Impact Factor
ISSN: 1449-1907
Int J Med Sci 2017; 14(10):1022-1030. doi:10.7150/ijms.18392 This issue Cite
Research Paper
LncRNA AFAP1-AS Functions as a Competing Endogenous RNA to Regulate RAP1B Expression by sponging miR-181a in the HSCR
1. State Key Laboratory of Reproductive Medicine, Institute of Toxicology, School of Public Health, Nanjing Medical University, Nanjing 211166, China;
2. Key Laboratory of Modern Toxicology (Nanjing Medical University), Ministry of Education, China;
3. Department of Pediatric Surgery, Children's Hospital of Nanjing Medical University.
* These authors contributed equally
Received 2016-11-17; Accepted 2017-3-14; Published 2017-9-3
Abstract
Background: Long noncoding RNAs (lncRNAs) have recently emerged as important regulators in a broad spectrum of cellular processes including development and disease. Despite the known engagement of the AFAP1-AS in several human diseases, its biological function in Hirschsprung disease (HSCR) remains elusive.
Methods: We used qRT-PCR to detect the relative expression of AFAP1-AS in 64 HSCR bowel tissues and matched normal intestinal tissues. The effects of AFAP1-AS on cell proliferation, migration, cell cycle, apoptosis and cytoskeletal organization were evaluated using CCK-8, transwell assay, flow cytometer analysis and immunofluorescence, in 293T and SH-SY5Y cell lines, respectively. Moreover, the competing endogenous RNA (ceRNA) activity of AFAP1-AS on miR-181a was investigated via luciferase reporter assay and immunoblot analysis.
Results: Aberrant inhibition of AFAP1-AS was observed in HSCR tissues. Knockdown of AFAP1-AS in 293T and SH-SY5Y cells suppressed cell proliferation, migration, and induced the loss of cell stress filament integrity, possibly due to AFAP1-AS sequestering miR-181a in HSCR cells. Furthermore, AFAP1-AS could down-regulate RAP1B via its competing endogenous RNA (ceRNA) activity on miR-181a.
Conclusions: These findings suggest that aberrant expression of lncRNA AFAP1-AS, a ceRNA of miR-181a, may involve in the onset and progression of HSCR by augmenting the miR-181a target gene, RAP1B.
Keywords: AFAP1-AS, Hirschsprung disease, Competing endogenous RNA, miR-181a.
Introduction
Hirschsprung disease (HSCR), or congenital megacolon, is the most common form of congenital digestive malformation characterized by the absence of ganglion cells [1]. This developmental disorder manifests as functional intestinal obstruction in neonates and children. HSCR has an incidence of around 1/5000 neonates alongside a 4:1 male: female gender rate [2]. As a neurocristopathy, any abnormality of the factors that affect proliferation, migration, or differentiation during the embryo development can lead to HSCR [3]. Numerous researchers have identified several crucial genes that participate in the occurrence of HSCR, containing RET and EDNRB [4]. Our previous study also showed that several genes were involved in HSCR [5-7]. However, the underlying genetic mechanisms for the pathogenesis of HSCR still remain elusive.
Long non-coding RNAs, also known as lncRNAs, have recently received wide attention due to their rising functions in development and diseases [8, 9]. Such RNA transcripts are characterized by more than 200 nucleotides that have no capacity of encoding proteins [10]. Increasing evidence has shown that lncRNAs are involved in several levels, including transcription and post-transcription [11-13]. Importantly, lncRNAs can interact with microRNA (miRNA) as a kind of competitive endogenous RNA (ceRNA) to alter the expression of target genes [14]. Recent studies have demonstrated that lncRNA AFAP1-AS mediates various cell biological processes of cancers, including cancer progression and metastasis [15-19]. However, the potential role for AFAP1-AS1 during the pathogenesis of HSCR remains unclear.
In this paper, we first identified AFAP1-AS that exhibited lower expression in HSCR than normal tissues. By down-regulating AFAP1-AS, we found a significant decrease of migration and proliferation in HSCR cell lines. Our results also demonstrated that AFAP1-AS acted as a ceRNA through binding to miR-181a and mediated the repression of RAP1B. Here, our results suggest AFAP1-AS plays a vital role during the progression of HSCR.
Material and Methods
Samples collection and ethics statement
In this study, we collected 64 HSCR samples from patients at Children's Hospital of Nanjing Medical University. All selected patients were confirmed by pathological examination through available biopsy samples. Sixty-four corresponding normal colon tissues were collected from patients without HSCR or other congenital anomalies. Immediately following removal, all tissues were stored at -80 °C before using. Each patient enrolled in the study has signed informed consent and this whole study was authorized by the Institutional Ethics Committee of Nanjing Medical University.
Cell lines and culture
We purchased human 293T and SH-SY5Y cells from the American Type Culture Collection (ATCC, Manassas VA, USA). All the cell lines were previously used as cell models for HSCR [20, 21]. The cell lines were cultured in DMEM medium (Hyclone, UT, USA) containing 10% FBS and 1% Penicillin-Streptomycin (Invitrogen) at 37˚C in a humidified incubator under an atmosphere of 5% carbon dioxide.
RNA extraction and quantitative real-time PCR (qRT-PCR)
Total RNA from tissues and cells were extracted using TRIzol reagent (Life Technologies, USA). For mRNA detection, each RNA sample was reverse transcribed into cDNAs using the reverse transcription kit (Takara, Tokyo, Japan). A cDNA library of miRNAs was reversed using the QuantiMir Kit (Takara). The qRT-PCR was employed to measure the levels of mRNAs and miRNAs using the comparative Ct method. GAPDH and U6 small nuclear RNA were considered as the normalization control for mRNA and miRNA, respectively. All primers for PCR were depicted in Supplement Table 1.
Cell transfection
Small interfering RNA (siRNA) duplexes, miR-181a mimics and miR-181a inhibitor were designed by GenePharma Co (Shanghai, China). Lipofectamine 2000 (Invitrogen, CA, USA) was used for transfection. Detailed sequences are depicted in the in Supplement Table 1.
Cell proliferation assays
For quantifying proliferation, cells transfected with NC or AFAP1-AS siRNA were incubated in the Cell Counting Kit-8 (Beyotime, Nantong, China) for 48h. Absorbance was detected at 450 nm using a microplate reader (Tecan, Mechelen, Belgium).
Cell migration assays
After transfection, 5×105 cells were incubated in 100 μl serum-free medium inside upper chamber (Millpore, MASS, USA) while 600 μl DMEM with 10%FBS were added into the lower chamber. At the end of the incubation period, transwell inserts were fixed with methanol followed by 0.1% crystal violet staining, and then photographed using a microscope at 20X magnification (five views per well). The relative number of stained cells was calculated using Image-pro Plus 6.0 (Media Cybernetics, USA).
Cell cycle and apoptosis assays
Cells after transfection were harvested by trypsinization 24 hours post-transfection. Cells were fixed for cell cycle analysis followed by the 70% cold ethanol at 4°C overnight and then incubated with propidium oxide (Sigma, USA). For analysis of apoptosis assay, cells were exposed to annexin-V/PI (BD Biopharmingen, NJ, USA). All experiments were conducted using a flow cytometer (FACScan; BD Biosciences, USA).
Immunofluorescence
Cells were fixed in 4% paraformaldehyde, exposed to 0.5% Triton X-100, and then incubated in a 1:1000 dilution of Rhodamine Phalloidin (Invitrogen) at 4 °C for 24 hours. After washing, DAPI was prepared for nuclei staining in a 1:1000 dilution. Images were captured with confocal laser scanning.
Subcellular fractionation location
Cytoplasmic and nuclear RNA was isolated with the PARIS Kit (Life Technologies, USA) as described in directions. Total RNA isolated from each fraction was determined by qRT-PCR. GAPDH and U6 were considered as cytoplasmic and nuclear markers, respectively.
Image-pro Plus 6.0 (Media Cybernetics,Silver Spring, MD, USA) to count migrated cells while cell numbers of normal control were normalized to 1.
Dual-luciferase reporter assay
The wild type (WT) or mutant (MUT) plasmids comprising the AFARl-AS or RAP1B 3'-UTR region including the miR-181a binding sites were constructed into the pGL3 promoter vector (Realgene, Nanjing, China). For luciferase reporter assay, cells were seeded in triplicate with a density of 5×105/well. Firefly luciferase (800 ng) and pRL-SV40 plasmid (5 ng) were co-transfected with 50nM miR-181a mimic or negative control. Firefly luciferase activity was measured 48 hours later and normalized to the Renilla value with Dual-Luciferase Reporter System (Promega, USA).
RNA Immunoprecipitation (RIP) assay
Immunoprecipitation assay was conducted in accordance with the kit instructions (Millipore, USA).Human 293T cells were lysed mixed with inhibitors of protease and RNase. RNAs magnetic beads were pre-incubated with 1:1000 anti-AGO2 antibody (Abcam, USA) or negative control anti-IgG (Millipore) and then immunoprecipitated RNAs were isolated from RNA-protein complexes. In addition, purified RNAs was extracted and subjected to qRT-PCR analysis using the corresponding primers.
Western blot
The process of protein samples were described before [22]. A primary antibody against Rap1B or GAPDH was purchased from Proteintech (1:1000, Chicago, IL, USA) followed by the goat anti-rabbit HRP conjugated antibody (1:1000, Nantong, China).
Statistical analysis
All experiments were independently repeated in triplicate. The expression of the tissue sample was treated by log transformation and plotted as box plot of the median using Wilcoxon rank-sum (Mann-Whiney). Expression differences between different groups were analyzed using unpaired t-test. Meanwhile chi-square test and multivariate regression analysis were used in the right place. Data were expressed as mean ± SE. The data is processed by STATA 9.2, and visualized by Graph PAD prism. P-value < 0.05 was considered to be statistically significant.
Results
Patient Characteristics and Expression level of AFAP1-AS RNA in HSCR and normal tissues
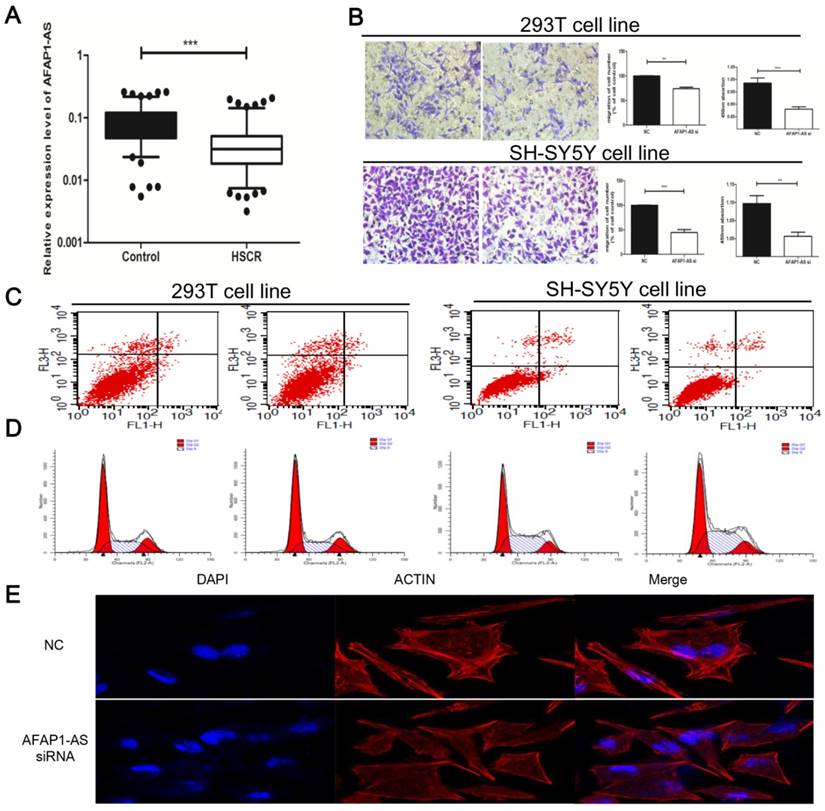
A total of 128 samples, including 64 HSCRs and 64 control tissues, were enrolled in the study. The clinical and demographic information was shown in Table 1. There was no statistically significant difference in age, weight, or gender between the HSCT patients and the control group. Next, we evaluated the expression of AFAP1-AS in HSCR and control colon tissues by qRT-PCR. The results noted that HSCR samples exhibited lower levels of AFAP1-AS compared to the normal tissues (Figure 1A).
Clinical characteristics of study population
| Variable | Control(n=64) | HSCR(n=64) | P |
|---|---|---|---|
| Age (months, mean, SE) | 3.91(0.43) | 4.61(0.33) | 0.1989a |
| Weight (kg, mean, SE) | 5.0(0.26) | 4.8(0.29) | 0.6085a |
| Sex (%) | |||
| Male | 44(68.75) | 51(79.69) | 0.1572b |
| Female | 20(31.25) | 13(20.31) |
aStudent's t-test.
bTwo-sided χ2 test.
Knockdown of AFAP1-AS decreased cell migration and proliferation and induced loss of cell stress filament integrity
To conduct subsequent functional and mechanistic researches, we developed SH-SY5Y and 293T cell lines with AFAP1-AS siRNA to down-regulate AFAP1-AS expression. Effects on cell migration and proliferation were assessed sequentially, and AFAP1-AS knockdown significantly suppressed the migration and viability of 293T and SH-SY5Y cells (Figure 1B). In addition, flow cytometer assays were implemented on cell cycle and apoptosis (Figure 1C, D). Between siAFAP1-AS and the control, we did not observe a significant difference in the cell cycle and apoptosis. As previous studies demonstrated that AFAP1-AS may participate in cytoskeletal organization [18]. Rhodamine-labeled phalloidin was used to characterize the cytoskeletal remodeling. The data indicated that F-actin was sturdy and assembled on the margin of SH-SY5Y cells; nevertheless, the cytoskeleton elements were attenuated after transfection with AFAP1-AS siRNA (Figure 1E).
AFAP1-AS was down-regulated and its cytobiology change transfected with AFAP1-AS siRNA. (A) The expression of AFAP1-AS in HSCR was significantly down-regulated compared with normal tissues. Data were presented as box plot of the median and range of log-transformed relative expression level. The top and bottom of the box represent the 75th and 25th percentile. The whiskers indicate the 10th and 90th points. (B) AFAP1-AS knockdown affected abilities of cell migration and proliferation. Representative images of migrated cells were visualized as shown (left panel). Quantifications of cell migration were presented as percentage migrated cell numbers (middle panel). Absorbance at 450 nm as measured by CCK8 was expressed as Mean ± SE (right panel). (C-D) Cycle and apoptosis assays were conducted after AFAP1-AS knockdowm by flow cytometer. (E) F-actin cytoskeleton of was visualized with Rhodamine Phalloidin staining in SH-SY5Y cells.
AFAP1-AS directly binds with miR-181a
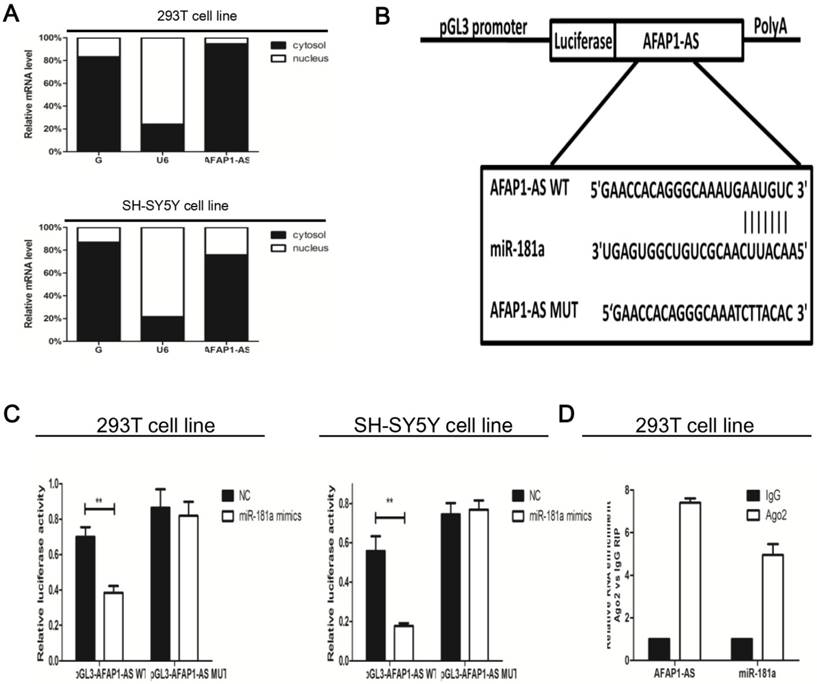
AFAP1-AS1 localizes to the antisense genomic DNA strand near the C-terminus of AFAP1, at the actin binding domain of AFAP1. The level of AFAP1 expression was not significantly altered after AFAP1-AS1 was knockdown (data was not shown), suggesting that functions of AFAP1-AS may involve an AFAP1-independent mechanism during the progression of HSCR.
To further study the molecular mechanism of AFAP1-AS involvement in HSCR progression, we determined the subcellular location of AFAP1-AS. Semi-quantitative PCR of nuclear and cytoplasmic fractions (Figure 2A) suggested that AFAP1-AS mainly located in the cytoplasm. Cytoplasmic lncRNAs are well known for modulating gene expression through interaction with miRNA. Recently, the competing endogenous RNAs (ceRNA) hypothesis has been reported to function by competitively binding common miRNAs. Bioinformatics prediction according to web server RegRNA (http://regrna.mbc.nctu.edu.tw/html/tutorial.html) suggested that four miR-181 family binding sites were found binding to AFAP1-AS with high scores [23]. Among the above four miRNAs (miR-181a/b/c/d), we mainly concentrated on miR-181a to study the interaction between AFAP1-AS and miR-181a in HSCR. To validate this hypothesis, a luciferase reporter including the wild type AFAP1-AS (pMIR-AFAP1-AS-WT) and mutant AFAP1-AS (pMIR-AFAP1-AS -MUT) was designed (Figure 2B). Based on the luciferase assay, we found that miR-181a significantly reduced luciferase activity for the wild-type reporter, while mutagenesis of the predicted miR-181a target sites abolished the previous suppressive effect (Figure 2C).To verify the physical interaction between miR-181a and AFAP1-AS at the endogenous level, RNA immunoprecipitation (RIP) assay was performed using a specific antibody against Ago2 protein. Compared with the control group, AFAP1-AS was preferentially enriched in Ago2-coating beads (Figure 2D). Together, our results revealed that miR-181a directly bind to AFAP1-AS.
AFAP1-AS acted as a ceRNA by biding miR-181a (A) Expression levels of AFAP1-AS in the nuclear and cytoplasm fractions were determined using qRT-PCR. GAPDH and U6 were used as cytoplasmic and nuclear markers, respectively. (B) The top and bottom regions are the sequences of AFAP1-AS wild-type binding to miR-181a and mutations in the 3'-UTR of AFAP1-AS, respectively. (C) The AFAP1-AS wild type or mutant vectors were co-transfected with miR-181a NC or miR-181a mimics. Firefly luciferase activities were then measured normalized to Renilla. (D) Extracts of 293T cells were applied for RNA binding protein immunoprecipitation (RIP). Relative RNA levels of AFAP1-AS and miR-181a were measured by qRT-PCR.
MiR-181a contributes to the development of HSCR
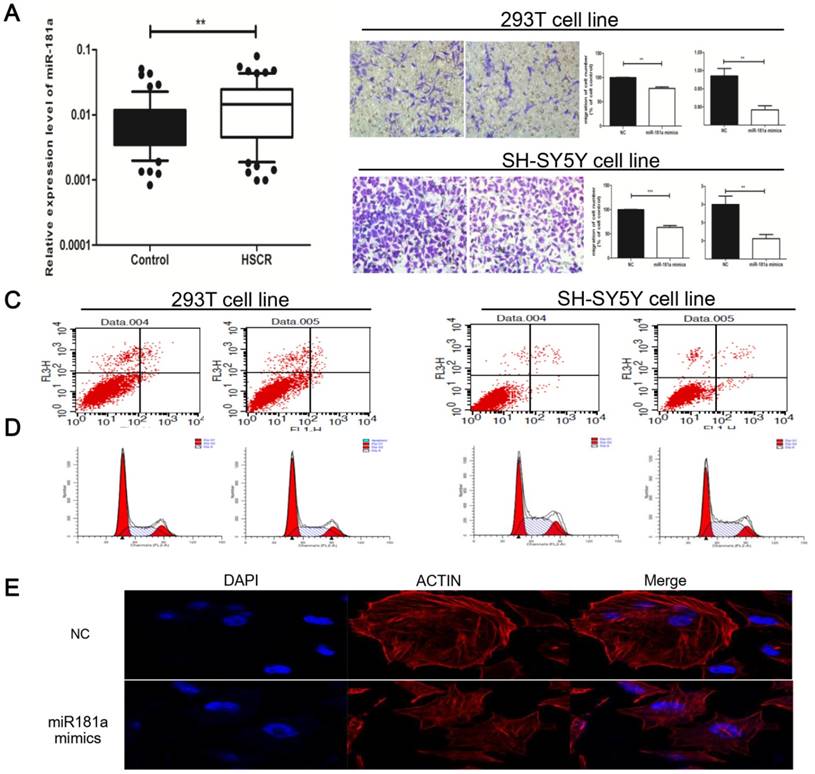
Based on the above results, miR-181a was likely to be involved in the pathogenesis of HSCR. Contrary to AFAP1-AS, miR-181a was remarkably up-regulated in HSCR compared to normal tissues, showing its potentiating effect on HSCR (Figure 3A). We then utilized 293T and SH-SY5Y cells to uncover the functions of miR-181a in HSCR. As was expected, CCK-8 assays revealed that cell viability was inhibited after transfection with miR-181a mimics. Meanwhile, cell migration was markedly reduced after transfection of miR-181a mimics both in 293T and SH-SY5Y cells (Figure 3B). No significance was detected in apoptosis and cell cycle (Figure 3C, D). Furthermore, the cytoskeleton was disrupted after transfection with miR-181a mimics (Figure 3E). In summary, these data presented the promoting effect of miR-181a on HSCR, in contrast to its ceRNA AFAP1-AS.
The functional assays of miR-181a were conducted in HSCR cells (A) The expression of miR-181a in HSCR was significantly over-expressed compared with normal tissues. Data were presented as box plot of the median and range of log-transformed relative expression level. The top and bottom of the box represent the 75th and 25th percentile. The whiskers indicate the 10th and 90th points. (B) Transwell analysis of 293T and SH-SY5Y cells transfected with negative control or miR-181a mimics (left panel); Migrated cells stained with crystal violet was shown (middle panel); Cell proliferation were evaluated by CCK8 assay(right panel).The data are presented as the mean ± SE. (C-D) Cycle and apoptosis assays were conducted after transfection with miR-181a mimics or negative control by flow cytometer. (E) F-actin cytoskeleton of was stained with Rhodamine Phalloidin staining after transfection with miR-181a mimics or negative control in SH-SY5Y cells.
AFAP1-AS acts as a ceRNA and regulates the miR-181a mRNA target, RAP1B
To identify the targets of miR-181a, we used miRsystem (http://mirsystem.cgm.ntu.edu.tw) which contains several algorithms and predicted numerous potential direct targets [24]. Gene ontology analysis revealed relative pathways for candidate genes. The Gene Ontology results showed that target genes of mir-181a were involved in multiple pathways, including MAPK signaling, regulation of the actin cytoskeleton, neurotrophin signaling and focal adhesion. Among these putative targets of miR-181a, we focused on RAP1B, shared by the above pathways. Previous study has reported that miR-181 targets RAP1B in glioblastoma cells [25]. As a small GTPaes, Rap1B is involved in cell adhesion which is mediated by integrin and cadherin, as well as the cytoskeleton during cell activation [26]. To confirm whether miR-181a targets RAP1B in HSCR, miRanda was applied to identify miR-181a recognition sites in the 3'-UTR of RAP1B (Figure 4A). Luciferase activity of wild RAP1B reporter was decreased, while mutant RAP1B reporter was not changed co-transfected with miR-181a (Figure 4B).
Relationship between AFAP1-AS and the miR-181a target, RAP1B (A) Top: Predicted binding sites between miR-181a and 3'UTR of RAP1B were constructed as well as mutations in the 3'-UTR of RAP1B. (B) The RAP1B wild type or mutant vectors were co-transfected with negative control or miR-181a mimics. Firefly luciferase activities were then measured normalized to Renilla. (C) Expression levels of RAP1B in the previous tissues were examined by qRT-PCR analysis. Data were presented as box plot of the median and range of log-transformed relative expression level. The top and bottom of the box represent the 75th and 25th percentile. The whiskers indicate the 10th and 90th points. (D) A correlation between AFAP1-AS and RAP1B was observed according to the qRT-PCR results (r2=0.4726; P<0.0001). (E) Western blots of RAP1B were measured in HSCR and normal tissues. (F) Western blot and qRT-PCR analysis of RAP1B were conducted after transfection with si-NC, si-AFAP1-AS, and inhibition of AFAP1-AS in combination with miR-181a inhibitor. GAPDH was used as control. All results were presented as mean ± SE.
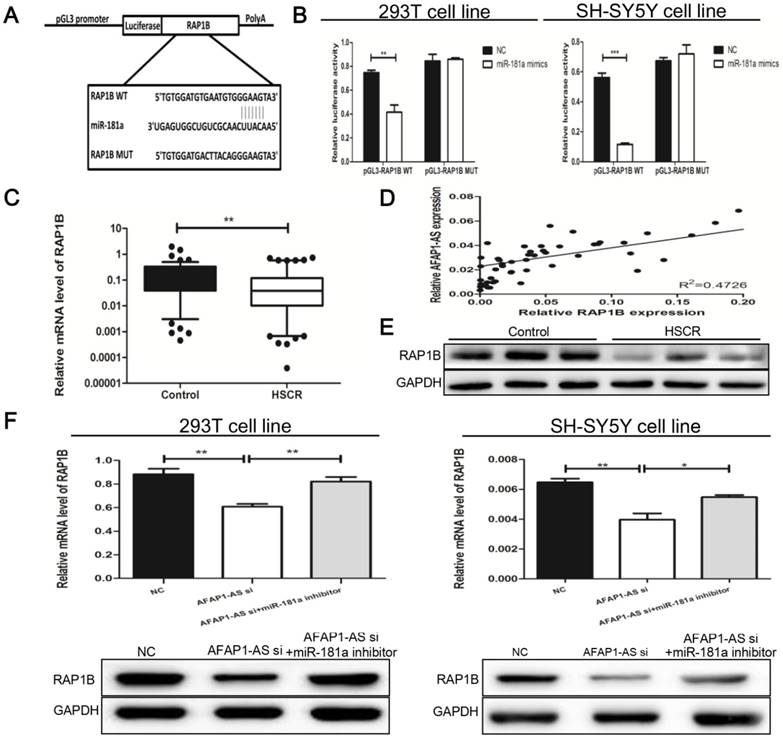
The above results strongly suggested that miR-181a directly targeted RAP1B, so we further explored the association of RAP1B with AFAP1-AS. Firstly, we measured the expression of RAP1B in the previous 128 tissues by qRT-PCR analysis and RAP1B was markedly decreased in HSCR tissue (Figure 4C). An inverse correlation between AFAP1-AS and RAP1B expression levels was observed (Figure 4D). The Western Blot results also revealed that RAP1B was down-regulated in HSCR tissues in comparison with normal tissues, which were consistent with the PCR result (Figure 4E).We further clarified the regulatory relationship between AFAP1-AS and RAP1B by AFAP1-AS knockdown against RAP1B in 293T and SH-SY5Y cells. PCR and Western Blot were carried out and showed that down-regulation of AFAP1-AS significantly inhibited RAP1B expression, whereas inhibition of AFAP1-AS in combination with miR-181a inhibitor did not affect RAP1B expression, suggesting that AFAP1-AS regulates RAP1B in a miR-181a-dependent manner (Figure 4F).
Together, these results revealed that through binding to miR-181a directly, AFAP1-AS acted as a ceRNA for RAP1B and modulated the expression of RAP1B in a post-transcriptional regulation.
Discussion
Increasing studies indicated that lncRNAs engaged in a broad spectrum of cellular processes, including tumorigenesis and embryogenesis. It has previously been reported that lncRNA AFAP1-AS was frequently dysregulated in several diseases, such as hepatocellular carcinoma, pancreatic ductal adenocarcinoma and nasopharyngeal carcinoma. However, the possible role of AFAP1-AS in HSCR remains to be clarified. In this study, we examined the expression of AFAP1-AS in HSCR samples and normal control tissues. The function of AFAP1-AS was further characterized by applying loss-of-function approaches in HSCR cell lines. We detected that AFAP1-AS was down-regulated in HSCR tissues in comparison with the normal tissues, and AFAP1-AS depletion inhibited cell migration and growth ability while no significant difference was observed about apoptosis and cell cycle. Furthermore, down-regulated AFAP1-AS contributed to the actin degradation and loss of cell polarity. Therefore, it is concluded that AFAP1-AS participated in the modulation of neural crest cell colonization. However, the inhibition of migration and growth capacity in HSCR by down-regulating AFAP1-AS needs further verification by in vivo experiments.
We also sought out the molecular mechanism of AFAP1-AS regulating neural crest cell colonization. AFAP1-AS exists in the cytoplasm, indicating that it can regulate gene expression at mRNA levels. As we known, ceRNAs are recognized as a novel regulatory mechanism of posttranscriptional gene expression by competing shared for miRNAs to suppress the expression of target genes. We integrated online bioinformatics database, and found that the mir-181a had the higher scores binding to AFAP1-AS. To further verify the relationship between miR-181a and AFAP1-AS, dual-luciferase reporter and RNA immunoprecipitation (RIP) assays were conducted. These results revealed that miR-181a can directly bind to the AFAP1-AS. As a well-known miRNA, miR-181a functions as a tumor suppressor in several cancers, such as breast cancer, glioma and gastric cancer [25, 27, 28]. Consistent with the above results, miR-181a was overexpression in HSCR tissues and negatively regulatd neural crest cell proliferation and migration through targeting RAP1B.Here we provide direct evidence that AFAP1-AS can function as a ceRNA, which sequesters miR-181a, thereby protecting RAP1B transcripts from miR-181a-mediated suppression. AFAP1-AS and RAP1B were validated as targets for miRNA-181a in our cellular models. Consistent with AFAP1-AS competing miRNA-181a with RAP1B, we found that while depression of AFAP1-AS could reduce expression of RAP1B. Besides, our studies found miRNA-181a inhibitor could rescue the down-regulation of RAP1B in AFAP1-AS knockdown cells, which support the ceRNA regulatory mechanism of RAP1B by AFAP1-AS.
Taken together, we reported that down-regulated lncRNA AFAP1-AS inhibits HSCR cell proliferation and migration by competitively binding the miR-181a, down-regulating RAP1B, and suppressing neural crest cell colonization. Our studies revealed a novel ceRNA regulatory network, AFAP1-AS/miR-181a/RAP1B, providing new insights into understanding the mechanisms of HSCR.
Abbreviations
HSCR: Hirschsprung disease; lncRNA: long non-coding RNA; miRNA: microRNA; ceRNA: competing endogenous RNA; CCK-8: Cell counting kit-8; DMEM: Dubelcco modified Eagle's medium; FBS: Fetal bovine serum; GAPDH: Glyceraldehyde-3-phosphate dehydrogenase; RIP: RNA immunoprecipitation
Acknowledgements
We thank Dr. Jie Zhang, HuanChen, Xiaofeng Lv, Weiwei Jiang, Wei Li and Changgui Lu (Children's Hospital of Nanjing Medical University) for sample collection. This study was supported by Natural Science Foundation of China (NSFC 81370473, NSFC 81400574, NSFC 81570467), Priority Academic Program Development of Jiangsu Higher Education Institutions (PAPD), Nanjing Medical Science and Technique Development Foundation (201405014) and Jiangsu Qing Lan Project.
Competing Interests
The authors have declared that no competing interest exists.
References
1. Borrego S, Ruiz-Ferrer M, Fernandez RM, Antinolo G. Hirschsprung's disease as a model of complex genetic etiology. Histol Histopathol. 2013;28:1117-36
2. Spouge D, Baird PA. Hirschsprung disease in a large birth cohort. Teratology. 1985;32:171-7
3. McKeown SJ, Stamp L, Hao MM, Young HM. Hirschsprung disease: a developmental disorder of the enteric nervous system. Wiley Interdiscip Rev Dev Biol. 2013;2:113-29
4. Butler Tjaden NE, Trainor PA. The developmental etiology and pathogenesis of Hirschsprung disease. Transl Res. 2013;162:1-15
5. Tang W, Tang J, Qin J, Geng Q, Zhou Z, Li B. et al. Involvement of down-regulated E2F3 in Hirschsprung's disease. J Pediatr Surg. 2013;48:813-7
6. Li H, Tang J, Lei H, Cai P, Zhu H, Li B. et al. Decreased MiR-200a/141 suppress cell migration and proliferation by targeting PTEN in Hirschsprung's disease. Cell Physiol Biochem. 2014;34:543-53
7. Tang W, Qin J, Tang J, Zhang H, Zhou Z, Li B. et al. Aberrant reduction of MiR-141 increased CD47/CUL3 in Hirschsprung's disease. Cell Physiol Biochem. 2013;32:1655-67
8. Lee S, Kopp F, Chang TC, Sataluri A, Chen B, Sivakumar S. et al. Noncoding RNA NORAD Regulates Genomic Stability by Sequestering PUMILIO Proteins. Cell. 2016;164:69-80
9. Shi X, Sun M, Liu H, Yao Y, Song Y. Long non-coding RNAs: a new frontier in the study of human diseases. Cancer Lett. 2013;339:159-66
10. Chen YA, Aravin AA. Non-Coding RNAs in Transcriptional Regulation: The review for Current Molecular Biology Reports. Curr Mol Biol Rep. 2015;1:10-8
11. Wan LB, Bartolomei MS. Regulation of imprinting in clusters: noncoding RNAs versus insulators. Adv Genet. 2008;61:207-23
12. Wan L, Sun M, Liu GJ, Wei CC, Zhang EB, Kong R. et al. Long Noncoding RNA PVT1 Promotes Non-Small Cell Lung Cancer Cell Proliferation through Epigenetically Regulating LATS2 Expression. Mol Cancer Ther. 2016;15:1082-94
13. Kitagawa M, Kitagawa K, Kotake Y, Niida H, Ohhata T. Cell cycle regulation by long non-coding RNAs. Cell Mol Life Sci. 2013;70:4785-94
14. Qi X, Zhang DH, Wu N, Xiao JH, Wang X, Ma W. ceRNA in cancer: possible functions and clinical implications. J Med Genet. 2015;52:710-8
15. Wu W, Bhagat TD, Yang X, Song JH, Cheng Y, Agarwal R. et al. Hypomethylation of noncoding DNA regions and overexpression of the long noncoding RNA, AFAP1-AS1, in Barrett's esophagus and esophageal adenocarcinoma. Gastroenterology. 2013;144:956-66 e4
16. Ye Y, Chen J, Zhou Y, Fu Z, Zhou Q, Wang Y. et al. High expression of AFAP1-AS1 is associated with poor survival and short-term recurrence in pancreatic ductal adenocarcinoma. J Transl Med. 2015;13:137
17. Zeng Z, Bo H, Gong Z, Lian Y, Li X, Li X. et al. AFAP1-AS1, a long noncoding RNA upregulated in lung cancer and promotes invasion and metastasis. Tumour Biol. 2016;37:729-37
18. Bo H, Gong Z, Zhang W, Li X, Zeng Y, Liao Q. et al. Upregulated long non-coding RNA AFAP1-AS1 expression is associated with progression and poor prognosis of nasopharyngeal carcinoma. Oncotarget. 2015;6:20404-18
19. Zhou XL, Wang WW, Zhu WG, Yu CH, Tao GZ, Wu QQ. et al. High expression of long non-coding RNA AFAP1-AS1 predicts chemoradioresistance and poor prognosis in patients with esophageal squamous cell carcinoma treated with definitive chemoradiotherapy. Mol Carcinog. 2016
20. Vargiolu M, Fusco D, Kurelac I, Dirnberger D, Baumeister R, Morra I. et al. The tyrosine kinase receptor RET interacts in vivo with aryl hydrocarbon receptor-interacting protein to alter survivin availability. J Clin Endocrinol Metab. 2009;94:2571-8
21. Kawamoto T, Ohira M, Hamano S, Hori T, Nakagawara A. High expression of the novel endothelin-converting enzyme genes, Nbla03145/ECEL1alpha and beta, is associated with favorable prognosis in human neuroblastomas. Int J Oncol. 2003;22:815-22
22. Peng H, Luo J, Hao H, Hu J, Xie SK, Ren D. et al. MicroRNA-100 regulates SW620 colorectal cancer cell proliferation and invasion by targeting RAP1B. Oncol Rep. 2014;31:2055-62
23. Huang HY, Chien CH, Jen KH, Huang HD. RegRNA: an integrated web server for identifying regulatory RNA motifs and elements. Nucleic Acids Res. 2006;34:W429-34
24. Lu TP, Lee CY, Tsai MH, Chiu YC, Hsiao CK, Lai LC. et al. miRSystem: an integrated system for characterizing enriched functions and pathways of microRNA targets. PLoS One. 2012;7:e42390
25. She X, Yu Z, Cui Y, Lei Q, Wang Z, Xu G. et al. miR-181 subunits enhance the chemosensitivity of temozolomide by Rap1B-mediated cytoskeleton remodeling in glioblastoma cells. Med Oncol. 2014;31:892
26. Schwamborn JC, Puschel AW. The sequential activity of the GTPases Rap1B and Cdc42 determines neuronal polarity. Nat Neurosci. 2004;7:923-9
27. Neel JC, Lebrun JJ. Activin and TGFbeta regulate expression of the microRNA-181 family to promote cell migration and invasion in breast cancer cells. Cell Signal. 2013;25:1556-66
28. Peng W, Si S, Zhang Q, Li C, Zhao F, Wang F. et al. Long non-coding RNA MEG3 functions as a competing endogenous RNA to regulate gastric cancer progression. J Exp Clin Cancer Res. 2015;34:79
Author contact
![]() Corresponding author: Weibing Tang, Department of Pediatric Surgery, Children's Hospital of Nanjing Medical University, Nanjing 210008, China. Tel: +86-25-83117354; E-mail: twbcnedu.cn; Fax: +86-25-86868427
Corresponding author: Weibing Tang, Department of Pediatric Surgery, Children's Hospital of Nanjing Medical University, Nanjing 210008, China. Tel: +86-25-83117354; E-mail: twbcnedu.cn; Fax: +86-25-86868427

 Global reach, higher impact
Global reach, higher impact