Impact Factor
ISSN: 1449-1907
Int J Med Sci 2017; 14(12):1197-1202. doi:10.7150/ijms.20933 This issue Cite
Research Paper
Association of HMGB1 Gene Polymorphisms with Lung Cancer Susceptibility and Clinical Aspects
1. Department of Thoracic Surgery, Affiliated Dongyang Hospital of Wenzhou Medical University, Dongyang, Zhejiang, China;
2. Graduate Institute of Biomedical Science, China Medical University, Taichung, Taiwan;
3. Department of Thoracic Surgery, Changhua Christian Hospital, Changhua, Taiwan;
4. Department of Nursing, National Taichung University of Science and Technology, Taichung, Taiwan;
5. Department of Biotechnology, College of Health Science, Asia University, Taichung, Taiwan;
6. Department of Biomedical Sciences Laboratory, Affiliated Dongyang Hospital of Wenzhou Medical University, Dongyang, Zhejiang, China;
7. Department of Respiratory Therapy, Fu-Jen Catholic University, New Taipei City, Taiwan;
8. School of Medicine, China Medical University, Taichung, Taiwan.
Received 2017-5-9; Accepted 2017-8-7; Published 2017-9-19
Abstract
Lung cancer is one of the most frequently diagnosed malignancies and is associated with a poor survival rate in the Chinese Han population. Analysis of genetic variants could lead to improvements in prognosis following lung cancer therapy. High-mobility group box 1 protein (HMGB1) is a ubiquitous nuclear protein found in eukaryotic cells that participates in several biological functions including immune response, cell survival, apoptosis and cancer development. We investigated the effects of HMGB1 gene polymorphisms on the risk of lung cancer progression in a Chinese Han population. Our sample of 751 participants included 372 patients with lung cancer and 379 healthy controls. Four single-nucleotide polymorphisms (SNPs) of the HMGB1 gene were examined by real-time polymerase chain reaction (RT-PCR). We found that the CT or CC+CT heterozygotes of the HMGB1 rs1045411 polymorphism reduced the risks for lung cancer, while the G/T/C haplotypes of three HMGB1 SNPs (rs1360485, rs1045411 and rs2249825) also reduced the risk for lung cancer by almost half (0.486-fold). The current study is the first to examine the risk factors associated with HMGB1 SNPs in lung cancer development in the Chinese Han population.
Keywords: HMGB1, Lung cancer, SNP, Susceptibility, Polymorphisms, Chinese Han.
Introduction
Lung cancer is one of the most frequently diagnosed cancers worldwide and is associated with a poor 5-year survival rate [1]. The specific mechanisms underlying lung cancer development and progression remain unclear. Although cigarette smoking and alcohol consumption are known to significantly increase the risk of lung cancer, various other environmental risks, such as exposure to air pollution, second-hand smoke, and genetic susceptibility, are also involved in the development of lung cancer [2]. Indeed, multiple genetic and epigenetic changes have been implicated in the development of lung cancer [2]. An increased understanding of genetic mechanisms, including DNA genotyping and heterogeneity, is required to improve our ability to predict disease risk and prognosis in lung cancer [3].
Genetic variation plays a key role in lung cancer susceptibility and development. Currently, genotyping single nucleotide polymorphism (SNPs) of a population and comparing the distribution frequency of SNPs among subgroups (for example, controls and patients) is frequently used to examine the risk and prognosis of a cancer [4]. Emerging reports indicate an association between SNPs in certain genes and lung cancer progression, as for instance, specific SNP variants within the vascular endothelial growth factor, the interleukin-32 (IL-32) and the PARK2 genes [5-7].
High-mobility group box 1 protein (HMGB1) is a ubiquitous nuclear protein that has been discovered in mammals [8, 9]. HMGB1 contains DNA binding domains and contributes to DNA repair and stabilization of nuclear homeostasis [10]. HMGB1 is also found in cytosol and is secreted into the extracellular space during necrosis. HMGB1 participates in both cell survival and death by regulating immune response, apoptosis and autophagy in cancer development [11-15].
SNPs of the HMGB1 gene influence gene expression, protein function and disease susceptibility in particular individuals [16, 17]. SNP had potential predictive significance risk of lung cancer in previous studies [18, 19]. However, little is known about the association between HMGB1 SNPs, lung cancer risk and disease progression. We therefore performed a case-control study of four HMGB1 SNPs to investigate the correlation between these SNPs and lung cancer susceptibility and progression.
Materials and Methods
Participants
Between 2014 and 2016, we collected 372 blood specimens from patients (cases) who had been diagnosed with lung cancer at Dongyang People's Hospital. The control group consisted of 379 healthy participants without a history of cancer. All patients and participants provided written informed consent, and the study was approved by the Ethics Committee of Dongyang People's Hospital. The pathological stages of lung cancer in all patients were examined according to their medical records and the Revised International System for Staging Lung Cancer. A standardized questionnaire and electronic medical record system were used to obtain data on age, sex, smoking history, and alcohol consumption.
Extraction of genomic DNA
We extracted genomic DNA from whole blood specimens using QIAamp DNA blood mini kits (Qiagen, Valencia, CA, USA), according to the manufacturer's instructions. DNA was eluted with TE buffer (10 mM Tris, 1 mM EDTA; pH 7.8) and kept at -20°C.
SNP selection
SNP rs2249825 (3814C/G; genomic number 31,037,903) near the exon, SNP rs1412125 (-1615T/C; genomic number 31,041,595) in the promoter region, rs1360485 (3′UTR, T/C; genomic number 31,031,884) in the 3′ untranslated region and rs1045411 (2262C/T; genomic number 31,033,232) in the exon were selected according to Chinese HapMap data and previous studies [17, 20]. The minor allele frequencies of these SNPs were all ≥5 %.
Real-time PCR
The four HMGB1 SNPs were investigated using the TaqMan SNP Genotyping Assay (Applied Biosystems, Warrington, UK), according to the manufacturer's procedures [21, 22].
Bioinformatic analysis of genomics data
We used data from the Genotype-Tissue Expression (GTEx) project to identify correlations between SNPs and levels of HMGB1 expression [23].
Statistical Analyses
Chi-square analysis was used to confirm that the genotype distribution of each SNP was in Hardy-Weinberg equilibrium (HWE). A Mann-Whitney U-test and a Fisher's exact test were used to compare demographic characteristics between patients and controls. Correlations between genotype frequencies, clinicopathological characteristics and lung cancer risk were examined using logistic regression analysis to estimate odds ratios (ORs) of association, controlling for other covariates. Statistical analysis of the haplotype structure was performed according to the method described by Barrett et al. [24]. Hardy-Weinberg equilibrium and linkage disequilibrium was estimated using SNPAnalyzer version 2.0 (Istech Corp., Korea). A p value of < 0.05 was considered statistically significant. Data were analyzed using SAS statistical software (Version 9.1, 2005; SAS Institute Inc., Cary, NC, USA).
Results
In this study, we evaluated differences in the general demographic characteristics of 372 patients with lung cancer and 379 cancer-free controls. All recruited subjects were of Chinese Han ethnicity (Table 1). The study groups did not differ significantly by gender (p = 0.63). Significant associations were seen between alcohol (p < 0.001) and tobacco consumption (p=0.05) with lung cancer risk. According to the American Joint Committee on Cancer (AJCC) TNM staging system, 261 patients with lung cancer had clinical stage I-II (70.2%) and 111 patients had clinical stage III-IV disease (29.8%). To reduce the possible interference of confounding variables, adjusted ORs (AORs) with 95% confidence intervals (CIs) were estimated by multiple logistic regression models that controlled for age, alcohol consumption, and tobacco use in each comparison.
Genotype distributions of SNPs rs1360485, rs1045411, rs2249825 and rs1412125 are presented in Table 2. In the healthy controls, all genotypic frequencies were consistent with Hardy-Weinberg equilibrium (p > 0.05). In both healthy controls and lung cancer patients, the highest distribution frequencies for rs1360485, rs1045411, rs2249825 and rs1412125, respectively, were homozygous for AA, CC, GG and TT (Table 2). After adjusting for confounders, patients carrying the CT or CC+CT genotypes heterozygous for the HMGB1 rs1045411 polymorphism had a 0.597-fold (95% CI: 0.385-0.928; p < 0.05) and a 0.603-fold (95% CI: 0.394-0.924-;, p < 0.05) lower risk, respectively, of developing lung cancer compared with patients carrying CC homozygotes. The incidence frequencies of the rs1360485, rs2249825 and rs1412125 polymorphisms did not differ significantly between patients and healthy controls.
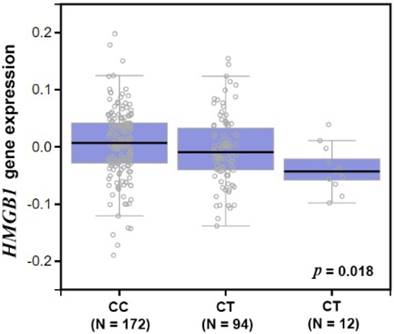
An analysis of the GTEx database investigated whether an association exists between rs1045411 and HMGB1 expression. Amongst individuals carrying a genotype with the variant T at rs1045411, a trend was observed for reduced expression of HMGB1, compared with the wild-type homozygous genotypes (p < 0.05; Fig. 1).
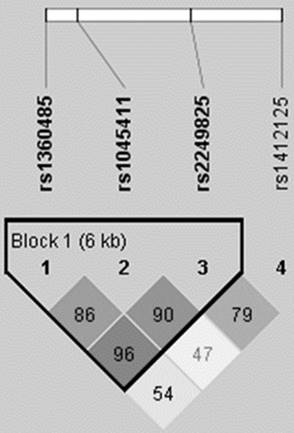
Haplotype analysis based on the HMGB1 rs1360485, rs1045411 and rs2249825 SNPs revealed that the most common haplotype in healthy controls was A/C/C (75.3%), which was therefore selected as the reference. The G/T/C HMGB1 haplotype significantly reduced the risk by nearly half (0.486-fold; 95% CI: 0.284-0.830; p < 0.05) (Table 3). The reconstructed linkage disequilibrium plot of the four SNPs is shown in Figure 2. Two polymorphisms (rs1360485 and rs2249811) showed very strong linkage disequilibrium (96%) in one haploblock. Similarly, we found strong linkage disequilibrium between rs1360485 and rs1045411 (86%), as well as between rs1045411 and rs2249825 (90%) (Fig. 2).
Demographic characteristics of the study population.
| Variable | Controls (N=379) | Patients (N=372) | p value |
|---|---|---|---|
| Age (yrs) | 45.15 ± 17.47 | 59.8 ± 10.38 | p <0.001* |
| Gender | |||
| Female | 187 (49.3%) | 190 (51.1%) | p = 0.63 |
| Male | 192 (50.1%) | 182 (48.9%) | |
| Alcohol consumption | |||
| No | 317 (83.6%) | 285 (76.6%) | p < 0.001* |
| Yes | 62 (16.4%) | 87 (23.4%) | |
| Tobacco consumption | |||
| No | 335 (88.4%) | 225 (60.5%) | p < 0.05* |
| Yes | 44 (11.6%) | 147 (39.5%) | |
| Stage | |||
| I-II | 261 (70.2%) | ||
| III-IV | 111 (29.8%) | ||
| Tumor T status | |||
| ≤T2 | 318 (85.5%) | ||
| >T2 | 54 (14.5%) | ||
| Lymph node status | |||
| N0 | 257 (69.1%) | ||
| >N0 | 115 (30.9%) | ||
| Metastasis | |||
| M0 | 301 (80.9%) | ||
| M1 | 71 (19.1%) |
Genotypic frequencies of the HMGB1 rs1360485, rs1045411, rs2249825 and rs1412125 SNPs in patients and controls and their association with the risk of lung cancer.
| Variable | Controls (N=187) n (%) | Patients (N=190) n (%) | OR (95% CI) | AOR (95% CI) |
|---|---|---|---|---|
| rs1360485 | ||||
| AA | 107 (57.2%) | 124 (65.3%) | 1 | 1 |
| AG | 68 (36.4%) | 56 (29.5%) | 0.711 (0.459-1.101) | 0.660 (0.424-1.028) |
| GG | 12 (6.4%) | 10 (5.3%) | 0.719 (0.299-1.730) | 0.709 (0.293-1.715) |
| AG+GG | 80 (42.8%) | 66 (34.7%) | 0.712 (0.470-1.079) | 0.671 (0.441-1.021) |
| rs1045411 | ||||
| CC | 109 (58.3%) | 130 (68.4%) | 1 | 1 |
| CT | 71 (38.0%) | 54 (28.4%) | 0.638 (0.412-0.986)* | 0.597 (0.385-0.928)* |
| TT | 7 (3.7%) | 6 (3.2%) | 0.719 (0.235-2.202) | 0.672 (0.219-2.164) |
| CT+TT | 78 (41.7%) | 60 (31.6%) | 0.645 (0.423-0.984)* | 0.603 (0.394-0.924)* |
| rs2249825 | ||||
| GG | 133 (71.1%) | 142 (74.7%) | 1 | 1 |
| GC | 50 (26.7%) | 46 (24.2%) | 0.862 (0.541-1.372) | 0.814 (0.510-1.300) |
| CC | 4 (2.1%) | 2 (1.1%) | 0.486 (0.084-2.599) | 0.442 (0.079-2.454) |
| GC+CC | 54 (28.9%) | 48 (25.3%) | 0.833 (0.528-1.312) | 0.786 (0.497-1.244) |
| rs1412125 | ||||
| TT | 107 (57.2%) | 109 (57.4%) | 1 | 1 |
| TC | 69 (36.9%) | 70 (36.8%) | 0.996 (0.650-1.525) | 0.963 (0.626-1.481) |
| CC | 11 (5.9%) | 11 (5.8%) | 0.982 (0.408-2.360) | 0.924 (0.383-2.228) |
| TC+CC | 80 (42.8%) | 81 (42.6%) | 0.994 (0.661-1.495) | 0.958 (0.634-1.446) |
OR = odds ratio; AOR = adjusted odds ratio; CI = confidence interval.
Haplotype frequencies of three HMGB1 polymorphisms (rs1360485, rs1045411 and rs2249825) and risk of lung cancer.
| Variable | Controls (N=758)n (%) | Patients (N=744)n (%) | OR (95% CI) | p value | ||
|---|---|---|---|---|---|---|
| rs1360485 A/G | rs1045411C/T | rs2249825C/G | ||||
| A | C | C | 571 (75.3%) | 588 (79.0%) | Reference | |
| G | T | G | 97 (12.8%) | 92 (12.4%) | 0.921 (0.677-1253) | 0.638 |
| G | T | C | 42 (5.5%) | 21 (2.8%) | 0.486 (0.284-0.830) | 0.009* |
| G | C | C | 27 (3.6%) | 20 (2.7%) | 0.719 (0.399-1.297) | 0.3 |
| A | T | C | 13 (1.7%) | 14 (1.9%) | 1.046 (0.487-2.244) | 1 |
| G | C | G | 4 (0.5%) | 8 (1.1%) | 1.942 (0.582-6.485) | 0.386 |
| A | C | G | 2 (0.3%) | 1 (0.1%) | 0.486 (0.044-5.369) | 0.62 |
| A | T | G | 2 (0.3%) | 0 (0.0%) | ----- |
Expression quantitative loci (eQTL) analysis of the lung cancer risk SNP rs1045411.
HMGB1 pairwise linkage disequilibrium patterns. The schematic presentation indicates the locations of the SNP polymorphisms and the pairwise linkage disequilibrium (measured as D') in one haploblock. The measure of D' is shown graphically according to a grey scale, where white represents low D' and dark represents high D'.
Discussion
Lung cancer is one of the most malignant cancers and is associated with severe morbidities and high mortality rates worldwide. Neither traditional chemotherapy nor molecular targeted therapy has proven efficacious in the treatment of lung cancer [14]. It is crucial that we perform genetic studies and investigations into signaling mechanisms, which might help to clarify a more appropriate strategy for lung cancer treatment. To the best of our knowledge, this current study is the first to examine the distribution of the rs1360485, rs1045411, rs2249825 and rs1412125 SNPs and their possible association with susceptibility to developing lung cancer. We also investigated the susceptibility to lung cancer when these HMGB1 SNPs were combined with environmental carcinogens. In analyses adjusted for confounding factors, individuals carrying the rs10454111 CT or CC+CT heterozygous polymorphisms had a significantly lower risk of developing lung cancer compared to those carrying the rs10454111 CC homozygous polymorphism (95% CI: 0.385-0.928, p < 0.05; and 95% CI: 0.394-0.924; p < 0.05).
Increasingly, reports are showing that cigarette smoking and alcohol consumption are risk factors for lung cancer [25, 26]. Our results showed that the ratios of cigarette smokers/nonsmokers in controls (88.4:11.6) and patients with lung cancer (60.5:39.5) were relatively normal, similar to the ratios of alcohol consumption/no alcohol consumption in controls and patients. Although our present results showed that smoking and alcohol consumption were risk factors for lung cancer, we failed to find a significant association after controlling for smoking and alcohol consumption in the Chinese Han population (data not shown) because of poor quality of data recording smoking and alcohol consumption in these patients. In addition, some patient survival data were unavailable because patients had only recently enrolled in the study. Further studies are needed using larger populations of patients to confirm the role of HMGB1 polymorphisms in lung cancer. The functional role of HMGB1 in metastasis in patients with lung cancer should also be evaluated.
HMGB1 regulates several functions inside and outside cells, including the immune response, DNA repair, chromatin stabilization, cell apoptosis and gene transcription. It has been suggested that HMGB1 plays a role in lung, oral, colon and breast cancer development [27-29]. Nevertheless, the correlation between HMGB1 polymorphisms and lung cancer development is uncertain. In the current study, we examined four HMGB1 SNPs in 372 lung cancer patients and in 379 healthy controls. Our data suggested that CT or CC+CT at the rs1045411 polymorphism is significantly correlated with a reduction in risk of lung cancer. However, the rs1360485, rs2249825 and rs1412125 polymorphisms do not reduce the lung cancer risk when compared with healthy controls. The polymorphisms in the 3ʹ-flanking region of a gene can control gene expression [30]. Notably, the rs1045411 polymorphism resides in the 3ʹ-flanking region and may therefore affect HMGB1 gene expression. Previous studies have also indicated that the rs1045411 polymorphism reduces the risk of oral squamous cell carcinoma and gastric cancer development [31, 32]. Findings from the GTEx database in the current study confirm that variant T at rs1045411 shows a reduced trend in HMGB1 expression, compared with the wild-type C homozygous genotypes.
The linkage disequilibrium is expressed across the human genome so therefore could be used as a genetic marker to locate adjacent variants that participate in the detection and treatment of disease. Haplotype analyses can also provide data on disease susceptibility [33]. We evaluated the impact of different haplotype combinations of three HMGB1 SNPs (rs1360485, rs1045411 and rs2249825) upon the risk of lung cancer and found that the G/T/C haplotype is associated with a low risk for lung cancer. It is possible that this haplotype is in linkage disequilibrium with other functional polymorphisms that are responsible for the susceptibility to lung cancer.
Taken together, our results demonstrate an association between HMGB1 gene variants and risk of lung cancer, with evidence showing that the HMGB1 rs1045411 polymorphism is associated with a significantly lower risk of lung cancer in the Chinese Han population. This study is the first to report a correlation between HMGB1 polymorphisms and lung cancer risk. Thus, HMGB1 could serve as a genetic prognostic marker for lung cancer therapy.
Acknowledgements
This work was supported by grants from the Ministry of Science and Technology of Taiwan (MOST 103-2628-B-039-002-MY3, MOST 106-2320-B-039-005) and China Medical University Hospital (DMU106-015).
Competing Interests
The authors have declared that no competing interest exists.
References
1. Torre LA, Bray F, Siegel RL, Ferlay J, Lortet-Tieulent J, Jemal A. Global cancer statistics, 2012. CA: a cancer journal for clinicians. 2015;65:87-108
2. Zhu J, Zeng Y, Xu C, Qin H, Lei Z, Shen D. et al. Expression profile analysis of microRNAs and downregulated miR-486-5p and miR-30a-5p in non-small cell lung cancer. Oncology reports. 2015;34:1779-86
3. Fang X, Yin Z, Li X, Xia L, Quan X, Zhao Y. et al. Multiple functional SNPs in differentially expressed genes modify risk and survival of non-small cell lung cancer in chinese female non-smokers. Oncotarget. 2017;8:18924-34
4. Shastry BS. SNP alleles in human disease and evolution. J Hum Genet. 2002;47:561-6
5. Naykoo NA, Dil A, Rasool R, Shah S, Ahangar AG, Bhat IA. et al. Single nucleotide polymorphisms, haplotype association and tumour expression of the vascular endothelial growth factor (VEGF) gene with lung carcinoma. Gene. 2017;608:95-102
6. Wang Y, Yang Y, Zhu Y, Li L, Chen F, Zhang L. Polymorphisms and expression of IL-32: impact on genetic susceptibility and clinical outcome of lung cancer. Biomarkers: biochemical indicators of exposure, response, and susceptibility to chemicals. 2017;22:165-70
7. Lee S, She J, Deng B, Kim J, de Andrade M, Na J. et al. Multiple-level validation identifies PARK2 in the development of lung cancer and chronic obstructive pulmonary disease. Oncotarget. 2016;7:44211-23
8. Mosevitsky MI, Novitskaya VA, Iogannsen MG, Zabezhinsky MA. Tissue specificity of nucleo-cytoplasmic distribution of HMG1 and HMG2 proteins and their probable functions. Eur J Biochem. 1989;185:303-10
9. Bustin M, Lehn DA, Landsman D. Structural features of the HMG chromosomal proteins and their genes. Biochim Biophys Acta. 1990;1049:231-43
10. Bustin M. Regulation of DNA-dependent activities by the functional motifs of the high-mobility-group chromosomal proteins. Mol Cell Biol. 1999;19:5237-46
11. Schmidt AM, Yan SD, Yan SF, Stern DM. The multiligand receptor RAGE as a progression factor amplifying immune and inflammatory responses. J Clin Invest. 2001;108:949-55
12. Tang D, Kang R, Zeh HJ 3rd, Lotze MT. High-mobility group box 1 and cancer. Biochim Biophys Acta. 2010;1799:131-40
13. Sims GP, Rowe DC, Rietdijk ST, Herbst R, Coyle AJ. HMGB1 and RAGE in inflammation and cancer. Annu Rev Immunol. 2010;28:367-88
14. Tang CH, Keng YT, Liu JF. HMGB-1 induces cell motility and alpha5beta1 integrin expression in human chondrosarcoma cells. Cancer Lett. 2012;322:98-106
15. Hou CH, Fong YC, Tang CH. HMGB-1 induces IL-6 production in human synovial fibroblasts through c-Src, Akt and NF-kappaB pathways. J Cell Physiol. 2011;226:2006-15
16. Wang B, Yeh CB, Lein MY, Su CM, Yang SF, Liu YF. et al. Effects of HMGB1 Polymorphisms on the Susceptibility and Progression of Hepatocellular Carcinoma. International journal of medical sciences. 2016;13:304-9
17. Wu HH, Liu YF, Yang SF, Lin WL, Chen SC, Han CP. et al. Association of single-nucleotide polymorphisms of high-mobility group box 1 with susceptibility and clinicopathological characteristics of uterine cervical neoplasia in Taiwanese women. Tumour biology: the journal of the International Society for Oncodevelopmental Biology and Medicine. 2016 [Epub ahead of print]
18. Benna C, Helfrich-Forster C, Rajendran S, Monticelli H, Pilati P, Nitti D. et al. Genetic variation of clock genes and cancer risk: a field synopsis and meta-analysis. Oncotarget. 2017;8:23978-95
19. Li R, Gu J, Heymach JV, Shu X, Zhao L, Han B. et al. Hypoxia pathway genetic variants predict survival of non-small-cell lung cancer patients receiving platinum-based chemotherapy. Carcinogenesis. 2017;38:419-24
20. Wang Y, Li XP, Yin JY, Zhang Y, He H, Qian CY. et al. Association of HMGB1 and HMGB2 genetic polymorphisms with lung cancer chemotherapy response. Clinical and experimental pharmacology & physiology. 2014;41:408-15
21. Lin YJ, Ho TJ, Lin TH, Hsu WY, Huang SM, Liao CC. et al. P-coumaric acid regulates exon 12 splicing of the ATP7B gene by modulating hnRNP A1 protein expressions. Biomedicine (Taipei). 2015;5:10
22. Li TC, Li CI, Liao LN, Liu CS, Yang CW, Lin CH. et al. Associations of EDNRA and EDN1 polymorphisms with carotid intima media thickness through interactions with gender, regular exercise, and obesity in subjects in Taiwan: Taichung Community Health Study (TCHS). Biomedicine (Taipei). 2015;5:8
23. Consortium GT. The Genotype-Tissue Expression (GTEx) project. Nature genetics. 2013;45:580-5
24. Barrett JC, Fry B, Maller J, Daly MJ. Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics. 2005;21:263-5
25. Bialous SA, Sarna L. Lung Cancer and Tobacco: What Is New? The Nursing clinics of North America. 2017;52:53-63
26. Sifaki-Pistolla D, Lionis C, Georgoulias V, Kyriakidis P, Koinis F, Aggelaki S. et al. Lung cancer and tobacco smoking in Crete, Greece: reflections from a population-based cancer registry from 1992 to 2013. Tobacco induced diseases. 2017;15:6
27. Ohmori H, Luo Y, Kuniyasu H. Non-histone nuclear factor HMGB1 as a therapeutic target in colorectal cancer. Expert Opin Ther Targets. 2011;15:183-93
28. Reismann M, Wehrmann F, Schukfeh N, Kuebler JF, Ure B, Gluer S. Carbon dioxide, hypoxia and low pH lead to overexpression of c-myc and HMGB-1 oncogenes in neuroblastoma cells. Eur J Pediatr Surg. 2009;19:224-7
29. Ahn MY, Kwon SM, Cheong HH, Park JH, Lee J, Min SK. et al. Toll-like receptor 7 agonist, imiquimod, inhibits oral squamous carcinoma cells through apoptosis and necrosis. J Oral Pathol Med. 2012;41:540-6
30. Duan ZX, Zhu PF, Dong H, Gu W, Yang C, Liu Q. et al. Functional significance of the TLR4/11367 polymorphism identified in Chinese Han population. Shock. 2007;28:160-4
31. Chen PC, Tang CH, Lin LW, Tsai CH, Chu CY, Lin TH. et al. Thrombospondin-2 promotes prostate cancer bone metastasis by the up-regulation of matrix metalloproteinase-2 through down-regulating miR-376c expression. J Hematol Oncol. 2017;10:33
32. Bao G, Qu F, He L, Zhao H, Wang N, Ji G. et al. Prognostic Significance of Tag SNP rs1045411 in HMGB1 of the Aggressive Gastric Cancer in a Chinese Population. PloS one. 2016;11:e0154378
33. Shifman S, Bronstein M, Sternfeld M, Pisante-Shalom A, Lev-Lehman E, Weizman A. et al. A highly significant association between a COMT haplotype and schizophrenia. American journal of human genetics. 2002;71:1296-302
Author contact
![]() Corresponding authors: Chih-Hsin Tang, PhD, Graduate Institute of Biomedical Science, China Medical University E-mail: chtangcmu.edu.tw Chia-Chia Chao, PhD, Department of Respiratory Therapy, Fu-Jen Catholic University E-mail: 095457fju.edu.tw
Corresponding authors: Chih-Hsin Tang, PhD, Graduate Institute of Biomedical Science, China Medical University E-mail: chtangcmu.edu.tw Chia-Chia Chao, PhD, Department of Respiratory Therapy, Fu-Jen Catholic University E-mail: 095457fju.edu.tw

 Global reach, higher impact
Global reach, higher impact