cells on the undersurface of the transwell membrane followed by cell lysis and measurement of absorbance value. Optical density measurements of the dye were directly proportional to numbers of migrated cells. A. Cell migration was significantly enhanced in L1-CHO cells after treatment with L1Ab (L1-CHO-L1Ab) versus L1Ab-treated non-transfected CHO cells (CHO-L1Ab). Photomicrographs illustrate the greater density of migrated L1-CHO cells treated with L1Ab (L1-CHO-L1Ab) on the undersurface of the membrane. B. Quantitative measurements from Figure 3A. C. To investigate the role of L1 in cell survival, MTT analysis was performed. Cell survival was significantly enhanced in the L1-CHO cells versus CHO cells. *: p<0.05, by Student's test.
L1 induced the upregulation of FUT9 and ST6Gal1 protein expressions, while L1-induced cell migration and survival are dependent on thePI3K and Erk signaling pathways
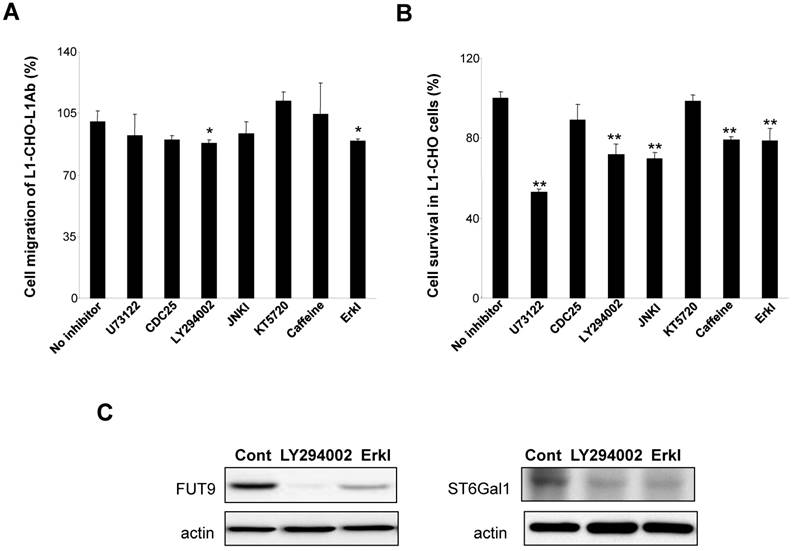
To understand the mechanism of L1 in modulating sialylation and fucosylation, we investigated the protein expressions of FUT9 and ST6Gal1 after inhibition of signal transduction pathways known to be activated by L1. In L1Ab-treated L1-CHO cells, both PI3K inhibitor (LY294002) and Erk inhibitor attenuated cell migration (Fig. 5A). PLCγ inhibitor, U73122, PI3K inhibitor, JNK inhibitor, caffoline and Erk inhibitor reduced cell survival in L1-CHO cells, whereas Cdc25 phosphatase inhibitor II and protein kinase A inhibitor (KT5720) had no effect on cell survival and migration (Fig. 5B). In addition, PI3K inhibitor and Erk inhibitor reduced both FUT9 and ST6Gal1 protein expressions (Fig. 5C). The down-regulation of FUT9 and ST6Gal1 protein expressions together with the reduction of L1-induced cell migration and survival were observed only when cells were treated with PI3K inhibitor and Erk inhibitor. Together, the data suggest that L1 regulates sialylation and fucosylation to modulate cell function via PI3K-dependent and Erk-dependent pathways.
Discussion
L1, a member of the cell adhesion molecule superfamily, consists of five fibronectin type Ⅲ repeats, six immunoglobulin-like domains, a single transmembrane region and a short cytoplasmic tail [19, 20]. The protein, encoded by the L1 gene, is a type of glycoprotein belonging to the immunoglobulin superfamily. L1 is able to bind to a number of other proteins or "homophilically" to itself [21]. Complex N-glycans are distinct and may regulate surface glycoprotein levels, cell differentiation and cell proliferation [22]. L1 plays an important role in the development of the nervous system by glycosylation [23]. Overexpression of L1 has been reported in a variety of cancers, including thyroid carcinoma, non-small cell lung cancer, gastric cancer, hepatocellular carcinoma, pancreatic cancer, colorectal cancer, ovarian cancer and endometrial cancer [24-27]. These findings implied that L1 also plays a role in human carcinogenesis. However, the mechanism in which L1 modulates cell migration, proliferation, invasion and apoptosis remains unknown [28, 29]. Previously, we have demonstrated that L1 interacted with several carbohydrates on cell surfaces and modulates cell function. In the present study, we show that L1 utilized the same mechanism together with the PI3K and Erk signal transduction systems to regulate cell survival and migration.
CHO cells produce recombinant glycoproteins via a glycosylation machinery which is similar to human cells. In this study, we have confirmed the expression of carbohydrates recognized with SNA or L5 antibody upon upregulation of L1 expression in CHO cells. SNA or L5 antibody can recognize terminal sialic acids or fucose, respectively. N-acetylglucosamine (GlcNAc) contributes to the function and structure of cells and is the precursor of N-acetylgalactosamine (GalNAc). GlcNAc is converted to sialic acid which is the terminal glycan in many glycosylated proteins. We have shown that ST6Gal1 was recognized by SNA and NeuAcα2 converted to GalNAc was increased in L1-CHO cells. Similarly, FUT9 was recognized by L5 antibody and α1Fuc converted to 4GlaNAc3 was augmentated in L1-CHO cells. This indicated that L1 may play a central role in modulation of sialylation and fucosylation by increasing the expression of ST6Gal1 and FUT9 on cell surfaces. Furthermore, we found that cell migration was significantly increased in L1-CHO cells treated with L1Ab, but not in L1Ab-treated CHO cells. Cell survival was also significantly enhanced in L1-CHO cells versus non-transfected CHO cells. Moreover, L1-induced cell migration and survival were repressed when sialylation and fucosylation were inhibited with specific sialyltransferase inhibitor or Tunicamycin which prevents N-glycosylation of fucosyltransferase. Tunicamycin has been shown to counteract GlcNAc from inducing the expression of E-cadherin and phosphorylation of β-catenin, it ultimately led to cell apoptosis. Therefore, the study emphasized the importance of N-glycosylation in cell survival [30]. Glycans have been recognized as important players in cell-cell interactions [31]. In cancer cells, malignant behaviors which depend on cell recognition- such as cell migration and survival- are mediated by distinct carbohydrate structures [1]. Since activated L1 can modulate sialylation and fucosylation, L1-induced specific glycosylation patterns may influence CHO cell survival and migration. This is in agreement with our previous studies in neuronal cells [3].
Inhibition of sialylation and fucosylation decreased cell migration and cell survival. A. Cell migration assay was performed in L1Ab-treated L1-CHO cells treated with Soyasaponin I (STI; 2.72mg/ml, 5.44mg/ml) or Tunicamycin (FUTI; 1μg/ml, 5μg/ml, 25μg/ml, 50μg/ml) versus L1-CHO. Cell migration was significantly inhibited after treatment with Soyasaponin I or Tunicamycin. B. MTT analysis of L1-CHO performed after treatment with Soyasaponin I (STI; 2.72mg/ml, 3.4mg/ml, 4.08mg/ml) or Tunicamycin (FUTI; 25μg/ml, 50μg/ml, 100μg/ml) showed a significant decrease of cell survival. C. Cell migration assay was performed in L1Ab-treated L1-CHO cells treated with Soyasaponin I (STI; 2.72mg/ml) and Tunicamycin (FUTI; 25μg/ml) together versus L1-CHO. Cell migration was significantly inhibited after treatment with Soyasaponin I and Tunicamycin together. D. MTT analysis of L1-CHO performed after treatment with Soyasaponin I (STI; 4.08mg/ml) and Tunicamycin (FUTI; 100μg/ml) together showed a significant decrease of cell survival. *: p<0.05; **: p<0.01, by Student's test.
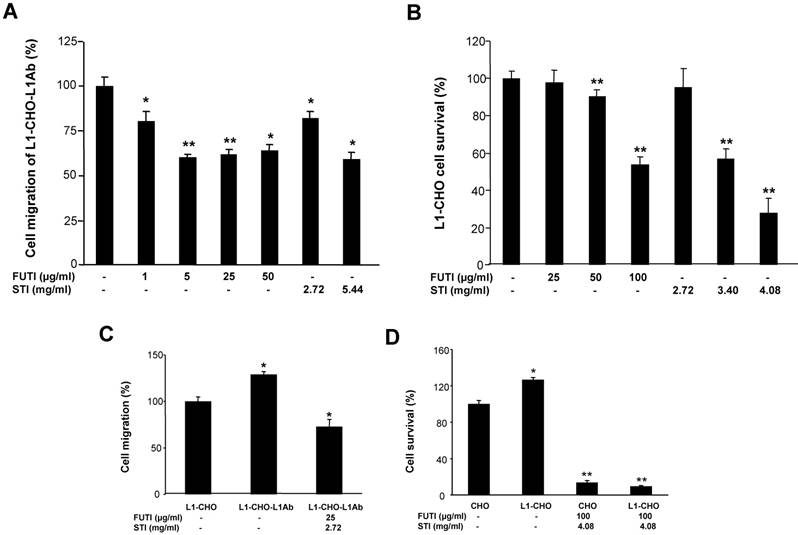
Effects of inhibitors of signal transduction pathways on cell migration, cell survival and the protein expressions of FUT9 and ST6Gal1. After incubation for 1 hour of L1-CHO cells treated with or without L1Ab, inhibitors of PLCγ (U73122, 10.5 μM), CDC25 phosphatase (CDC25 inhibitor II, 1.05 μM), PI3K (LY294002, 16.5 μM), Erk (Erk inhibitor, 50 μM), JNK (JNK inhibitor, 200nM), ATM/ATR (Caffeine, 1mM) or PKA inhibitor (KT5720, 280 nM) were added into the culture medium and the cells were incubated further for 24 hours. A. After treatment with inhibitors of the signal transduction pathways, cell migration assay was performed. Cell migration was significantly inhibited in L1Ab-treated L1-CHO cells by LY294002 and Erk inhibitor. B. After treatment with inhibitors of the signal transduction pathways, cell survival was quantified by MTT assay. U73122, LY294002, JNK inhibitor, Caffeine and Erk inhibitor significantly repressed cell survival in L1-CHO cells. C. Western blot was used to detect the expression of transferases. The protein expressions of ST6Gal1 and FUT9 were significantly downregulated in L1-CHO cells treated with LY294002 and Erk inhibitor. *: p<0.05; **: p<0.01, by Student's test comparison with the no inhibitor control.
L1 has been known to modulate the activation of AKT, MAPK, Erk, PI3K and FAK intracellular and extracellular signaling pathways [32-36]. To explore the mechanism by which L1 induces sialylation and fucosylation, we investigated the protein expressions of ST6Gal1 and FUT9 after cells were treated with inhibitors of signaling pathways known to be activated by L1. We show that L1 regulated sialylation and fucosylation to modulate CHO cell migration and survival via the PI3K-dependent and Erk-dependent pathways. The Erk pathway activated by L1 has been linked to a motile phenotype in carcinomas [37]. Additionally, Erk signaling has been reported to be involved in diverse cellular processes in cancer, including proliferation, survival, differentiation and migration [38, 39]. We propose that L1 may interact with transmembrane binding cell adhesion molecules, resulting in activation of Erk-dependent genes which induce cell surface sialylation and fucosylation and subsequently, promote cell migration and survival. PI3K/Akt signaling pathway has been reported to contribute significantly to cellular transformation and cancer development, and appears to have a serine/threonine kinase activity [40]. L1 has been reported to bind to integrin and PI3K is a classical component of the integrin signaling system. L1 acts as an integrin ligand, which activates the phosphorylation of downstream targets of the PI3K pathway. Therefore, the role of L1 in promoting cell migration and survival by regulating cell surface sialylation and fucosylation via the PI3K and Erk signaling pathways could be a promising therapeutic intervention in cancer.
Materials and Methods
Antibodies and inhibitors
Polyclonal goat anti-mouse, anti-rat and anti-human FUT9 antibodies and polyclonal goat anti-mouse, anti-rat and anti-human ST6Gal1 antibodies were purchased from Santa Cruz Biotechnology (Santa Cruz, CA, USA). Polyclonal mouse anti-β-actin antibody was purchased from Sigma-Aldrich (St. Louis, MO, USA). R-Phycoerythin (R-PE)-conjugated mouse anti-rat monoclonal, PerCP-CY5.5-conjugated rat anti-mouse IgM monoclonal and R-PE-conjugated rat anti-mouse IgM monoclonal antibodies were purchased from BD Biosciences (Franklin Lakes, NJ, USA). Biotinylated lectins and Texas Red avidin D were purchased from Vector Laboratories, Inc. (Burlingame, CA, USA). Soyasaponin I (Wako Pure Chem. Ind. Co. Ltd., Japan), Tunicamycin (Calbiochem, CA, USA), U73122 (Calbiochem, CA, USA), Cdc25 phosphatase inhibitor II (Calbiochem, CA, USA), LY294002 (Sigma, MO, USA), Erk inhibitor (3-(2-Aminoethyl)-5-((4-ethoxyphenyl) methylene) -2,4-thiazolidinedione, HCl; Calbiochem, CA, USA) and KT5720 (Sigma, MO, USA) are available commercially. L3, L4 and L5 antibodies were produced as previously described and presented by Melitta in Hamberg University (Kucherer et al., 1987; Fahrig et al., 1990; Streit et al., 1990; Streit et al., 1996) [42-45].
Cell culture
All cell culture reagents were purchased from Invitrogen Life Technologies (Merelbeke, Belgium) unless indicated otherwise. The CHO cells and CHO cells transfected with L1 were done as previously described [46]. The cells were maintained in DMEM (Dulbecco′s Modified Eagle Medium) supplemented with 10% FBS, 0.5 U/mL penicillin and 0.5 U/mL streptomycin. Cells were passaged every 2 or 3 days using 0.05% trypsin/0.04% EDTA. The single cells obtained were used for flow cytometry assay.
Flow cytometry assay
Cell surface carbohydrate expression was assessed by indirect immunofluorescence detected by flow cytometry to provide a quantitative percentage binding and measure of the fluorescence intensity of carbohydrate antibodies and lectins. CHO cells were harvested into single cell suspensions by trypsinization as described above. Cells were placed in sterile conical tubes in aliquots of 500,000 cells each and stained with one of the 14 carbohydrate antibodies and lectins. Cells were washed 3 times with PBS and then stained with secondary antibody. Unstained cells and cells stained with secondary antibody alone were used as controls.
Migration assay
Costar transwell polycarbonate filters (5.0um pore size) were used for the migration assay [40]. The undersurfaces of the 6.5 mm transwell membranes were coated with anti-L1 antibody in PBS overnight at 4oC, then blocked with 2% BSA. Next 2.5x10E5 cells/mL were plated in culture medium into the upper chamber and allowed to migrate through the pores onto the coated undersurfaces at 37oC in a CO2 incubator. After 24 hours, cells from the inner surface of the insert were gently wiped out with cotton-tipped swabs, and the inserts were fixed and stained. After a final wash with PBS, the cells were examined under a microscope to confirm proper morphology, and the dye was extracted. The absorbance was measured using a microplate reader. Dye levels are directly proportional to the numbers of cells. Data are presented as mean ± S.E.
MTT assay
Cells were cultured in 96 well plates for 24 hours. 10 µl 5mg/mL MTT solution was then added and cultured at 37oC for 4 hours. After washing with PBS, 20% SDS was added in to lyse the cells and dissolve the crystal. The OD (absorbance) was determined with aElx800 universal microplate reader (BioTek Instruments, Inc., Winooski, VT, USA) and the percentage of cell survival was compared [47].
Western blot assay
Cells were harvested and lysed in RIPA buffer (1% Triton X-100, 150 mM NaCl, 25 mM Tris, pH 7.5, 0.5% sodium deoxycholate, 0.1% SDS, 5 mM pyrophosphate, and 50 mM NaF). Samples containing equal amounts of protein were resolved using SDS-PAGE and transferred to polyvinylidene difluoride (PVDF) membranes (Millipore), followed by incubation with primary antibodies (anti-FUT9 antibody, anti-ST6Gal1 antibody, anti-PLCγ antibody) and secondary antibodies. Chemiluminescent detection was done using the ECL kit (Amersham Biosciences, Pittsburgh, PA, USA).
Data analysis
All data were expressed as mean ± S.E. Statistical evaluations were done by one-way analysis of variance and Student's t-test. Differences were considered to be significant when p<0.05.
Acknowledgements
This work was supported by the National Natural Science Foundation of China (31070728), Liaoning Doctoral Scientific Research Initial Foundation (201601417), and Liaoning Natural Science Foundation (2014020102) and National Natural Science Foundation (81672427).
Competing Interests
The authors have declared that no competing interest exists.
References
1. Haltiwanger RS, Lowe JB. Role of glycosylation in development. Annu Rev Biochem. 2004;73:491-537
2. Kawakami S, Hashida M. Glycosylation-mediated targeting of carriers. J Control Release. 2014;190:542-55
3. Li YL, Wu GZ, Dawe GS. et al. Cell surface sialylation and fucosylation are regulated by L1 via phospholipase Cgamma and cooperate to modulate neurite outgrowth, cell survival and migration. PLoS One. 2008;3:e3841
4. Kurz S, King JG, Dinglasan RR. et al. The fucomic potential of mosquitoes: Fucosylated N-glycan epitopes and their cognate fucosyltransferases. Insect Biochem Mol Biol. 2015;68:52-63
5. Zhang YM, Zhang YY, Bulbul A. et al. Baicalin promotes embryo adhesion and implantation by upregulating fucosyltransferase IV (FUT4) via Wnt/beta-catenin signaling pathway. FEBS Lett. 2015;589:1225-33
6. Li J, Guillebon AD, Hsu JW. et al. Human fucosyltransferase 6 enables prostate cancer metastasis to bone. Br J Cancer. 2013;109:3014-22
7. Homrich M, Gotthard I, Wobst H. et al. Cell Adhesion Molecules and Ubiquitination-Functions and Significance. Biology (Basel). 2015;23:5 (1)
8. Huang X, Hu J, Li Y. et al. The cell adhesion molecule L1 regulates the expression of FGF21 and enhances neurite outgrowth. Brain Res. 2013;1530:13-21
9. Ben Q, An W, Fei J. et al. Downregulation of L1CAM inhibits proliferation, invasion and arrests cell cycle progression in pancreatic cancer cells. Exp Ther Med. 2014;7:785-790
10. Chen DL, Zeng ZL, Yang J. et al. L1cam promotes tumor progression and metastasis and is an independent unfavorable prognostic factor in gastric cancer. J Hematol Oncol. 2013;6:43
11. Dippel V, Milde-Langosch K, Wicklein D. et al. Influence of L1-CAM expression of breast cancer cells on adhesion to endothelial cells. J Cancer Res Clin Oncol. 2013;139:107-21
12. Fang QX, Lu LZ, Yang B. et al. L1, beta-catenin, and E-cadherin expression in patients with colorectal cancer: correlation with clinicopathologic features and its prognostic significance. J Surg Oncol. 2010;102:433-42
13. Hai J, Zhu CQ, Bandarchi B. et al. L1 cell adhesion molecule promotes tumorigenicity and metastatic potential in non-small cell lung cancer. Clin Cancer Res. 2012;18:1914-24
14. Li YL, Wu GZ, Zeng L. et al. Cell surface sialylation and fucosylation are regulated by the cell recognition molecule L1 via PLCgamma and cooperate to modulate embryonic stem cell survival and proliferation. FEBS Lett. 2009;583:703-10
15. Ha TK, Kim YG, Lee GM. et al. Understanding of altered N-glycosylation-related gene expression in recombinant Chinese hamster ovary cells subjected to elevated ammonium concentration by digital mRNA counting. Biotechnol Bioeng. 2015;112:1583-93
16. Chung CY, Yin B, Wang Q. et al. Assessment of the coordinated role of ST3GAL3, ST3GAL4 and ST3GAL6 on the alpha2,3 sialylation linkage of mammalian glycoproteins. Biochem Biophys Res Commun. 2015;463:211-5
17. Silletti S, Yebra M, Perez B. et al. Extracellular signal-regulated kinase (ERK)-dependent gene expression contributes to L1 cell adhesion molecule-dependent motility and invasion. J Biol Chem. 2004;279:28880-8
18. Doberstein K, Pfeilschifter J, Gutwein P. The transcription factor PAX2 regulates ADAM10 expression in renal cell carcinoma. Carcinogenesis. 2011;32:1713-23
19. Jacob J, Haspel J, Kane-Goldsmith N. et al. L1 mediated homophilic binding and neurite outgrowth are modulated by alternative splicing of exon 2. J Neurobiol. 2002;51:177-89
20. Moos M, Tacke R, Scherer H. et al. Neural adhesion molecule L1 as a member of the immunoglobulin superfamily with binding domains similar to fibronectin. Nature. 1988;334:701-3
21. Wei CH, Ryu SE. Homophilic interaction of the L1 family of cell adhesion molecules. Exp Mol Med. 2012;44:413-23
22. Lau KS, Partridge EA, Grigorian A. et al. Complex N-glycan number and degree of branching cooperate to regulate cell proliferation and differentiation. Cell. 2007;129:123-34
23. Li Y, Huang X, An Y. et al. Cell recognition molecule L1 promotes embryonic stem cell differentiation through the regulation of cell surface glycosylation. Biochem Biophys Res Commun. 2013;440:405-12
24. Salvo E, Garasa S, Dotor J. et al. Combined targeting of TGF-beta1 and integrin beta3 impairs lymph node metastasis in a mouse model of non-small-cell lung cancer. Mol Cancer. 2014;13:112
25. Suh DH, Kim MA, Kim HS. et al. L1 cell adhesion molecule expression is associated with pelvic lymph node metastasis and advanced stage in diabetic patients with endometrial cancer: a matched case control study. J Cancer Prev. 2014;19:231-9
26. Yoon H, Min JK, Lee DG. et al. L1 cell adhesion molecule and epidermal growth factor receptor activation confer cisplatin resistance in intrahepatic cholangiocarcinoma cells. Cancer Lett. 2012;316:70-6
27. Zeimet AG, Reimer D, Huszar M. et al. L1CAM in early-stage type I endometrial cancer: results of a large multicenter evaluation. J Natl Cancer Inst. 2013;105:1142-50
28. Altevogt P, Doberstein K, Fogel M. L1CAM in human cancer. Int J Cancer. 2016;138(7):1565-76
29. Siesser PF, Maness PF. L1 cell adhesion molecules as regulators of tumor cell invasiveness. Cell Adh Migr. 2009;3:275-7
30. Shang J, Korner C, Freeze H. et al. Extension of lipid-linked oligosaccharides is a high-priority aspect of the unfolded protein response: endoplasmic reticulum stress in Type I congenital disorder of glycosylation fibroblasts. Glycobiology. 2002;12:307-17
31. Pinho SS, Reis CA. Glycosylation in cancer: mechanisms and clinical implications. Nat Rev Cancer. 2015;15:540-55
32. Ben-Shmuel A, Shvab A, Gavert N. et al. Global analysis of L1-transcriptomes identified IGFBP-2 as a target of ezrin and NF-kappaB signaling that promotes colon cancer progression. Oncogene. 2013;32:3220-30
33. Ito T, Yamada S, Tanaka C. et al. Overexpression of L1CAM is associated with tumor progression and prognosis via ERK signaling in gastric cancer. Ann Surg Oncol. 2014;21:560-8
34. Kiefel H, Bondong S, Pfeifer M. et al. EMT-associated up-regulation of L1CAM provides insights into L1CAM-mediated integrin signalling and NF-kappaB activation. Carcinogenesis. 2012;33:1919-29
35. Schmid RS, Midkiff BR, Kedar VP. et al. Adhesion molecule L1 stimulates neuronal migration through Vav2-Pak1 signaling. Neuroreport. 2004;15:2791-4
36. Watanabe H, Yamazaki M, Miyazaki H. et al. Phospholipase D2 functions as a downstream signaling molecule of MAP kinase pathway in L1-stimulated neurite outgrowth of cerebellar granule neurons. J Neurochem. 2004;89:142-51
37. Cheng L, Lemmon S, Lemmon V. RanBPM is an L1-interacting protein that regulates L1-mediated mitogen-activated protein kinase activation. J Neurochem. 2005;94:1102-10
38. Han S, Bui NT, Ho MT. et al. Dexamethasone Inhibits TGF-beta1-induced Cell Migration by Regulating the ERK and AKT Pathways in Human Colon Cancer Cells Via CYR61. Cancer Res Treat. 2016;48(3):1141-53
39. Liao JC, Lee KT, You BJ. et al. Raf/ERK/Nrf2 signaling pathway and MMP-7 expression involvement in the trigonelline-mediated inhibition of hepatocarcinoma cell migration. Food Nutr Res. 2015;59:29884
40. Faes S, Dormond O. PI3K and AKT: Unfaithful Partners in Cancer. Int J Mol Sci. 2015;16:21138-52
41. Zhang X, Dong W, Zhou H. et al. alpha-2,8-Sialyltransferase Is Involved in the Development of Multidrug Resistance via PI3K/Akt Pathway in Human Chronic Myeloid Leukemia. IUBMB Life. 2015;67:77-87
42. Fahrig T, Schmitz B, Weber D. et al. Two monoclonal antibodies recognizing carbohydrate epitopes on neural adhesion molecules interfere with cell interactions. Eur J Neurosci. 1990;2:153-161
43. Kucherer A, Faissner A, Schachner M. The novel carbohydrate epitope L3 is shared by some neural cell adhesion molecules. J Cell Biol. 1987;104:1597-1602
44. Streit A, Faissner A, Gehrig B. et al. Isolation and biochemical characterization of a neural proteoglycan expressing the L5 carbohydrate epitope. J Neurochem. 1990;55:1494-1506
45. Streit A, Yuen CT, Wendy Loveless R. et al. The Lex carbohydrate sequence is recognized by antibody to L5, a functional antigen in early neural development. J Neurochem. 1996;66:834-844
46. Chen S, Mantei N, Dong L. et al. Prevention of neuronal cell death by neural adhesion molecules L1 and CHL1. J Neurobiol. 1999;38:428-39
47. Mosmann T. Rapid colorimetric assay for cellular growth and survival: application to proliferation and cytotoxicity assays. J Immunol Methods. 1983;65:55-63
Author contact
![]() Corresponding authors: Email: shigangcom (GS), yalilipapercom (YLL) and liufangcom (FL)
Corresponding authors: Email: shigangcom (GS), yalilipapercom (YLL) and liufangcom (FL)

 Global reach, higher impact
Global reach, higher impact