3.2
Impact Factor
ISSN: 1449-1907
Int J Med Sci 2017; 14(11):1118-1129. doi:10.7150/ijms.19814 This issue Cite
Research Paper
The Crosstalk between HDPSCs and HUCMSCs on Proliferation and Osteogenic Genes Expression in Coculture System
1. School of Stomatology, Shandong University, Jinan, China;
2. Shandong provincial key laboratory of oral tissue regeneration, Jinan, China;
3. Qilu hospital of Shandong University, Jinan, China.
* These two authors contributed equally to this work and should be considered as co-first authors.
Received 2017-2-26; Accepted 2017-6-19; Published 2017-9-4
Abstract
Objectives: The present study established a non-contact coculture system in vitro, aiming to investigate the crosstalk between human dental pulp stem cells (hDPSCs) and human umbilical cord mesenchymal stem cells (hUCMSCs) on proliferation activity and osteogenic genes expression through paracrine.
Materials and methods: The stemness of hDPSCs and hUCMSCs were identified by flow cytometric analysis and multipotential differentiation assays. With the help of transwell inserts, the non-contact coculture system in vitro was established between hDPSCs and hUCMSCs. EdU labeling analysis and Western Blot were used to detect the proliferation activity. The mRNA and protein levels of osteogenic genes were evaluated by RT-PCR and Western Blot. The expression of elements in Akt/mTOR signaling pathway were detected by Western Blot.
Results: Both hDPSCs and hUCMSCs were positive to MSCs specific surface markers and had multi-differentiation potential. The proportion of EdU-positive cells increased and the expression of CDK6 and CYCLIN A were up-regulated in cocultured hDPSCs. Both prior coculture and persistent coculture improved mRNA and protein levels of osteogenic genes in hDPSCs. While in cocultured hUCMSCs, no statistical differences were observed on proliferation and osteogenesis. The phosphorylation of Akt and mTOR was up-regulated in cocultured hDPSCs.
Conclusions: The crosstalk between hDPSCs and hUCMSCs in coculture system increased the proliferation activity and enhanced osteogenic genes expression in hDPSCs. Akt/mTOR signaling pathway might take part in the enhancing effects in both cell proliferation and gene expression.
Keywords: human dental pulp stem cells, human umbilical cord mesenchymal stem cells, proliferation, osteogenesis, crosstalk.
Introduction
As a common disease in adult, periodontitis, which is the inflammation of periodontal supporting tissues caused by local stimulation, can lead to the progressive destruction and loss of periodontium [1]. The ultimate goal of periodontal treatment is to achieve the regeneration of the lost tissues. The rapid-developing tissue engineering provides more approaches to realize the goal, among which the application of adult mesenchymal stem cells (MSCs) has become a hotspot [2]. As “seed cells”, MSCs possess the capacities of self-renewal and multi-lineage differentiation, and have the potential to rebuild tissues with the support of suitable scaffolds and signaling molecules [3]. Up to now, various kinds of MSCs from different sources have been studied by scholars. Among dental-derived MSCs, dental pulp stem cells (DPSCs), which are separated from dental pulp tissues, are proved to promote tissue reparation and regeneration in both animal periodontal defect models [4-6] and human alveolar bone resorption cases [7]. However, the limited source of DPSCs makes it difficult to meet the requirements of cell quantity in a short time for regenerative treatment [8]. While among non-dental-derived stem cells, umbilical cord mesenchymal stem cells (UCMSCs), which come from Wharton's jelly of umbilical cord, are abundant at cell source and exhibit a strong ability to proliferate [9], and were also applied in periodontal regeneration [10]. Nevertheless, a few studies indicated that the osteogenic differentiation potential of UCMSCs was not as strong as dental-derived stem cells, which might be a weakness in the application of UCMSCs for periodontal regeneration [11, 12]. Based on the above facts, it's necessary to explore feasible methods to optimize the biological properties of existing stem cells for oral tissue engineering.
Cells could secrete different factors to regulate the surrounding microenvironment and change features of cells nearby, and this phenomenon is called paracrine [13, 14]. Although the mechanism is not clear enough, several studies have proved that both DPSCs and UCMSCs are capable of altering biological behaviors of surrounding cells through paracrine. For example, UCMSCs could inhibit growth and promote apoptosis of HepG2 hepatocellular carcinoma cells [15]; the viability and the secretion function were significantly increased in damaged mouse hepatocytes when they were indirect cocultured with UCMSCs [16]; UCMSCs could efficiently suppress the proliferation and cytotoxicity of T cells and B cells in vitro [17, 18]. As to DPSCs, some researches proved that they could affect biological characters of lymphocytes [19, 20] and Malassez cells [21] in coculture condition. In recent years, a few scholars found that the paracrine effects also existed among different kinds of stem cells, which meant that the proliferation or differentiation abilities of stem cells could be modified by other kinds of stem cells [22-26]. Since both DPSCs and UCMSCs were proved to have paracrine abilities, we speculated that there was crosstalk between these two kinds of stem cells, and the proliferation or differentiation ability of each other could be altered.
With this in mind, we established a non-contact coculture system between human DPSCs (hDPSCs) and human UCMSCs (hUCMSCs) in vitro, to investigate the crosstalk between them on proliferation activity and osteogenic genes expression. The findings may provide new ideas to optimize characteristics of existing stem cells and offer guidelines for the application of stem cells in tissue engineering.
Materials and Methods
Isolation and culture of hDPSCs and hUCMSCs
All the following procedures were approved by Ethics Committee of Shandong University. The hDPSCs were isolated and cultured according to previous study [27]. In brief, healthy third molars or premolars extracted for orthodontic reason were collected from several healthy voluntary donors (16-25 years old) in Stomatological Hospital of Shandong University. After separated from the split teeth, the dental pulp tissues were cut into small pieces (1mmx1mmx1mm) and digested in the solution containing 1.5 mg/ml collagenase I (Sigma) and 2 mg/ml dispase (Sigma) for 1 h at 37℃. Then the solution containing single cells was obtained by passing through a 70 μm strainer. The hUCMSCs were isolated from umbilical cords of several cesarean-delivered full-term neonates in Qilu Hospital of Shandong University. According to the previous study [9], umbilical cords were gently rinsed with phosphate-buffered saline (PBS) and cut into several short sections. After two umbilical arteries and one umbilical vein were removed, the gel mesenchymal tissue named Wharton's jelly was separated and chopped, and digested in the solution containing 1 mg/ml collagenase I (Sigma) at 37℃ for 8 h with gentle soft shaking. Then the single cells solution was obtained by passing through a 70 μm strainer. Both hDPSCs and hUCMSCs were seeded separately in 10 cm dishes with the complete culture medium containing Dulbecco's Modified Eagle's Medium/F12 (DMEM/F12, Hyclone), 10% fetal bovine serum (FBS) (BI), 100 U/ml penicillin G and 0.1 mg/ml streptomycin (Beyotime) at 37℃ in 5% CO2 incubator. The medium was refreshed every 3 d. Cells at passages 3-6 were used for the following studies.
Phenotyping analysis by flow cytometry
The immunophenotype of hDPSCs and hUCMSCs were analyzed by flow cytometry at passages 3. Briefly, after being trypsinized and washed with PBS, cells were incubated with monoclonal antibodies conjugated with fluorescent dyes in the dark at 4 ℃ for 20 min. The following antibodies were used: CD90 FITC, CD44 PE, CD105 PerCP-Cy, CD73 APC, PE-negative cocktail (CD34PE, CD11b PE, CD19 PE, CD45 PE and HLA-DR PE) [28, 29]. Then the cells were washed with PBS and analyzed by flow cytometry (BD Biosciences). The results were analyzed by software FlowJo.
Multipotent differentiation assays
The hDPSCs and hUCMSCs were seeded in 6-well dishes at 1 × 10^5 cells/well and cultured in complete culture medium. When cells reached 90% confluence, the medium was changed to induced medium. For osteogenic differentiation assays, cells were exposed to osteogenic medium (DMEM/F12 containing 10% FBS [BI], 100 U/ml penicillin G and 0.1 mg/ml streptomycin [Beyotime], 10nmol/l dexamethasone [Solarbio], 10 mmol/l β-glycerophosphate [Biosharp], 50 mg/l ascorbic acid [Solarbio]) [6]. The medium was refreshed every 3 d. After 4 weeks, Alizarin Red staining (Sigma) was used to detect the formation of mineralized nodule. For adipogenic differentiation assays, cells were exposed to adipogenic medium (DMEM/F12 containing 10% FBS [BI], 100 U/ml penicillin G and 0.1 mg/ml streptomycin [Beyotime], 2 μmol/l dexamethasone [Solarbio], 0.2 mmol/l indomethacin [Sigma], 0.01g/l insulin [Sigma], 0.5 mmol/l isobutyl-methylxanthine [IBMX] [Sigma]) [30]. The medium was refreshed every 3 d. After 2 weeks, the cells were stained with oil red O (Cyagen).
Establish the hDPSCs-hUCMSCs coculture system in vitro
The coculture systems were established through 6-well plate and 3 μm pore size transwell inserts (Corning) [21]. The hDPSCs were seeded in 6-well plate at the amount of 5×10 ^4 cells/well and the same quantity of hUCMSCs were seeded in the transwell inserts located in neighboring wells. After cells attached to the wall firmly (about 24 h), the transwell inserts with hUCMSCs were moved to the wells containing hDPSCs so that the hDPSCs-hUCMSCs coculture system was established, and the hDPSCs in this coculture system were regarded as coculture groups. In control groups, both plate wells and transwell inserts were seeded with hDPSCs. Another coculture system exchanged the position of hDPSCs and hUCMSCs, which meant hDPSCs were seeded in the transwell inserts and hUCMSCs were seeded in lower 6-well plate, so that the hUCMSCs in this coculture system were regarded as coculture groups. In control groups, both wells and transwell inserts were seeded with hUCMSCs. For convenience, aforementioned cells located in lower plates would be regarded as research objects in the following experiments.
Analysis of proliferation activity
After the coculture groups and the control groups were established, cells were cultured in the complete culture medium at 37℃in 5% CO2 for 3 d and 5 d. Then 5-ethynyl-2'-deoxyuridine (EdU) labeling [31] staining and Western Blot were used to analyze the proliferation activity. According to the instructions of EdU detection kit (Ribobio), cells were incubated with 50 μm EdU labeling medium at 37℃ for 2 h. After immobilization, staining with Apollo®567 solution and Hoechst33342 solution, cells were observed under the fluorescence microscope and more than 6 random fields per well were captured. Image-Pro Plus (IPP) was used to calculate the percentage of EdU-positive cells (identified by Apollo®567 staining) in total cells (identified by Hoechst33342 staining). The expression levels of CDK6 and CYCLIN A in hDPSCs and hUCMSCs were detected by Western Blot, and the procedures were mentioned in a later section.
Analysis of osteogenic genes expression
To study the effects of prior coculture on osteogenic differentiation of hDPSCs and hUCMSCs, the coculture groups and the control groups were established in 6-well plates, and all cells were cultured in the complete culture medium for 7 d. Subsequently, the transwell inserts were relieved, and all cells located in lower palate were induced in the osteogenic medium mentioned above for another 7 d. Then the osteogenesis related mRNAs including collagen type I (COL I), runt-related transcription factor 2 (RUNX2), osteocalcin (OCN), were analyzed by quantitative real-time PCR (QRT-PCR). The osteogenesis related proteins including COL I, RUNX2, and osteopontin (OPN) were analyzed by Western Blot.
To study the effects of persistent coculture on osteogenic differentiation of hDPSCs and hUCMSCs, when the coculture groups and the control groups were established, all cells were exposed to the osteogenic medium directly. After 7 d and 14 d, the expression levels of aforementioned mRNA and proteins in hDPSCs and hUCMSCs were analyzed by QRT-PCR and Western Blot.
Total protein isolation and Western Blot
The cells were washed 3 times with ice-cold PBS and lysed with RIPA buffer (solarbio) containing phosphatase inhibitor on ice for 30 min. After ultrasonic lysis and centrifugation at 12 000 rpm at 4 ℃ for 15 min, the supernatant lysate with proteins was collected. Then proteins were separated by SDS-PAGE and transferred to polyvinylidene difluoride membranes. The membrane was blocked with 5% nonfat-dried milk solution at room temperature for 1 h and incubated with primary antibodies overnight at 4 ℃. The following primary antibodies were used: CDK6 (1:1000, CST), CYCLIN A (1:1000, CST), Akt (pan) (1:1000, CST), phospho-Akt (Thr308) (p-Akt) (1:1000 CST), mTOR (1:1000, CST), phospho-mTOR (p-mTOR) (1:1000, CST), COL I (1:1000, abcam), Runx2 (1:1000, CST), OPN (1:1000, Santa), glyceraldehyde-3-phosphate dehydrogenase (GAPDH) (1:10000, Proteintech). Then the membrance was incubated with anti-rabbit or anti-mouse secondary antibody conjugated with horseradish peroxidase for 1 h at room temperature. Finally, the protein bands were detected by enhanced chemiluminescence (Millipore) and band intensities were analyzed by Image J software.
Total RNA isolation and QRT-PCR
Total RNA was isolated from cells by RNAios Plus reagent (Takara) according to the instructions. Total 1 μg RNA (in 10 μl reaction volume) was reverse transcribed to cDNA using PrimeScript ™ RT reagent Kit with gDNA Eraser (Takara). QRT-PCR was carried out in a reaction volume of 10 μl of SYBR® Premix Ex Taq™ (Takara) by Roche LightCycler® 480II as follows: an initial denaturation at 95℃ for 30 s, followed by 55 cycles of 95℃ for 5 s, 60℃ for 35 s, and extension at 72℃ for 1 min, finally at 40℃ for 30 s. The results were normalized against the internal control GAPDH and calculated by the 2-△△Ct method (△△Ct=(CT target-CT GAPDH) cocuture-(CT target-CT GAPDH) control). The expression levels of COL I, Runx2 and OCN were analyzed and primers used in this study were as follows:
COL I forward 5'-GCTGATGATGCCAATGTGGTT-3',
COL I reverse 5'-CCAGTCAGAGTGGCACATCTTG-3',
Runx2 forward 5'-GTTTCACCTTGACCATAACCGT-3',
Runx2 reverse 5'-GGGACACCTACTCTCATACTGG-3',
OCN forward 5'- AATCCGGACTGTGACGAGTTG-3',
OCN reverse 5'- CAGCAGAGCGACACCCTAGAC-3'.
Statistical analysis
All experiments were performed independently and at least three times and all data were presented as mean±standard deviations. Independent-sample t tests or t' tests were used to determine statistical differences between coculture and control values. The statistical analyses were conducted by SPSS 19.0 and differences at P < 0.05 were considered to be statistically significant.
Results
Isolation, culture and identification of hDPSCs and hUCMSCs
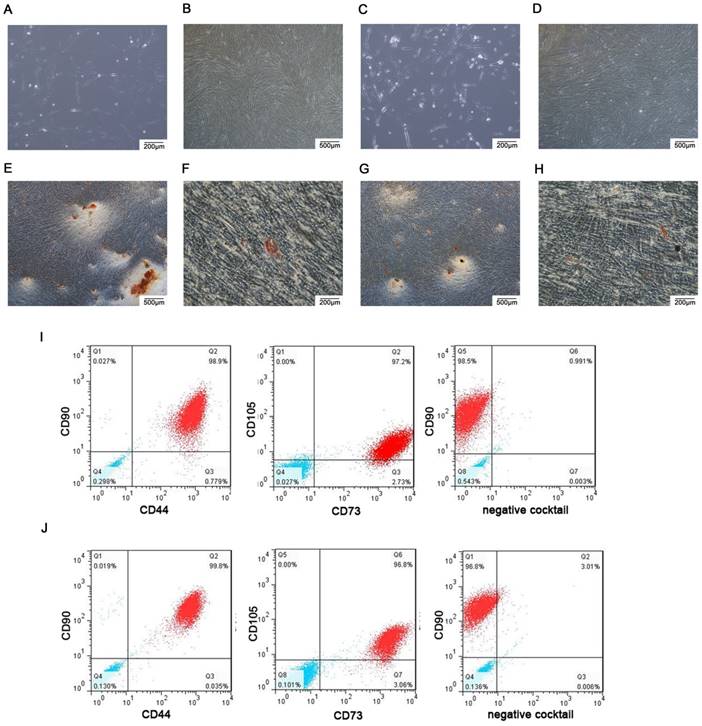
Both hDPSCs and hUCMSCs were observed under phase contrast microscope. The shapes of hDPSCs were similar to fibroblasts which presented spindle-shaped morphology (Fig. 1A) and could form spiral arrangement (Fig. 1B). The hUCMSCs were fusiform or polygonal-shaped (Fig. 1C) and also tended to form spiral arrangement (Fig. 1D). For multipotent differentiation assays, mineralized nodules were detected as red nodules in both hDPSCs (Fig. 1E) and hUCMSCs (Fig. 1G) by Alizarin red staining after osteogenic induction, and lipid droplets were demonstrated as red drops in both hDPSCs (Fig. 1F) and hUCMSCs (Fig. 1H) by oil red O staining after adipogenic induction. The results of flow cytometry indicated that both hDPSCs (Fig. 1I) and hUCMSCs (Fig.1J) were positive to MSCs specific surface markers (CD90, CD44, CD105, CD73), but negative to hematopoietic and endothelial cell-specific markers (CD34, CD11b, CD19, CD45, HLA-DR). These immunophenotype results confirmed hDPSCs and hUCMSCs as MSCs.
Establish the hDPSCs -hUCMSCs coculture system in vitro
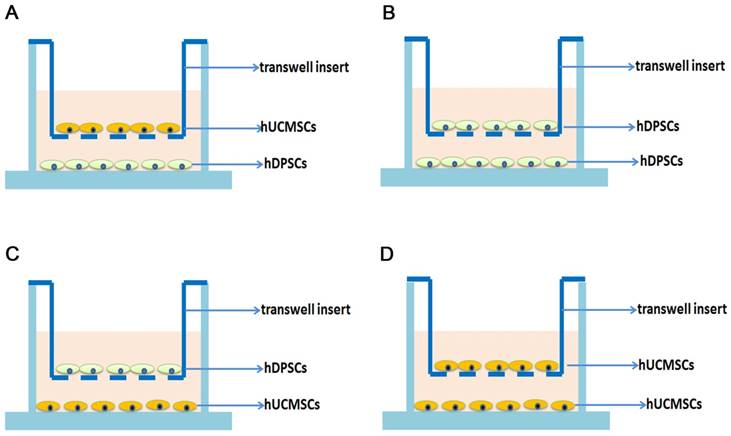
The coculture groups and control groups were established as shown in figure 2. To study the impact of coculture system on hDPSCs, hDPSCs cocultured with hUCMSCs were regarded as coculture groups (Fig. 2A) and the hDPSCs cultured without hUCMSCs were regarded as control groups (Fig. 2B). To study the impact of coculture system on hUCMSCs, hUCMSCs cocultured with hDPSCs were regarded as coculture groups (Fig. 2C) and the hUCMSCs cultured without hDPSCs were regarded as control groups (Fig. 2D).
Effects of coculture on the proliferation of hDPSCs and hUCMSCs
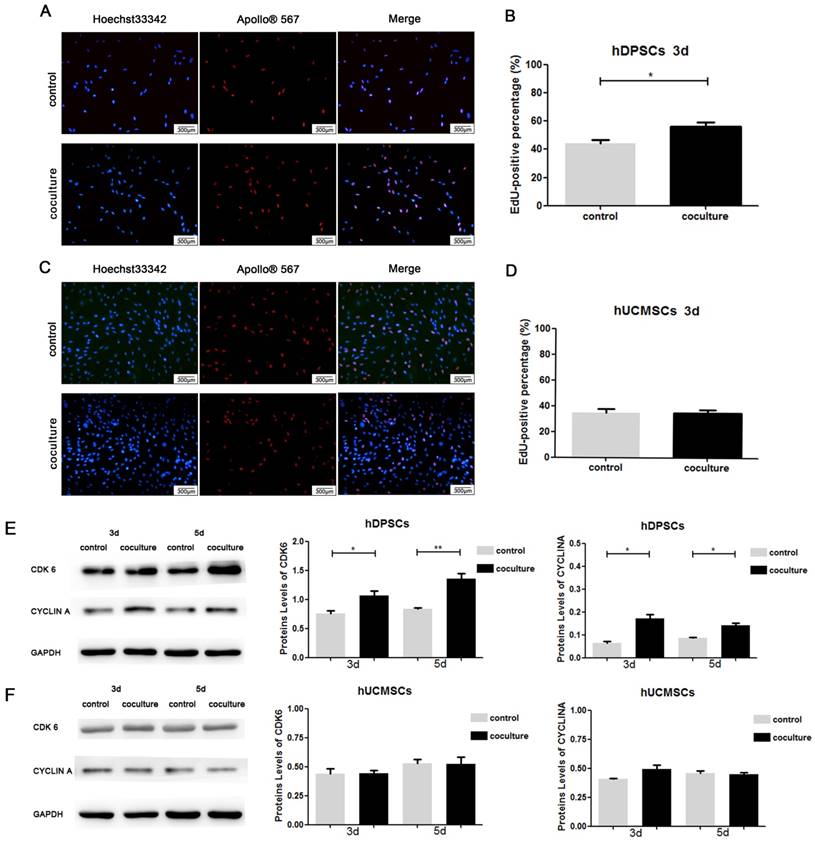
After coculture for 3 d, the proliferation activity of hDPSCs and hUCMSCs were measured by EdU staining. For hDPSCs, the average percentage of EdU-positive cells in coculture groups revealed a statistically significant increase compared to control groups on 3d (Fig.3A, 3B) (P<0.05). Since EdU-positive staining represented DNA replication of cell cycle, it suggested that coculture system promoted hDPSCs to proliferate. While in hUCMSCs, the average percentages of EdU-positive cells were similar in coculture groups and control groups, and the difference was not statistically significant (Fig. 3C, 3D) (P>0.05). The Western Blot bands and analysis of bands intensities after coculture for 3 d and 5 d were shown in Fig. 3E and Fig. 3F. The expression of CDK6 and CYCLIN A of coculture groups was higher than that of control groups in hDPSCs (Fig. 3E) (P<0.01), which indicated the acceleration of cell cycle in cocultured hDPSCs. For hUCMSCs, there was little difference in the expression levels of CDK6 and CYCLIN A between the coculture groups and control groups (Fig.3F) (P>0.05). These results demonstrated that coculture for 3 d and 5 d could improve the proliferation activity of hDPSCs, but had little impact on that of hUCMSCs.
Effects of coculture on osteogenic genes expression in hDPSCs and hUCMSCs
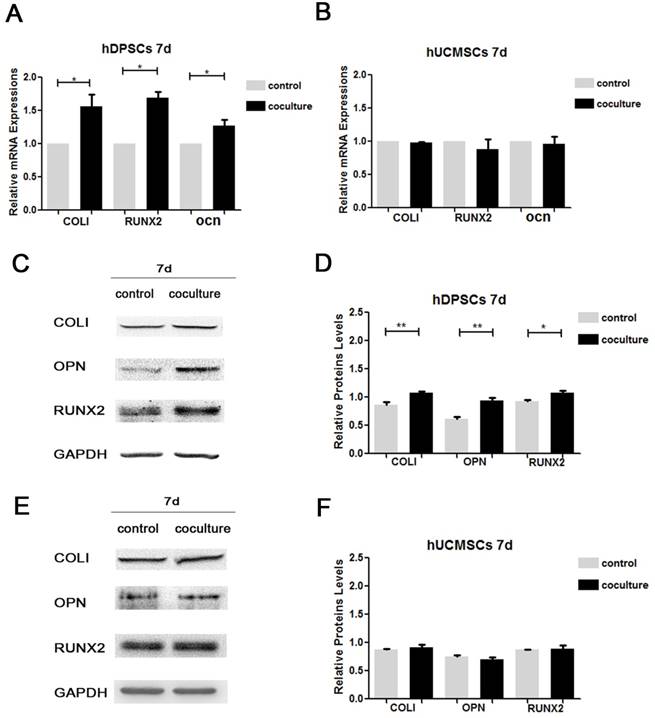
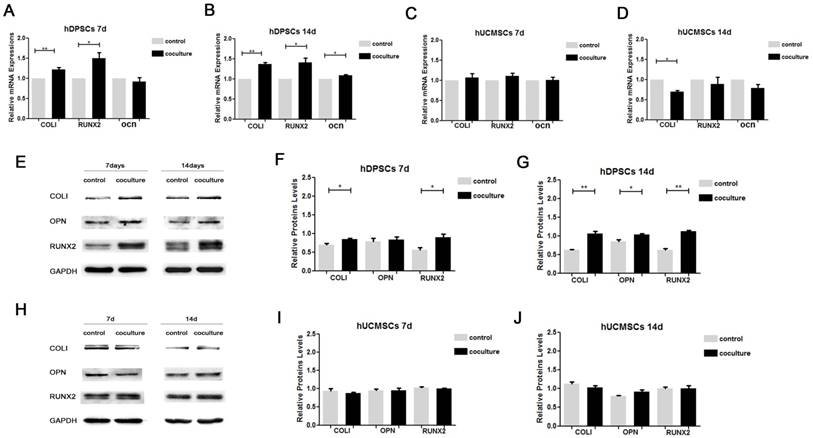
The detection results about effects of prior coculture on osteogenic differentiation were shown in Figure 4. For hDPSCs, the QRT-PCR results showed that mRNA of Col I, RUNX2 and OCN expressed more in coculture groups than in control groups, which had significant differences (Fig. 4A). For hUCMSCs, no statistical difference was observed in the mRNA expression of Col I, RUNX2 and OCN between coculture groups and control groups (Fig. 4B) (P>0.05). To further investigate, the expression of osteogenic proteins, including COLI, RUNX2 and OPN were detected by Western Blot, and the bands intensities were analyzed by Image J software. Various degrees of elevation on these proteins were evaluated in coculture groups compared with control groups in hDPSCs (Fig.4C, 4D), while the expression levels of these proteins showed little difference between coculture and control groups in hUCMSCs (Fig.4E, 4F). To sum up, our experiment results suggested that the prior hDPSCs-hUCMSCs coculture enhanced the expression of osteogenic mRNA and proteins in hDPSCs, but had little impact on the osteogenic differentiation of hUCMSCs.
Culture and identification of hDPSCs and hUCMSCs. Cells were observed under phase contrast microscope. A.B. The hDPSCs (P3) presented spindle-shaped morphology and formed spiral arrangement. C.D. The hUCMSCs (P3) were fusiform or polygonal-shaped and formed spiral arrangement. E. Osteogenic differentiation of hDPSCs was demonstrated as red mineralized nodules by Alizarin red staining. F. Adipogenic differentiation of hDPSCs was demonstrated as red oil drops by oil red O staining. G. Osteogenic differentiation of hUCMSCs was demonstrated as red mineralized nodules by Alizarin red staining. H. Adipogenic differentiation of hUCMSCs was demonstrated as red oil drops by oil red O staining. I.J. Both hDPSCs (I) and hUCMSCs (J) were positive to MSC specific surface markers (CD90, CD44, CD105, CD73), but negative to hematopoietic and endothelial cell-specific markers (CD34, CD11b, CD19, CD45, HLA-DR). (The blue drops respected isotype control)
Establish the hDPSCs -hUCMSCs coculture system. The transwell coculture system was established in 6-well plate with same quantity of hDPSCs and hUCMSCs. To study the influence of coculture system on hDPSCs, hDPSCs in the coculture system were regarded as coculture groups (A), and hDPSCs without coctulture were regarded as control groups (B). To study the influence of coculture system on hUCMSCs, hUCMSCs cocultured with hDPSCs were regarded as coculture groups (C) and the hUCMSCs without coculture were regarded as control groups (D).
The detection results about effects of persistent coculture on osteogenic differentiation were shown in Figure 5. For hDPSCs, it was found that the mRNA expression levels of Col I and RUNX2 in coculture groups were up-regulated after coculture for 7 and 14 d, and OCN mRNA was up-regulated after coculture for 14 d (Fig. 5A, 5B). While in hUCMSCs, the mRNA of RUNX2 and OCN in coculture groups had no statistical difference with control groups on day 7 or 14, and COLI mRNA reduced on day 14 (Fig. 5C, 5D). The results of Western Blot and intensities analysis were quite consistent with that of QRT-PCR, which meant that the hDPSCs-hUCMSCs coculture system promoted protein expressions of COLI, RUNX2 and OPN in hDPSCs on day 7 and 14 (Fig. 5E, 5F, 5G), but didn't change the expression pattern of them in hUCMSCs (Fig. 5H, 5I, 5J).
Effects of coculture on Akt/mTOR signaling pathway of hDPSCs
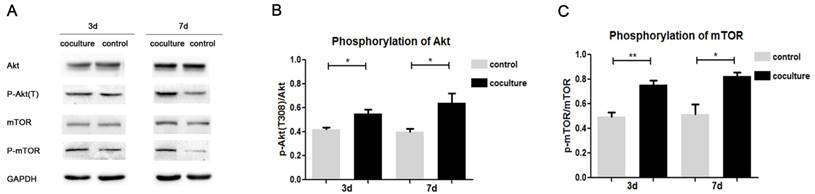
Above results showed that the proliferation and osteogenic differentiation abilities of hDPSCs were enhanced in coculture system, but the mechanism was not clear. After hDPSCs were cocultured for 3 and 7 d, the expression of elements in Akt/mTOR signaling pathway were detected by Western Blot, including Akt, p-Akt(T308), mTOR, p-mTOR. As shown in Fig. 6A, the expression of p-Akt and p-mTOR were up-regulated both on day 3 and 7. Intensities analysis results indicated that the phosphorylation degree of Akt and mTOR increased with the extension of coculture (Fig. 6B, 6C), which suggested Akt/mTOR signaling pathway was actived in cocultured hDPSCs.
Discussion
Communication between cells takes part in the regulation of cell biological behaviors [32]. Simply speaking, it includes direct communication under cell contact and indirect communication through chemical, physical or other signals without contact. Different mesenchymal stem cells have been proved to secrete paracrine factors (such as growth factors, cytokines, and hormones) to influence characteristics of other cells, which was regarded as a form of indirect communication [14]. Either hUCMSCs or hDPSCs had been proved to influence biological characteristics of tumor cells [15], somatic cells [16, 33, 34] and immune cells [17, 18] through paracrine, but little was known about the crosstalk between these two kinds of stem cells. In this study, we established the non-contact coculture system in vitro with the help of transwell inserts to study the indirect communication between hDPSCs and hUCMSCs. The 3μm pore size microporous membrane of transwell inserts could separate cells from upper and lower spaces, but allow small molecules to pass through freely. Compared with other methods of studying cell communication such as conditioned media [35] and direct coculture [36], transwell inserts are more convenient to operate, making a more realistic simulation of cell communication environment, and could study the changes of two kinds of cells separately. It was the first time to explore the indirect communication between hDPSCs and hUCMSCs, and our experiment results indicated that these two kinds of stem cells indeed had crosstalk on proliferation and differentiation.
Effects of coculture on the proliferation of hDPSCs and hUCMSCs A. EdU staining of hDPSCs in cocultured groups and control groups after 3 d. The nucleus of EdU-positive cells were identified by Apollo®567 staining and total cell's nucleus were identified by Hoechst33342 staining. B. For hDPSCs, the average percentage of EdU-positive cells in control groups was 44.38±2.26%, while that in cocultured hDPSCs groups was 56.02±3.23%. C. EdU staining of hUCMSCs in cocultured groups and control groups after 3 d. D. For hUCMSCs, the average percentage of EdU-positive cells in control groups was 34.85±2.78%, while that in cocultured hUCMSCs was 34.76±2.54%. E. The expression of CDK6 and CYCLIN A in hDPSCs was detected by Western Blot after coculture for 3 d and 5 d. Grey value of protein bands in hDPSCs was measured based on three independent experiments. Data were normalized by GAPDH. F. The expression of CDK6 and CYCLIN A in hUCMSCs was detected by Western Blot after coculture for 3 d and 5 d. Grey value of protein bands in hUCMSCs was measured based on three independent experiments. Data were normalized by GAPDH. (*p<0.05, **p<0.01)
Effects of prior coculture for 7 d on osteogenic genes expression of hDPSCs and hUCMSCs. A. The expression of Col I, RUNX2 and OCN mRNA in prior cocultured and control hDPSCs. B. The expression of Col I, RUNX2 and OCN mRNA in prior cocultured and control hUCMSCs. C. The expression of Col I, OPN and RUNX2 proteins in prior cocultured and control hDPSCs. D. Grey value of protein bands in hDPSCs was measured based on three independent experiments. Data were normalized by GAPDH. E. The expression of Col I, OPN and RUNX2 proteins in prior cocultured and control hUCMSCs. F. Grey value of protein bands in hUCMSCs was measured based on three independent experiments. Data were normalized by GAPDH. (*p<0.05, **p<0.01)
To detect the proliferation activity, EdU labeling analysis was used for it reflected the ability of DNA replication in cell cycle [31], and the results proved that higher proportion of hDPSCs engaged in DNA replication in cocultured group than in control group. At the same time, we detected the expression of CDK6 and CYCLIN A by Western Blot, which were essential regulators of cell cycle progression. CDK6 was important for G1 phase progression and promoted G1/S transition in cell cycle [37], while CYCLIN A was involved in both S phase and the G2/M transition [38]. Western Blot results displayed that the expression levels of CDK6 and CYCLIN A were enhanced in cocultured hDPSCs, suggesting the process of cell cycle were accelerated. All above results indicated that paracrine factors secreted by hUCMSCs promoted proliferation of hDPSCs. This finding was similar with previous study, whose results proved that indirect coculture with hUCMSCs enhanced the proliferation ability of injured hepatocytes [16]. In the application of periodontal tissue engineering, the acquirement in amount of stem cells was quite urgent, and our study proved coculture with hUCMSCs to be a useful means to promote the expansion of hDPSCs in vitro. As to hUCMSCs, no obvious alteration in EdU labeling or CDK6 and CYCLIN A expression were observed between cocultured group and control group, which suggested the proliferation of hUCMSCs changed little when they were cocultured with hDPSCs for 3 to 5d.
Effects of persistent coculture for 7 d and 14 d on osteogenic genes expression of hDPSCs and hUCMSCs. A.B. The expression of Col I, RUNX2 and OCN mRNA in persistent cocultured and control hDPSCs after induction for 7 d and 14 d. C.D. The expression of Col I, RUNX2 and OCN mRNA in persistent cocultured and control hUCMSCs after induction for 7 d and 14 d. E. The expression of Col I, OPN and RUNX2 proteins in persistent cocultured and control hDPSCs after induction for 7 d and 14 d. F.G. Grey value of protein bands in hDPSCs after induction for 7 d and 14 d was measured based on three independent experiments. Data were normalized by GAPDH. H. The expression of Col I, OPN and RUNX2 proteins in persistent cocultured and control hUCMSCs after induction for 7 d and 14 d. I.J. Grey value of protein bands in hUCMSCs after induction for 7 d and 14 d was measured based on three independent experiments. Data were normalized by GAPDH. (*p<0.05, **p<0.01)
Effects of coculture on Akt/mTOR signaling pathway of hDPSCs. A. After coculture for 3 d and 7 d, the expression of Akt, p-Akt(T308), mTOR, and p-mTOR proteins was detected in hDPSCs. B.C. Phosphorylation degree of Akt and mTOR in hDPSCs after cococulture for 3 d and 7 d was measured based on three independent Western Blot results. (*p<0.05, **p<0.01)
To explore the osteogenesis potential of hDPSCs and hUCMSCs, the expressions of related genes such as COLI, RUNX2, OCN and related proteins such as COLI, RUNX2, OPN were detected. Among these items, COL I is considered to be an early marker of osteoblast differentiation [39]; Runx2 is an osteogenesis specific transcription factor which can promote the expression of osteogenenic genes in MSCs [40-42]; OPN [43] and OCN [44] are important markers of mineralization. In order to investigate the paracrine pattern and effects on osteogenesis, we established two kinds of coculture system — the prior and persistent coculture system. In the prior coculture studies, hDPSCs and hUCMSCs were cocultured for 7 d in complete culture medium, during which the paracrine factors released by cells could affect osteogenic differentiation potential of each other. After that, the coculture environment was removed and cells were induced in osteogenic medium for another 7 d. The detection results showed that prior coculture with hUCMSCs promoted the expression of osteogenic genes in hDPSCs effectively. While in persistent coculture studies, cells were exposed to coculture and osteogenic induction environment simultaneously, and we wondered whether cells would change their paracrine pattern or not when they were located in osteogenic medium. The final results showed that the contribution of hUCMSCs paracrine to osteogenesis potential of hDPSCs didn't disappear in osteogenic medium, which meant persistent coculture could promote the expression of osteogenic genes in hDPSCs as well. What's more, with time prolonging, the expression level of osteogenic genes remained higher in cocultured hDPSCs than in control hDPSCs throughout the experiment time. These findings suggested that paracrine effects of hUCMSCs on osteogenesis potential of hDPSCs were persistent and relatively stable both in complete culture medium and osteogenic culture medium. In fact, some previous studies have proved the relationship between coculture and multi-differentiation ability among different stem cells. For example, human amnion-derived mesenchymal stem cells [26] and adipose-derived stromal cells [45] could promote osteogenic differentiation of bone marrow mesenchymal stem cells in coculture systems, periodontal ligament stem cells could enhance osteogenic gene expression in de-differentiated fat cells though paracrine [46], conditioned media from craniofacial bone marrow stem cells could influence mineralization of periodontal ligament stem cells [23] and so on. Our experiment was the first to identify that the paracrine of hUCMSCs enhanced osteogenesis potential of hDPSCs, and coculture was considered to be a practical method to promote osteogenesis of stem cells in tissue engineering. Turn attention to hUCMSCs, neither prior nor persistent coculture brought significant changes to the expression of COLI, RUNX2, OPN or OCN in hUCMSCs, which suggested that coculture with hDPSCs had little effects on osteogenesis of hUCMSCs under the aforementioned conditions.
The mechanism of non-contact coculture system promoted hDPSCs to proliferate and express osteogenic gene could be complicated, for the paracrine factors secreted by hUCMSCs were not clear enough. In previous studies, the cytokines released in paracrine were various in kinds, and could activate different signaling pathways related with different biological characteristics in effector cells. For instance, UCMSCs inhibited growth and promoted apoptosis of HepG2 hepatocellular carcinoma cells by downregulating the mRNA and protein expression of α‑fetoprotein (AFP), Bcl‑2 and Survivin in HepG2 cells [15]; UCMSCs suppressed proliferation and differentiation of B cell, which may be related with the change in phosphorylation pattern of Akt and p38 pathways of B cell [18]; human amnion-derived mesenchymal stem cells promoted osteogenic differentiation of human bone marrow mesenchymal stem cells by influencing the ERK1/2 signaling pathway [26]. Our study found that the phosphorylations of Akt and mTOR were increased in hDPSCs after they were cocultured with hUCMSCs, and these effects were enhanced with the extension of coculture time. Since Akt/mTOR signaling pathway was proved to be related with proliferation [47-49] and osteogenic differentiation [50, 51] in many studies, we suggested that the change of Akt/mTOR signaling pathway was associated with the improvement of proliferation and differentiation in cocultured hDPSCs. Although the factors secreted by hUCMSCs need to be further studied, our experiments indicated that Akt/mTOR signaling pathway might take part in the regulation effects of hUCMSCs paracrine on hDPSCs.
Remarkably, although results in this study showed that the coculture had little influence on proliferation and osteogenesis potential of hUCMSCs, it was worth noting that we didn't deny the paracrine effects of hDPSCs on hUCMSCs, but stressed on the crosstalk between these two kinds of stem cells under certain coculture condition (the initial amount of hDPSCs and hUCMSCs was 1:1, and coculture time was 3 d, 5d, 7d or 14 d) in aspect of proliferation and osteogenesis potential. Since different coculture condition would change the amount of paracrine cyctokines and length of paracrine time [15, 26], the crosstalk between hDPSCs and hUCMSCs could be various in forms and effects. For further understading, more investigation and analysis were needed.
The requirement in the amount and differentiation potential of seed cells in tissue engineering is still a problem for the application of adult stem cells [52]. Our studies found that hDPSCs-hUCMSCs coculture system could promote the proliferation and osteogenic genes expression in hDPSCs, provided hDPSCs and hUCMSCs with better abilities for the application in tissue engineering. Also, the non-contact coculture system could be used as an exploring method for the optimization of biological characteristics of seed cells.
Conclusions
This study demonstrated that the hDPSCs-hUCMSCs non-contact coculture system in vitro increased the proliferation activity and enhanced osteogenic genes expression in hDPSCs, while little effect was observed on that of hUCMSCs. Also, Akt/mTOR signaling pathway might take part in the enhancing effects of hDPSCs-hUCMSCs coculture system on hDPSCs. Our findings may provide new ideas to optimize the characteristics of hDPSCs and hUCMSCs in tissue engineering as seed cells, and offer guidelines for the application of stem cells in tissue regeneration.
Acknowledgements
This work was supported by grants from the National Natural Science Foundation of China (Grant No. 81300885 and 81402150), Shandong Provincial key research and development program (Grant No. 2015GSF118122, 2015GSF118183, 2016GSF201115 and 2016GSF201220), China Postdoctoral Science Foundation (Grant No. 2017M610432), Young Scholars Program of Shandong University (Grant No. 2015WLJH53) and the Construction Engineering Special Fund of Taishan Scholars (Grant No. ts201511106).
Competing Interests
The authors have declared that no competing interest exists.
References
1. Chapple IL, Van der Weijden F, Doerfer C. et al. Primary prevention of periodontitis: managing gingivitis. J Clin Periodontol. 2015;42(Suppl 16):S71-6
2. Izumi Y, Aoki A, Yamada Y. et al. Current and future periodontal tissue engineering. Periodontol 2000. 2011;56:166-187
3. Ashri NY, Ajlan SA, Aldahmash AM. Dental pulp stem cells. Biology and use for periodontal tissue engineering. Saudi Med J. 2015;36:1391-1399
4. Jahanbin A, Rashed R, Alamdari DH. et al. Success of Maxillary Alveolar Defect Repair in Rats Using Osteoblast-Differentiated Human Deciduous Dental Pulp Stem Cells. J Oral Maxillofac Surg. 2016;74:829 e1-9
5. Huojia M, Wu Z, Zhang X. et al. Effect of Dental Pulp Stem Cells (DPSCs) in Repairing Rabbit Alveolar Bone Defect. Clin Lab. 2016;61:1703-1708
6. Liu HC, E LL, Wang DS. et al. Reconstruction of alveolar bone defects using bone morphogenetic protein 2 mediated rabbit dental pulp stem cells seeded on nano-hydroxyapatite/collagen/poly(L-lactide). Tissue Eng Part A. 2011;17:2417-2433
7. d'Aquino R, De Rosa A, Lanza V. et al. Human mandible bone defect repair by the grafting of dental pulp stem/progenitor cells and collagen sponge biocomplexes. Eur Cell Mater. 2009;18:75-83
8. Requicha JF, Viegas CA, Munoz F. et al. Periodontal tissue engineering strategies based on nonoral stem cells. Anat Rec (Hoboken). 2013;297:6-15
9. Schugar RC, Chirieleison SM, Wescoe KE. et al. High harvest yield, high expansion, and phenotype stability of CD146 mesenchymal stromal cells from whole primitive human umbilical cord tissue. J Biomed Biotechnol. 2009;2009:789526
10. Li Y, Hou R, Wang Y. et al. Fundamental study of application of umbilical cord mesenchymal stem cells to the periodontium to aid healing after autotransplantation of teeth. Br J Oral Maxillofac Surg. 2014;52:501-506
11. Ren H, Sang Y, Zhang F. et al. Comparative Analysis of Human Mesenchymal Stem Cells from Umbilical Cord, Dental Pulp, and Menstrual Blood as Sources for Cell Therapy. Stem Cells Int. 2016;2016:3516574
12. Yu S, Long J, Yu J. et al. Analysis of differentiation potentials and gene expression profiles of mesenchymal stem cells derived from periodontal ligament and Wharton's jelly of the umbilical cord. Cells Tissues Organs. 2012;197:209-223
13. Ahmed M, Ffrench-Constant C. Extracellular Matrix Regulation of Stem Cell Behavior. Curr Stem Cell Rep. 2016;2:197-206
14. Spees JL, Lee RH, Gregory CA. Mechanisms of mesenchymal stem/stromal cell function. Stem Cell Res Ther. 2016;7:125
15. Tang YM, Bao WM, Yang JH. et al. Umbilical cord-derived mesenchymal stem cells inhibit growth and promote apoptosis of HepG2 cells. Mol Med Rep. 2016;14:2717-2724
16. Zhang S, Chen L, Liu T. et al. Human umbilical cord matrix stem cells efficiently rescue acute liver failure through paracrine effects rather than hepatic differentiation. Tissue Eng Part A. 2012;18:1352-1364
17. Liu X, Feng T, Gong T. et al. Human Umbilical Cord Mesenchymal Stem Cells Inhibit the Function of Allogeneic Activated Vgamma9Vdelta2 T Lymphocytes In Vitro. Biomed Res Int. 2015;2015:317801
18. Che N, Li X, Zhou S. et al. Umbilical cord mesenchymal stem cells suppress B-cell proliferation and differentiation. Cell Immunol. 2012;274:46-53
19. Ding G, Niu J, Liu Y. Dental pulp stem cells suppress the proliferation of lymphocytes via transforming growth factor-beta1. Hum Cell. 2015;28:81-90
20. Demircan PC, Sariboyaci AE, Unal ZS. et al. Immunoregulatory effects of human dental pulp-derived stem cells on T cells: comparison of transwell co-culture and mixed lymphocyte reaction systems. Cytotherapy. 2011;13:1205-1220
21. Farea M, Husein A, Halim AS. et al. Cementoblastic lineage formation in the cross-talk between stem cells of human exfoliated deciduous teeth and epithelial rests of Malassez cells. Clin Oral Investig. 2015;20:1181-1191
22. Jiang ZM, Ji PH, Liu J. et al. [Induction of mouse embryonic stem cells forming odontoblast-like cells by co-culture with pulp fibroblast]. Shanghai Kou Qiang Yi Xue. 2006;15:653-656
23. Jin Z, Feng Y, Liu H. Conditioned media from differentiating craniofacial bone marrow stromal cells influence mineralization and proliferation in periodontal ligament stem cells. Hum Cell. 2016;29:162-175
24. Zhang H, Liu S, Zhu B. et al. Composite cell sheet for periodontal regeneration: crosstalk between different types of MSCs in cell sheet facilitates complex periodontal-like tissue regeneration. Stem Cell Res Ther. 2016;7:168
25. Xie H, Liu H. A novel mixed-type stem cell pellet for cementum/periodontal ligament-like complex. J Periodontol. 2012;83:805-815
26. Wang Y, Jiang F, Liang Y. et al. Human Amnion-Derived Mesenchymal Stem Cells Promote Osteogenic Differentiation in Human Bone Marrow Mesenchymal Stem Cells by Influencing the ERK1/2 Signaling Pathway. Stem Cells Int. 2016;2016:4851081
27. Gronthos S, Mankani M, Brahim J. et al. Postnatal human dental pulp stem cells (DPSCs) in vitro and in vivo. Proc Natl Acad Sci U S A. 2000;97:13625-13630
28. El Omar R, Beroud J, Stoltz JF. et al. Umbilical cord mesenchymal stem cells: the new gold standard for mesenchymal stem cell-based therapies? Tissue Eng Part B Rev. 2014;20:523-544
29. Cui X, Chen L, Xue T. et al. Human umbilical cord and dental pulp-derived mesenchymal stem cells: biological characteristics and potential roles in vitro and in vivo. Mol Med Rep. 2015;11:3269-3278
30. Saben J, Thakali KM, Lindsey FE. et al. Distinct adipogenic differentiation phenotypes of human umbilical cord mesenchymal cells dependent on adipogenic conditions. Exp Biol Med (Maywood). 2014;239:1340-1351
31. Wen Q, Zhou L, Zhou C. et al. Change in hepatocyte growth factor concentration promote mesenchymal stem cell-mediated osteogenic regeneration. J Cell Mol Med. 2012;16:1260-1273
32. Zullo J, Matsumoto K, Xavier S. et al. The cell secretome, a mediator of cell-to-cell communication. Prostaglandins Other Lipid Mediat. 2015;120:17-20
33. Mead B, Logan A, Berry M. et al. Paracrine-mediated neuroprotection and neuritogenesis of axotomised retinal ganglion cells by human dental pulp stem cells: comparison with human bone marrow and adipose-derived mesenchymal stem cells. PLoS One. 2014;9:e109305
34. Li X, Hou J, Wu B. et al. Effects of platelet-rich plasma and cell coculture on angiogenesis in human dental pulp stem cells and endothelial progenitor cells. J Endod. 2014;40:1810-1814
35. Jeong D, Han C, Kang I. et al. Effect of Concentrated Fibroblast-Conditioned Media on In Vitro Maintenance of Rat Primary Hepatocyte. PLoS One. 2016;11:e0148846
36. Li X, Duan L, Liang Y. et al. Human Umbilical Cord Blood-Derived Mesenchymal Stem Cells Contribute to Chondrogenesis in Coculture with Chondrocytes. Biomed Res Int. 2016;2016:3827057
37. Kawasaki Y, Komiya M, Matsumura K. et al. MYU, a Target lncRNA for Wnt/c-Myc Signaling, Mediates Induction of CDK6 to Promote Cell Cycle Progression. Cell Rep. 2016;16:2554-2564
38. Desdouets C, Sobczak-Thepot J, Murphy M. et al. Cyclin A: function and expression during cell proliferation. Prog Cell Cycle Res. 1995;1:115-123
39. Franceschi RT, Iyer BS. Relationship between collagen synthesis and expression of the osteoblast phenotype in MC3T3-E1 cells. J Bone Miner Res. 1992;7:235-2346
40. Xu J, Li Z, Hou Y. et al. Potential mechanisms underlying the Runx2 induced osteogenesis of bone marrow mesenchymal stem cells. Am J Transl Res. 2016;7:2527-2535
41. Vimalraj S, Arumugam B, Miranda PJ. et al. Runx2: Structure, function, and phosphorylation in osteoblast differentiation. Int J Biol Macromol. 2015;78:202-208
42. Bruderer M, Richards RG, Alini M. et al. Role and regulation of RUNX2 in osteogenesis. Eur Cell Mater. 2014;28:269-286
43. Shigetani Y, Ohkura N, Yoshiba K. et al. GaAlAs laser-induced pulp mineralization involves dentin matrix protein 1 and osteopontin expression. Oral Dis. 2016;22:399-405
44. Campos JM, Prati AJ, Cirano FR. et al. Smoking Modulates Gene Expression of Type I Collagen, Bone Sialoprotein, and Osteocalcin in Human Alveolar Bone. J Oral Maxillofac Surg. 2015;73:2123-2131
45. Kim KI, Park S, Im GI. Osteogenic differentiation and angiogenesis with cocultured adipose-derived stromal cells and bone marrow stromal cells. Biomaterials. 2014;35:4792-4804
46. Tansriratanawong K, Tamaki Y, Ishikawa H. et al. Co-culture with periodontal ligament stem cells enhances osteogenic gene expression in de-differentiated fat cells. Hum Cell. 2014;27:151-161
47. Sun W, Huang H, Ma S. et al. Akirin2 could promote the proliferation but not the differentiation of duck myoblasts via the activation of the mTOR/p70S6K signaling pathway. Int J Biochem Cell Biol. 2016;79:298-307
48. Lim W, Yang C, Bazer FW. et al. Luteolin Inhibits Proliferation and Induces Apoptosis of Human Placental Choriocarcinoma Cells by Blocking the PI3K/AKT Pathway and Regulating Sterol Regulatory Element Binding Protein Activity. Biol Reprod. 2016;95:82
49. Yu JS, Cui W. Proliferation, survival and metabolism: the role of PI3K/AKT/mTOR signalling in pluripotency and cell fate determination. Development. 2016;143:3050-3060
50. Hu XK, Yin XH, Zhang HQ. et al. Liraglutide attenuates the osteoblastic differentiation of MC3T3E1 cells by modulating AMPK/mTOR signaling. Mol Med Rep. 2016;14:3662-3668
51. Feng XM, Huang D, Lu XH. et al. Insulin-like growth factor 1 can promote proliferation and osteogenic differentiation of human dental pulp stem cells via mTOR pathway. Development Growth & Differentiation. 2014;56:615-624
52. Liu S, Zhou J, Zhang X. et al. Strategies to Optimize Adult Stem Cell Therapy for Tissue Regeneration. Int J Mol Sci. 2016:17
Author contact
![]() Corresponding authors: Yong Wen (wenyongedu.cn),No. 44-1, Wenhua Xi Road, Jinan, Shandong, 250012 P.R. China Tel./Fax: +86-531-88382923 Xin Xu (xinxuedu.cn),No. 44-1, Wenhua Xi Road, Jinan, Shandong, 250012 P.R. China Tel./Fax: +86-531-88382923
Corresponding authors: Yong Wen (wenyongedu.cn),No. 44-1, Wenhua Xi Road, Jinan, Shandong, 250012 P.R. China Tel./Fax: +86-531-88382923 Xin Xu (xinxuedu.cn),No. 44-1, Wenhua Xi Road, Jinan, Shandong, 250012 P.R. China Tel./Fax: +86-531-88382923

 Global reach, higher impact
Global reach, higher impact