3.2
Impact Factor
ISSN: 1449-1907
Int J Med Sci 2017; 14(11):1088-1093. doi:10.7150/ijms.20171 This issue Cite
Research Paper
Association of IFNL3 Genotype with Hepatic Steatosis in Chronic Hepatitis C Patients Treated with Peginterferon and Ribavirin Combination Therapy
1. Department of Gastroenterology, Graduate School of Medicine, Chiba University, Japan.
2. Department of Molecular Virology, Graduate School of Medicine, Chiba University, Japan.
Received 2017-3-20; Accepted 2017-7-5; Published 2017-9-4
Abstract
Background: Genetic variation near the interferon lambda 3 (IFNL3) is known to be associated with response to pegylated interferon (pegIFN) and ribavirin combination therapy in patients with chronic hepatitis C virus (HCV) infection which is often accompanied by hepatic steatosis.
Aims: We examined whether this genetic variation is associated with host lipids and treatment response.
Methods: A total of 101 Japanese patients who had underwent liver biopsy before treatment with pegIFN and ribavirin for HCV genotype 1b infection were retrospectively analyzed for association between IFNL3 genotypes (rs8099917) and clinical factors including histopathological features of the liver. The presence of >5% steatosis in the liver specimen was defined as hepatic steatosis.
Results: Forty patients (40%) had liver steatosis before therapy. Patients with IFNL3 minor genotype (non-TT) showed lower low-density lipoprotein cholesterol level (p=0.0045), higher γ-glutamyl transpeptidase level (p=0.0003) and higher prevalence of hepatic steatosis (p=0.0002). Advanced fibrosis [odds ratio (OR) 4.63, p=0.03] and IFNL3 major genotype (OR 0.13, p=0.001) were 2 independent factors for determining the presence of hepatic steatosis. Among the factors associated with sustained virological response, IFNL3 genotype was the most significant predictor, as per multivariate analysis.
Conclusions: Our results confirmed that IFNL3 genotype is associated with hepatic steatosis as well as IFN response.
Keywords: Hepatitis C virus, Interferon lambda 3, Peginterferon, Ribavirin, Hepatic steatosis
Introduction
Hepatitis C virus (HCV) affects an estimated 170 million individuals worldwide and is one of the major causes of chronic hepatitis, cirrhosis, and hepatocellular carcinoma (HCC) [1]. Although direct-acting antivirals against HCV (DAAs) have been developed, a combination treatment of pegylated interferon (pegIFN) plus ribavirin is still an important option for such patients as failed to respond to DAAs-based therapy [2]. IFN-based therapy is effective in 50% of the patients with HCV genotype 1 infection, which is the most commonly encountered genotype in Japan, Europe, and the US.
Recent studies have shown that single nucleotide polymorphisms (SNPs) located upstream of the IFN lambda 3 (IFNL3) gene were strongly associated with treatment response for HCV [3-7]. These SNPs include rs12979860 and rs8099917, which are in strong linkage disequilibrium. Homozygous for the major allele of these SNPs is associated with good response. The IFNL3 gene encodes IFN-lambda, which has been shown to have antiviral activity like type I IFNs (alpha and beta). The mechanism through which IFNL3-related SNPs affect treatment response is unclear; however, the differential expression or activity of IFN-lambda between these genotypes is a possible reason [4,5].
Host lipid metabolism has been shown to be associated with treatment responsiveness [8] and HCC [9] as well as cardiovascular events. HCV modulates the pathway of host lipid metabolism according to its lifecycle: cellular entry, replication, assembly, and secretion [10]. HCV infection causes dyslipidemia; pretreatment serum total and low-density lipoprotein cholesterol (LDL-C) levels have been associated with treatment response [11]. Low serum lipid levels in HCV infection have been correlated with steatosis, which is associated with more severe liver disease and poorer IFN response [8]. It has been reported that IFNL3-related SNPs may be associated with serum cholesterol levels and hepatic steatosis, which suggests the role for the IFNL3 genotype in the altered lipid metabolism of HCV-infected patients [12-14]. Indeed, IFN treatment has been reported to affect serum cholesterol levels during the therapy [15]. A recent study also suggests that antiviral immune response to HCV induce lipogenesis [16].
In the present study, we examined the relationship between IFNL3 genotype and host lipids including serum cholesterol levels and hepatic steatosis, and the relationship between IFNL3 genotype and treatment response in patients infected with HCV genotype 1.
Results
Baseline characters according to IFNL3 genotypes
IFNL3 genotypes were determined in 100 out of 101 patients (Table 1). Of these, 62 (62%) patients were homozygous for the major IFNL3 allele (TT: major genotype), and 38 (38%) had non-TT allele (TG or GG: minor genotype). Most factors were similar between genotypes, except that IFNL3 minor genotype showed lower LDL-C level (p = 0.0045), higher γ-glutamyl transpeptidase (γ-GT) level (p = 0.0003), and higher prevalence of hepatic steatosis (p = 0.0002).
Clinical factors associated with hepatic steatosis
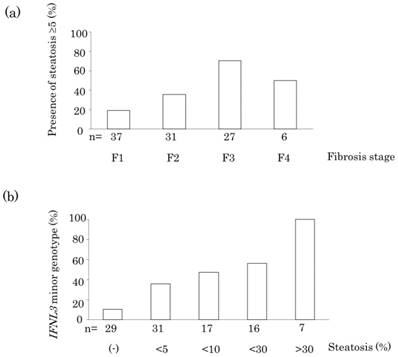
Clinical factors associated with hepatic steatosis were analyzed in Table 2. Univariate analysis showed that the presence of hepatic steatosis was significantly associated with increased body mass index (BMI; p = 0.02), advanced fibrosis (p = 0.0001), increased activity grade (p = 0.02), increased alanine aminotransferase (ALT) level (p = 0.001), increased aspartate aminotransferase (AST) level (p = 0.001), increased γ-GT level (p = 0.04), and IFNL3 minor genotype (p = 0.0002). Multivariate analysis showed that advanced fibrosis (p = 0.03), and IFNL3 minor genotype (p = 0.001) were independently associated with hepatic steatosis. Patients showed an increasing trend for the prevalence of hepatic steatosis with fibrosis stage (Figure 1 (a), p = 0.0001). Finally, patients showed an increasing trend for the prevalence of IFNL3 minor genotype with the grade of hepatic steatosis (Figure 1 (b), p < 0.0001).
Association between treatment response and lipid profile
Treatment response was evaluated in 99 of 101 patients; 45 (45%) achieved sustained virological response (SVR). Patients with SVR had higher level of LDL-C (99 mg/dL vs. 88 mg/dL, p=0.04) while total cholesterol level did not differ between groups (p=0.29). The prevalence of hepatic steatosis tended to be lower in SVR patients (29% or 13/45) than in non-SVR patients (46% or 25/54) (p=0.076). Multivariate analysis showed that neither LDL nor steatosis was associated with SVR while IFNL3 major genotype (OR 6.3, 1.7-22.5, p=0.005) and age (OR 0.93, 0.89-0.99, p=0.01) were good predictors of treatment response.
Baseline characteristics of patients according to IFNL3 genotypes
| IFNL3 minor genotype n = 38 | IFNL3 major genotype n = 62 | p value | |
|---|---|---|---|
| Age | 54 ± 13 | 55 ± 11 | NS |
| Sex (M/F) | 15/23 | 35/27 | NS |
| BMI (kg/m2) | 23.3 ± 3.4 | 23.3 ± 3.3 | NS |
| Treatment naive (%) | 15 (41) | 15 (25) | NS |
| Fibrosis stage ≥3 (%) | 15 (39) | 18 (29) | NS |
| Activity grade ≥2 (%) | 25 (66) | 33 (53) | NS |
| Steatosis ≥5% (%) | 24 (63) | 16 (26) | 0.0002 |
| ALT (IU/L) | 66 ± 43 | 72 ± 72 | NS |
| AST (IU/L) | 61 ± 34 | 60 ± 72 | NS |
| Hemoglobin (g/dL) | 14.1 ± 1.5 | 14.4 ± 1.1 | NS |
| Platelet (× 104/mm3) | 16.1 ± 6.1 | 17.5 ± 6.9 | NS |
| WBC (/mm3) | 5000 ± 1400 | 5300 ± 1300 | NS |
| HCV RNA (log IU/mL) | 6.2 ± 0.6 | 6.4 ± 0.6 | NS |
| Total cholesterol (mg/dL) | 166 ± 38 | 175 ± 28 | NS |
| LDL-C (mg/dL) | 87 ± 31 | 98 ± 24 | 0.0045 |
| γ-GT (IU/L) | 63 ± 49 | 35 ± 25 | 0.0003 |
IFNL3 genotypes were determined based on rs8099917. BMI: body mass index; ALT: alanine aminotransferase; AST: aspartate aminotransferase; WBC: white blood cell; LDL-C: low density lipoprotein cholesterol; γ-GT: γ-glutamyl transpeptidase; IFNL3: interferon lambda 3; NS: not statistically significant
Clinical factors associated with the presence of hepatic steatosis
| Steatosis <5% n = 61 | Steatosis ≥5% n = 40 | Univariate analysis p value | Multivariate analysis Steatosis ≥5% vs. <5% OR (95% CI), p value | |
|---|---|---|---|---|
| Age | 53 ± 12 | 56 ± 10 | NS | - |
| Sex (M/F) | 32/29 | 18/22 | NS | - |
| BMI(kg/m2) | 22.7 ± 3.3 | 24.2 ± 3.2 | 0.02 | NS |
| Treatment naive (%) | 15 (25) | 15 (65) | NS | - |
| Fibrosis stage ≥3 (%) | 11 (18) | 22 (55) | 0.0001 | 4.63 (1.12-19.2), p = 0.03 |
| Activity grade ≥2 (%) | 30 (49) | 29 (73) | 0.02 | NS |
| ALT (IU/L) | 53 ± 35 | 94 ± 84 | 0.001 | NS |
| AST (IU/L) | 44 ± 21 | 84 ± 87 | 0.001 | NS |
| Hemoglobin (g/dL) | 14.2 ± 1.3 | 14.6 ± 1.2 | NS | - |
| Platelet (× 104/mm3) | 17.9 ± 6.8 | 15.6 ± 6.0 | NS | - |
| WBC (/mm3) | 5100 ± 1300 | 5300 ± 1300 | NS | - |
| HCV RNA (log IU/mL) | 6.3 ± 0.6 | 6.2 ± 0.6 | NS | - |
| Total cholesterol (mg/dL) | 174 ± 29 | 168 ± 36 | NS | - |
| LDL-C (mg/dL) | 94 ± 24 | 93 ± 32 | NS | - |
| γ-GT (IU/L) | 39 ± 41 | 55 ± 30 | 0.04 | NS |
| IFNL3 major genotype (%) | 46 (77) | 16 (40) | 0.0002 | 0.13 (0.038-0.45), p = 0.001 |
IFNL3 genotypes were determined based on rs8099917. BMI: body mass index; ALT: alanine aminotransferase; AST: aspartate aminotransferase; WBC: white blood cell; LDL-C: low density lipoprotein cholesterol; γ-GT: γ-glutamyl transpeptidase; IFNL3: interferon lambda 3; OR: odds ratio; 95%CI: 95% confidence interval; NS: not statistically significant
(a) Association between the presence of steatosis and fibrosis stage in the liver of a patient infected with HCV genotype 1. (b) Association between IFNL3 minor genotype and grade of hepatic steatosis.
Discussion
Several host and viral factors have been reported to be associated with the efficacy of pegIFN plus ribavirin combination therapy in chronic hepatitis C patients. These factors include age, gender, ethnicity, viral genotypes, stage of liver disease, metabolic factors, and variations in host genes involved in the IFN pathway and viral genome sequence [17,18]. Among them, IFNL3 genotype has been reported to be one of the strongest predictors of treatment response [19,20]. In the present study, we assessed the effect of hepatic steatosis and IFNL3 genotype on treatment response in Japanese patients infected with HCV genotype 1b. Consistent with previous reports [12-14], we confirmed that host IFNL3 genotype is strongly associated with treatment response.
Li et al. [12] reported that IFNL3 genotype was associated with serum LDL-C levels and suggested a relationship between IFN-lambda and lipid metabolism. Moreover, Clark et al. [13] reported an association between the IFNL3 minor genotype, low LDL-C and hepatic steatosis. Recently, Cai et al. [14] reported HCV genotype-specific association between IFNL3 genotype and hepatic steatosis in a European cohort; HCV non-3 genotypes, especially HCV genotype 1, had this association while HCV genotype 3 did not.
In the present study, we confirmed that IFNL3 minor genotype is associated with low LDL-C levels and with hepatic steatosis in HCV genotype-1 infection. The effect of IFNL3 genotype on hepatic steatosis remained significant after multivariate modeling. Moreover, a positive correlation seems to exist between the prevalence of IFNL3 minor genotype and the degree of hepatic steatosis. When IFNL3 genotype is included in the multivariate analysis, pretreatment serum cholesterol levels and/or hepatic steatosis is no longer associated with treatment response.
Consequently, it appears that the effects of hepatic steatosis and serum lipid levels on treatment response may be linked to host IFNL3 genotype. For instance, IFNs can activate signal transducer and activators of transcription [18,21,22] which is involved in adipogenesis through the peroxisome-proliferator-activated receptor-gamma (PPARG) activation pathway. Cai et al. [14] reported the association of steatosis with SNPs in PPARG as well as in patatin-like phospholipase domain-containing 3 and IFNL3.
Previous reports describing the relationship between IFNL3 genotype and liver fibrosis are conflicting [23-25]. Fabris et al. [22] reported that the IFNL3 minor genotype is more prevalent in cirrhotic patients and in patients with HCC, when compared to patients with chronic hepatitis. Abe et al. [23] reported that inflammatory activity and stage of liver fibrosis are significantly more severe in patients homozygous for the IFNL3 major genotype. In the current study, and as reported by Thompson et al. [25], neither liver fibrosis nor inflammatory activity grades were associated with IFNL3 genotype. If the IFNL3 major genotype is associated with severe inflammation and progression of fibrosis, clinical course of patients according to IFNL3 genotype was affected by IFN treatment. Long-term outcome of HCV patients according to IFNL3 genotype needs to be investigated in future studies.
The mechanism of lipid metabolism is complex in HCV patients. Several host factors, such as ALT level, fibrosis stage, or metabolic factors like body mass index, have been well-reported to be associated with hepatic steatosis in HCV genotypes 1 and non-3 [8,26,27]. In addition, viral factors such as HCV core amino acid variations have been associated with IFNL3 genotype and hepatic steatosis [28]. In our study, although the association was not significant, mutations at HCV core amino acid at position 70 tended to be lower in IFNL3 major genotype than in minor genotype (37% vs. 56%, p=0.08), and lower in patients without steatosis than in patients with steatosis (39% vs. 51%, p=0.22) (data not shown). On the other hand, amino acid mutations at core position 91 was significantly lower in major genotype (28% vs. 58%, p=0.004), and lower in patients without hepatic steatosis (26% vs. 59%, p=0.001). Viral factors may affect the association between IFNL3 genotype and hepatic steatosis. Limitations of our study include retrospective nature of the study and small sample size. Furthermore, HCV genotype and ethnicity could affect the results [29,30]. Recently, HCV treatment by IFN-free DAA regimen has been shown to affect host lipid metabolism, suggesting the role of HCV replication and/or core protein on lipid homeostasis [31,32]. Further studies will be needed to clarify the role of host genotype, viral factors, and antiviral therapy on lipid metabolism and outcome of HCV infection.
In conclusion, we examined the relationship between IFNL3 genotype, liver histopathology, and predictive markers with response to the combination therapy of pegIFN and ribavirin in HCV genotype 1b-infected Japanese patients. Hepatic steatosis and LDL-C levels may not have any independent clinical significance beyond that of using IFNL3 genotype to predict treatment response. However, considering that IFNL3 minor genotype could be associated with hepatic steatosis, early intervention combined with potent antiviral therapy appear reasonable. A greater understanding of the relationship between host genotype and viral pathogenesis will help to better manage HCV infection.
Patients and methods
Study population
This retrospective study included 101 HCV genotype 1b-infected patients who had undergone liver biopsy and had received combination therapy of pegIFN alfa-2a (Pegasys, Chugai, Tokyo, Japan) or alpha-2b (Pegintron, MSD, Tokyo, Japan) plus ribavirin [Copegas (Chugai) or Rebetol (MSD)] between January 2005 and November 2009 at Chiba University Hospital. Patients with chronic hepatitis B, autoimmune hepatitis, primary biliary cirrhosis, hemochromatosis, Wilson disease, or alcoholic liver disease were excluded from this study.
Patients received a standard dose of pegIFN alfa-2a or alpha-2b subcutaneously each week plus oral ribavirin daily for more than 48 weeks. Weight-based ribavirin were administered (600 mg for ≤60 kg, 800 mg for >60 kg and ≤80 kg, and 1000 mg for >80 kg). Table 1 shows the clinical background of the patients at the start of combination therapy. Sixty of them (59%) were previously reported [33].
This study was approved by ethics committee of Chiba University, Graduate School of Medicine (No. 406/582/1753). Participation in the study was posted at our institutions. Written informed consent was obtained from all the patients before liver biopsy.
Definition of treatment response
Sustained virological response (SVR) was defined as HCV RNA negativity at 6 months after the end of treatment.
Laboratory tests
HCV RNA levels were measured quantitatively before treatment and during the 12-week therapy by Taqman PCR (Cobas Taqman HCV, Roche, Tokyo, Japan). RNA levels were expressed using log10 IU/mL of viral loads, with the lower limit of 1.2 log IU/mL. HCV RNA genotype was determined as per the method of Ohno et al [34].
Histopathological examination
Liver biopsy specimens were obtained percutaneously, and the specimens were histopathologically assessed, according to the criteria of Desmet et al. [35]. Hepatic fibrosis was staged as F1-2 for mild to moderate fibrosis and F3-4 for severe fibrosis to cirrhosis. Inflammatory activity of the liver was graded as A1 for mild activity and A2-3 for moderate-severe activity. Hepatic steatosis was graded as follows: <1%, ≥1% to <5%, ≥5% to <10%, ≥10% to <30%, and ≥30%. Hepatic steatosis was confirmed when >5% steatosis were observed in a hepatic tissue specimen [36]. Among 101 patients, 40 (40%) had liver steatosis before therapy.
Determination of SNP rs8099917 associated with IFNL3
Human genomic DNA was extracted from serum samples using the QIAamp Circulating Nucleic Acid kit (QIAGEN, Tokyo, Japan). The rs8099917 SNP was genotyped by PCR and restriction fragment length polymorphism, as previously described [5,37]. We analyzed IL28B rs8099917 TT as major genotype and TG and GG as minor genotypes in this study.
Statistical analysis
Comparisons between groups were made by the χ2 or Fisher's exact test for categorical variables and Student's t-test for quantitative variables. Quantitative data are expressed as mean and standard deviation. Univariate analysis was performed for the variables listed in Table 1. Multivariate logistic regression analyses were used to determine the predictors of treatment response. Variables with p < 0.05 after univariate analysis were subjected to multivariate analysis. The Cochran-Armitage trend test was used to analyze the association between the prevalence of steatosis and liver fibrosis in the subjects. Statistical significance was defined at p < 0.05.
Abbreviations
HCV: hepatitis C; HCC: hepatocellular carcinoma; DAA: direct-acting antiviral; IFNL3: interferon lambda 3; SNP: single nucleotide polymorphism; LDL-C: low density lipoprotein cholesterol; γ-GT: γ-glutamyl transpeptidase; BMI: body mass index; ALT: alanine aminotransferase; AST: aspartate aminotransferase; SVR: sustained virological response; PPARG: peroxisome-proliferator-activated receptor-gamma; WBC: white blood cell; NS: not statistically significant; OR: odds ratio; CI: confidence interval.
Acknowledgements
This work was supported by the Grant for Scientific Research 22200701 from the Ministry of Education, Culture, Sports, Science, and Technology, Japan (OY).
Tatsuo Kanda received research grants from Merck Sharp and Dohme (MSD), Chugai Pharm and AbbVie. The founding sponsors had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript, or in the decision to publish the results. The other authors declare no conflicts of interest. We thank Prof. Naoya Kato and Dr. Hitoshi Maruyama at Department of Gastroenterology, Chiba University for valuable discussions.
Author Contributions
Shingo Nakamoto, Fumio Imazeki, Osamu Yokosuka and Hiroshi Shirasawa conceived and designed the experiments; Shingo Nakamoto and Shuang Wu performed the experiments; Shingo Nakamoto and Fumio Imazeki analyzed the data; Masato Nakamura, Shin Yasui, Akinobu Tawada, Rintaro Mikata, Tetsuhiro Chiba, Makoto Arai, Tatsuo Kanda, Fumio Imazeki and Osamu Yokosuka saw the patients. Shingo Nakamoto, Tatsuo Kanda and Fumio Imazeki wrote the paper.
Competing Interests
The authors have declared that no competing interest exists.
References
1. Seeff LB. Natural history of chronic hepatitis C. Hepatology. 2002;36:S35-46
2. Di Bisceglie AM, Hoofnagle JH. Optimal therapy of hepatitis C. Hepatology. 2002;36:S121-127
3. Ge D, Fellay J, Thompson AJ. et al. Genetic variation in IL28B predicts hepatitis C treatment-induced viral clearance. Nature. 2009;461:399-401
4. Tanaka Y, Nishida N, Sugiyama M. et al. Genome-wide association of IL28B with response to pegylated interferon-alpha and ribavirin therapy for chronic hepatitis C. Nat Genet. 2009;41:1105-1109
5. Suppiah V, Moldovan M, Ahlenstiel G. et al. IL28B is associated with response to chronic hepatitis C interferon-alpha and ribavirin therapy. Nat Genet. 2009;41:1100-1104
6. Mangia A, Thompson AJ, Santoro R. et al. An IL28B polymorphism determines treatment response of hepatitis C virus genotype 2 or 3 patients who do not achieve a rapid virologic response. Gastroenterology. 2010;139:821-827
7. Yu ML, Huang CF, Huang JF. et al. Role of interleukin-28B polymorphisms in the treatment of hepatitis C virus genotype 2 infection in Asian patients. Hepatology. 2011;53:7-13
8. Poynard T, Ratziu V, McHutchison J. et al. Effect of treatment with peginterferon or interferon alfa-2b and ribavirin on steatosis in patients infected with hepatitis C. Hepatology. 2003;38:75-85
9. Kurosaki M, Hosokawa T, Matsunaga K. et al. Hepatic steatosis in chronic hepatitis C is a significant risk factor for developing hepatocellular carcinoma independent of age, sex, obesity, fibrosis stage and response to interferon therapy. Hepatol Res. 2010;40:870-877
10. Syed GH, Amako Y, Siddiqui A. Hepatitis C virus hijacks host lipid metabolism. Trends Endocrinol Metab. 2009;21:33-40
11. Harrison SA, Rossaro L, Hu KQ. et al. Serum cholesterol and statin use predict virological response to peginterferon and ribavirin therapy. Hepatology. 2010;52:864-874
12. Li JH, Lao XQ, Tillmann HL. et al. Interferon-lambda genotype and low serum low-density lipoprotein cholesterol levels in patients with chronic hepatitis C infection. Hepatology. 2010;51:1904-1911
13. Clark P, Thompson A, Zhu M. et al. IL28B genetic polymorphism has genome wide significant associations with serum low density lipoprotein levels and hepatic steatosis in patients with genotype 1 chronichepatitis C (CHC). Hepatology. 2010;52:439A
14. Cai T, Dufour JF, Muellhaupt B. et al. Viral genotype-specific role of PNPLA3, PPARG, MTTP, and IL28B in hepatitis C virus-associated steatosis. J Hepatol. 2011;55:529-535
15. Ramcharran D, Wahed AS, Conjeevaram HS. et al. Associations between serum lipids and hepatitis C antiviral treatment efficacy. Hepatology. 2010;52:854-863
16. Li Q, Pene V, Krishnamurthy S. et al. Hepatitis C virus infection activates an innate pathway involving IKK-alpha in lipogenesis and viral assembly. Nat Med. 2013;19:722-729
17. Asselah T, Estrabaud E, Bieche I. et al. Hepatitis C: viral and host factors associated with non-response to pegylated interferon plus ribavirin. Liver Int. 2010;30:1259-1269
18. Wesoly J, Szweykowska-Kulinska Z, Bluyssen HA. STAT activation and differential complex formation dictate selectivity of interferon responses. Acta Biochim Pol. 2007;54:27-38
19. McCarthy JJ, Li JH, Thompson A. et al. Replicated association between an IL28B gene variant and a sustained response to pegylated interferon and ribavirin. Gastroenterology. 2010;138:2307-2314
20. Hayes CN, Kobayashi M, Akuta N. et al. HCV substitutions and IL28B polymorphisms on outcome of peg-interferon plus ribavirin combination therapy. Gut. 2010;60:261-267
21. Brand S, Zitzmann K, Dambacher J. et al. SOCS-1 inhibits expression of the antiviral proteins 2',5'-OAS and MxA induced by the novel interferon-lambdas IL-28A and IL-29. Biochem Biophys Res Commun. 2005;331:543-548
22. Pazienza V, Vinciguerra M, Andriulli A. et al. Hepatitis C virus core protein genotype 3a increases SOCS-7 expression through PPAR-gamma in Huh-7 cells. J Gen Virol. 2010;91:1678-1686
23. Fabris C, Falleti E, Cussigh A. et al. IL-28B rs12979860 C/T allele distribution in patients with liver cirrhosis: role in the course of chronic viral hepatitis and the development of HCC. J Hepatol. 2011;54:716-722
24. Abe H, Ochi H, Maekawa T. et al. Common variation of IL28 affects gamma-GTP levels and inflammation of the liver in chronically infected hepatitis C virus patients. J Hepatol. 2010;53:439-443
25. Thompson A, Clark P, Fellay J. et al. IL28B genotype is not associated with advanced hepatic fibrosis in chronic hepatitis C patients enrolled in the IDEAL study. Hepatology. 2010;52:437A
26. Hui JM, Kench J, Farrell GC. et al. Genotype-specific mechanisms for hepatic steatosis in chronic hepatitis C infection. J Gastroenterol Hepatol. 2002;17:873-881
27. Patton HM, Patel K, Behling C. et al. The impact of steatosis on disease progression and early and sustained treatment response in chronic hepatitis C patients. J Hepatol. 2004;40:484-490
28. Tachi Y, Katano Y, Honda T. et al. Impact of amino acid substitutions in the hepatitis C virus genotype 1b core region on liver steatosis and hepatic oxidative stress in patients with chronic hepatitis C. Liver Int. 2010;30:554-559
29. Eslam M, Booth DR, George J. et al. Interaction of IFNL3 with insulin resistance, steatosis and lipid metabolism in chronic hepatitis C virus infection. World J Gastroenterol. 2013;19:7055-7061
30. Sato M, Kondo M, Tateishi R. et al. Impact of IL28B genetic variation on HCV-induced liver fibrosis, inflammation, and steatosis: a meta-analysis. PLoS One. 2014;9:e91822
31. Meissner EG, Lee YJ, Osinusi A. et al. Effect of sofosbuvir and ribavirin treatment on peripheral and hepatic lipid metabolism in chronic hepatitis C virus, genotype 1-infected patients. Hepatology. 2015;61:790-801
32. Hashimoto S, Yatsuhashi H, Abiru S. et al. Rapid Increase in Serum Low-Density Lipoprotein Cholesterol Concentration during Hepatitis C Interferon-Free Treatment. PLoS One. 2016;11:e0163644
33. Nakamoto S, Imazeki F, Arai M. et al. Effect of Hepatitis C Virus Genotype 1b Core and NS5A Mutations on Response to Peginterferon Plus Ribavirin Combination Therapy. Int J Mol Sci. 2015;16:21177-21190
34. Ohno O, Mizokami M, Wu RR. et al. New hepatitis C virus (HCV) genotyping system that allows for identification of HCV genotypes 1a, 1b, 2a, 2b, 3a, 3b, 4, 5a, and 6a. J Clin Microbiol. 1997;35:201-207
35. Desmet VJ, Gerber M, Hoofnagle JH. et al. Classification of chronic hepatitis: diagnosis, grading and staging. Hepatology. 1994;19:1513-1520
36. Neuschwander-Tetri BA, Caldwell SH. Nonalcoholic steatohepatitis: summary of an AASLD Single Topic Conference. Hepatology. 2003;37:1202-1219
37. Nakamoto S, Kanda T, Imazeki F. et al. Simple assay based on restriction fragment length polymorphism associated with IL28B in chronic hepatitis C patients. Scand J Gastroenterol. 2011;46:955-961
Author contact
![]() Corresponding author: Shingo Nakamoto, Department of Molecular Virology, Graduate School of Medicine, Chiba University, 1-8-1 Inohana, Chuo-Ward, Chiba City, Chiba 260-8670, Japan. Tel.: +81 43 226 2083; Fax: +81 43 226 2088; E-mail address: nakamotoerchiba-u.jp.
Corresponding author: Shingo Nakamoto, Department of Molecular Virology, Graduate School of Medicine, Chiba University, 1-8-1 Inohana, Chuo-Ward, Chiba City, Chiba 260-8670, Japan. Tel.: +81 43 226 2083; Fax: +81 43 226 2088; E-mail address: nakamotoerchiba-u.jp.

 Global reach, higher impact
Global reach, higher impact