Impact Factor
ISSN: 1449-1907
Int J Med Sci 2017; 14(7):622-628. doi:10.7150/ijms.18972 This issue Cite
Research Paper
MicroRNA Expression Analysis of Centenarians and Rheumatoid Arthritis Patients Reveals a Common Expression Pattern
1. Department of Biomedical Sciences, University of Sassari, vl. San Pietro 43b, Sassari 07100, Italy;
2. Associazione “L'Isola dei Centenari”, Via Milano 4, Sassari 07100, Italy;
3. Istituto Zooprofilattico Sperimentale della Sardegna, Via Vienna 2, Sassari 07100, Italy;
4. Internal Medicine 1, A.O.U.University of Sassari, Sassari, Italy;
5. Surgical Pathology, A.O.U. University of Sassari, Sassari, Italy.
Received 2016-12-29; Accepted 2017-3-29; Published 2017-6-14
Abstract
Micro-RNA (miRNA) are a family of small non-coding ribonucleic acids that inhibits post-transcriptionally the expression of their target messenger RNA (mRNA). We are interested in studying the involvement of miRNA in longevity and autoimmune diseases. In this study we compared the different expression of seven microRNAs between human plasma healthy controls, plasma samples of centenarians and samples from patients with rheumatoid arthritis. We used the Life Technologies' protocol to quantify seven miRNAs from 62 plasma samples: 20 healthy human controls, 14 centenarians, 28 patients with rheumatoid arthritis. TaqMan MicroRNA assays were used to analyze the expression profiles of miR-125b-5p, miR-425-5p, miR-200b5p, miR-200c-3p, miR-579-3p, miR-212-3p, miR-21-5p and miR-126-3p. The relative expression of mature miRNAs was analyzed using software REST. Our results show that miR-425-5p, miR-21 and miR-212 significantly decreased in centenarians and in patients with rheumatoid arthritis compared with controls. Furthermore in this work we highlight a connection between corticosteroid treatment and miRNAs expression.
Keywords: miRNA, centenarians, autoimmune diseases, glucocorticoids
Introduction
Micro-RNAs (miRNAs) are a family of small non-coding ribonucleic acids that inhibits post-transcriptionally the expression of their target messenger RNA (mRNA), which then acts as general repressor gene [1]. miRNAs seem to act in highly complex regulatory networks that control batteries of target genes [2]. Human aging is an extremely complex process and miRNAs have proven to play a role in modulating the duration and the aging process. Recently, miRNAs have emerged as critical regulators of gene expression and a link between multiple miRNAs, longevity [2, 3] and aging [4] has been demonstrated in C. elegans, implicating their role in regulation of lifespan and in the aging process. Since a significant number of miRNAs are evolutionarily conserved [5, 6], it is conceivable that miRNAs play a role in human aging and longevity but more information are needed to understand their whole role. Particularly, few studies are still available on miRNAs' pattern in centenarians, persons who live to or beyond the age of 100 years. miRNAs also circulate in a cell-free form in body fluids including serum and plasma [7]. These cell-free miRNAs are highly stable and resistant to harsh conditions including heat, pH changes, freeze/thaw cycles and extended storage [8]. Altered expression of circulating miRNAs has been associated with age-related diseases including cancer, inflammatory and autoimmune diseases. Autoimmune diseases (ADs) are featured by body's immune responses being directed towards its own specific target organs or multiple organ systems, causing persistent inflammation and consequent tissue damage. Because both this groups (autoimmune disease patients and old people) are characterized by an increase of inflammatory processes and we are interested in studying the involvement of miRNAs in longevity and autoimmunity, in this study we compared the different expression of seven microRNAs between 20 human plasma healthy controls, 14 plasma samples of centenarians and 28 samples from patients with rheumatoid arthritis. All samples were recruited from the biobank longevity Akea project (project approved by the local bioethics).
Results
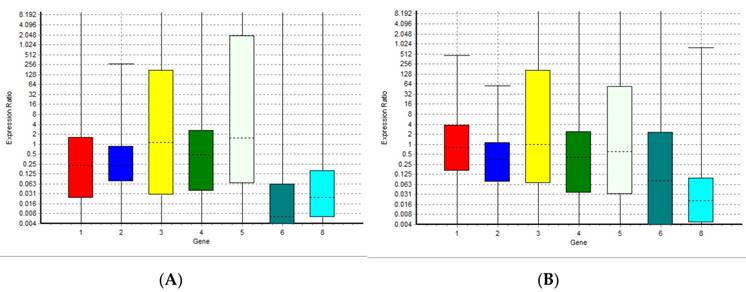
The results of the analysis performed with normfinder software suggested miRNA-126-3p as the best normaliser. The statistical analysis showed that data for all targets resulted not normally distributed. The Kruskal-Wallis test revealed no significant differences between groups for miRNA-126-3p, confirming the results of normfinder software, because of the miRNA-126-3p greater stability between groups of samples. We analyzed eight miRNAs: miR-125b-5p; miR-425-5p; miR-200b-5p; miR-200c-3p; miR-579-3p; miR-212-3p; miR-126-3p; and miR-21-5p identified with a number from 1 to 8 in all graphics and tables, using miRNA miR-126-3p (number 7) as a normaliser. This miRNA is not present in the graphs but only in the tables. We performed a comparison between the level of miRNAs in human plasma healthy controls and both the level of miRNAs in patients with rheumatoid arthritis and in plasma samples of centenarians. Figure 1A and Table 1 show the relative expression of the miRNAs analyzed in this study in plasma samples from patients with rheumatoid arthritis compared with healthy controls. Figure 1B and Table 2 show the relative expression of the miRNAs analyzed in this study in plasma samples of centenarians compared with healthy controls. The results show that there are no significant differences in the levels of miR-125b-5p, miR-200c-3p, miR-200b-5p, miR-579-3p and miR-126-3p between patients with rheumatoid arthritis and healthy controls. Instead, the expression level of miR-425, miR-21 and miR-212-3p is significantly lower in patients with rheumatoid arthritis compared to healthy controls. In the same manner, the comparison between plasma samples of centenarians and healthy controls shows no significant differences in the levels of miR-125b-5p, miR-200c-3p, miR-200b-5p, miR-579-3p and miR-126-3p while the expression level of miR-425, miR-21 and miR-212-3p is significantly lower in centenarians compared to healthy controls. - Furthermore the comparison between rheumatoid arthritis patients and centenarians doesn't show significant differences (data not shown).
Boxplots representing the expression variation of the miRNAs analyzed in this study in rheumatoid arthritis patients and centenarians compared with healthy controls. The graph (A) shows the expression variation in autoimmune disease patients; the graph (B) shows the expression variation in centenarians. 1 = miR-125b-5p; 2 = miR-425-5p; 3 = miR-200b-5p; 4 = miR-200c-3p; 5 = miR-579-3p; 6 = miR-212-3p; 8 = miR-21-5p.
Relative expression, standard error, 95% confidence interval (C.I.), and p-value of miRNAs analyzed in this study (rheumatoid arthritis patients compared with healthy controls). p-value (≥0.05) shows that there are no significant differences. 1 = miR-125b-5p; 2 = miR-425-5p; 3 = miR-200b-5p; 4 = miR-200c-3p; 5 = miR-579-3p; 6 = miR-212-3p; 7 = miR-126-3p; 8 = miR-21-5p.
| Gene | Type | Reaction Efficiency | Expression | Std. Error | 95% C.I. | P(H1) | Result |
|---|---|---|---|---|---|---|---|
| 1 | TRG | 1.0 | 0.228 | 0.005 - 5.223 | 0.000 - 870.638 | 0.067 | |
| 2 | TRG | 1.0 | 0.229 | 0.040 - 1.687 | 0.001 - 16.943 | 0.003 | DOWN |
| 3 | TRG | 1.0 | 4.471 | 0.008 - 7.628 | 0.000 - 14.404 | 0.340 | |
| 4 | TRG | 1.0 | 0.597 | 0.011 - 7.964 | 0.001 - 162.782 | 0.616 | |
| 5 | TRG | 1.0 | 6.872 | 0.007 - 15.484 | 0.000 - 441.260 | 0.180 | |
| 6 | TRG | 1.0 | 0.013 | 0.000 - 0.302 | 0.000 - 90.867 | 0.000 | DOWN |
| 7 | REF | 1.0 | 1.000 | ||||
| 8 | TRG | 1.0 | 0.020 | 0.000 - 0.433 | 0.000 - 36.225 | 0.000 | DOWN |
Relative expression, standard error, 95% confidence interval (C.I.), and p-value of miRNAs analyzed in this study (centenarians compared with healthy controls). p-value (≥0.05) shows that there are no significant differences. 1 = miR-125b-5p; 2 = miR-425-5p; 3 = miR-200b-5p; 4 = miR-200c-3p; 5 = miR-579-3p; 6 = miR-212-3p; 7 = miR-126-3p; 8 = miR-21-5p.
| Gene | Type | Reaction Efficiency | Expression | Std. Error | 95% C.I. | P(H1) | Result |
|---|---|---|---|---|---|---|---|
| 1 | TRG | 1.0 | 0.692 | 0.069 - 8.580 | 0.002 - 94.403 | 0.599 | |
| 2 | TRG | 1.0 | 0.000 | 0.002 - 1.874 | 0.000 - 7.908 | 0.004 | DOWN |
| 3 | TRG | 1.0 | 7.958 | 0.015 - 153.955 | 0.001 - 25.642. | 0.242 | |
| 4 | TRG | 1.0 | 0.462 | 0.006 - 4.394 | 0.001 - 120.067 | 0.509 | |
| 5 | TRG | 1.0 | 1.571 | 0.004 - 1.569. | 0.000 - 102.350 | 0.744 | |
| 6 | TRG | 1.0 | 0.048 | 0.000 - 15.842 | 0.000 - 1.696 | 0.027 | DOWN |
| 7 | REF | 1.0 | 1.000 | ||||
| 8 | TRG | 1.0 | 0.020 | 0.002 - 0.218 | 0.000 - 14.026 | 0.001 | DOWN |
Discussion
Our study showed a similar miRNA expression pattern in centenarians and rheumatoid arthritis patients treated with glucocortioids, the comparison of both groups to healthy controls showed a low expression of miR-425-p, miR-21-5p and miR-212-3p.
miR-425-5p and miR-21-5p are implicated in chronic-degenerative inflammations like those associated with autoimmune and age-related diseases [9], they have a well-known role in the progression of cancer diseases. The low expression of miR-425-5p, miR-21-5p and the high expression of miR-212-3p seems to be associated with antitumor and therapeutic activity [10].
miR-425
Jun Ma et al. have investigated the effect of miR-425 on tumorigenicity in vivo [11]. The tumors treated with anti-miR-425 showed increased levels of the phosphatase and tensin homolog (PTEN) protein, a gene identified as a tumor suppressor that is mutated in a large number of cancers at high frequency. PTEN is also regulated by miR-21.
miR-21
Down regulation of miR-21 leads to increased PTEN expression both endogenously and in a reporter gene assay [(12]. Microarray analysis showed that the knockdown of miR-21 significantly alters the expression of 169 genes involved in nine cell-cycle and signaling pathways. MiR-21 was recently reported to target multiple genes associated with the immunological antimicrobial response, providing an effective mechanism to escape from the antimicrobial pathway [13]. Previous research showing correlation between miR-21 expression and IL-6 and IL-8 levels suggests a potential role of miR-21 in inflammatory gene expression [14]. Increased miR-21 expression has also been reported in diseases characterized by chronic inflammation including colitis, atherosclerosis, type 2 diabetes, and SLE. MiR-21 ablation in some of these models, including colitis and psoriasis [15] has actually shown protection from disease, indicating that in these diseases, miR-21 activity promotes inflammation. In some cancer models, miR-21 expression itself is associated with inflammatory activation. It can promote Nuclear Factor kB (NFκB) activation in breast cancer cells and TNF and IFNγ production in activated T-cells [15] clearly acting to induce inflammation in transformed tumor-cells and activated T-cells. miR-21 can regulate numerous processes involved in correct cell function, survival and proliferation, which if interrupted, can predispose to cellular transformation.
Nuclear Factor kB (NFκB)
NF-kB is a transcriptional factor that regulates a battery of genes that are critical to innate and adaptive immunity, cell proliferation, inflammation, and tumor development. NF-κB positively regulates miR-21, mir-212 and mir-425 expression. Consistent with published data these miRNAs are a potential target of NF-κB [16]. Some of the transcription factors that are inhibited by glucocorticoids, such as NF-κB, are also targets for novel anti-inflammatory therapies [17].
Glucocorticoids
Glucocorticoids are effective anti-inflammatory drugs and potent immunosuppressive agents. The pharmacology of glucocorticoids is very complex and is the subject of continuous studies in an attempt to clarify the mechanisms by which these activities are exercised. Glucocorticoids penetrate the cell and bind to their cytoplasmatic receptor (GR). The glucocorticoid-GR complex, in addition, is able to block the way of the NF-kB; probably, most of the anti-inflammatory actions are identifiable at this level. The NF-kB is normally present in the cytoplasm in an inactive form.
Lower expression of miR-21-5p, miR-425-5p, miR-212-3p
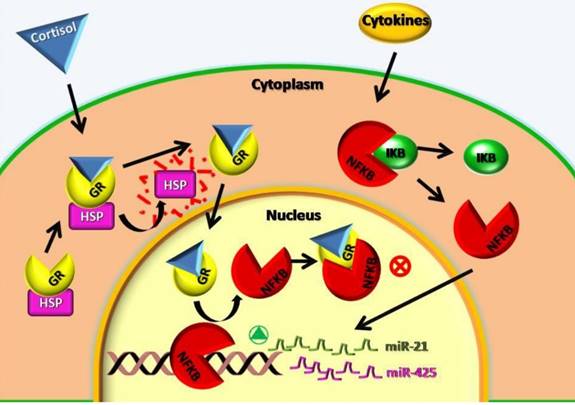
The analysis of the miRNA expression performed in this study showed a lower expression of mir-21-5p, mir-212-3p and mir-425-5p in contrast to other authors that found an increase of mir-21-5p in autoimmune diseases and increase of mir-425-5p in inflammation and cancer diseases [11]. Concerning our patients with autoimmune diseases this could be explained with the corticosteroids treatment. These subjects have a low expression of mir-21-5p, mir-212-3p and mir-425-5p because the corticosteroids inhibit the nuclear factor NF-KB and its absence blocks the expression of mir-21-5p, mir-212-3p and mir-425-5p. The figure 2 could better explain the cellular mechanisms above mentioned. Furthermore, if up-regulated, miR-425-5p and miR-21-5p directly target PTEN and negatively regulate its expression, which promotes cell survival through the apoptosis inhibition [11].
Our results show that in centenarians 4 out 7 miRNAs analysed don't show significant differences in their expression level respect to the controls, but the extreme variation of the expression ratio does not allow us to reach definitive conclusions.
Cellular mechanisms involved in miR-21 and miR-425 low expression after cortisol treatment. GR: cortisol receptor; HSP: Heat Shock Protein; NFKB: Nuclear Factor NFκB; IKB: NFKB receptor; ⊗: block of metabolic way;  : activation of metabolic way.
: activation of metabolic way.
The low expression of miR-425-5p, miR-21-and miR-212.3p seems to be a centenarians' characteristic. Some authors showed that inhibitors of miR-425 might low blood pressure by depressing the atrial natriuretic peptide expression [19]. This finding is very interesting and, if confirmed, could demonstrate a protective effect of mir-425-5p low expression, in particular in centenarians.
The lack of differences between the centenarians and the autoimmune group and the similarities of both these groups compared with healthy controls should be further investigated. Our data are not sufficient to establish if the two groups are characterized by some of the same regulatory mechanisms. However, we can speculate that the observed lower expression of the miR-425-5p, miR-21-5p and miR-212-3p in centenarians could guarantee a lower level of inflammatory processes and this could be protective for several diseases. In centenarians this miRNAs pattern could protect against inflammation like in patients treated with glucocorticoids but without the negative effects of glucocorticoids. Another study suggests that miRNA deregulation, particularly mir-21-5p, may limit cardiovascular repair responses and result in an altered function and differentiation of cardiovascular progenitor cells and endothelial progenitor cells (EPCs), modulating endothelial regeneration and cardiomyocyte homeostasis and playing a crucial role in cardiovascular disease (CVD) [20]. This suggests the importance of studying the mechanism of the drugs on miRNAs because it could open new frontiers for the treatment of many diseases.
Materials and Methods
Human Plasma Samples
We used the Life Technologies' protocol to analyze eight miRNAs from 62 plasma samples. 20 healthy human controls aged between 30 and 50 years, 14 healthy non medicated centenarians exceeding 100 years, 28 patients with rheumatoid arthritis aged between 30 and 60 years, were analysed.
The patients with rheumatoid arthritis were in treatment with glucocorticoids and in the remission phase of the pathology. The drug used was Prednisone, a synthetic drug similar to cortisone, used to relieve rheumatic and allergic conditions and to treat leukemia. The dosage was 2,5-5 mg of prednisone oral administration. The donors were volunteers recruited from the longevity AKeA Project [21] (project approved by the Institutional Local Ethics Committee,Azienda Sanitaria Locale n°1 di Sassari, Italy,N° 398/L.) and ubjects signed a written consent prior to blood sampling. Blood samples were collected early in the morning (to reduce the biological variability) by venipucture into a vacutainer (Greiner Bio-One, Monroe, NC, USA) containing K2 EDTA as an anticoagulant. Then we proceeded to the separation of the plasma by centrifugation at 2500 g for 15 min at room temperature. The supernatant containing the plasma was divided into a defined number of aliquots of 200 µL each and stored at -80°C until RNA extraction under a temperature monitoring system.
RNA Extraction
The RNA was extracted from plasma fractions using miRNeasy kits Serum / Plasma (50) (Qiagen, Milan, Italy) according to the manufacturer's instructions, with the final elution volume of 15 µl. Eight individual miRNAs (miR-125b-5p, miR-425-5p, miR-200b-5p, miR-200c-3p, miR-579-3p, miR-126-3p, miR-212-3p, and miR-21-5p) were selected based on their expression in the plasma of healthy donors and previously conducted studies on centenarians and patients with autoimmune diseases [22]. The U6 RNA was analyzed in order to verify its use as data normaliser [24, 25].
qPCR Analysis
The expression level of mature miRNAs was tested by quantitative real-time PCR (qPCR) using TaqMan® MicroRNA Reverse Transcription Kit, Life Technologies (Carlsbad, CA, USA), for the reverse transcription. TaqMan® Universal Master Mix II, Life Technologies, was used for the PCR according to the manufacturer's instructions; 45 amplification cycles were performed. MiRNA expression levels were quantified using the IQ5, BIORAD, instrument (Milan, Italy). miR-126-3p was selected for the data normalization [23, 24, 25]. Real-time PCR was done in duplicate. The miRNAs analyzed in this study were identified with progressive numbers from 1 to 8 (Table 6). The sequences and the identification symbols were retrieved from miRbase and are reported in Table 6 [26].
Statistical Analysis and Real-Time PCR Data Analysis
All the miRNAs and U6sRNA were checked to use as normaliser using both the Normfinder software [23] and classical statistical analysis. The raw Ct values for each miRNA and U6snRNA were checked for normal distribution. The Kruskal-Wallis test was applied to compare the groups (centenarians, rheumatoid arthritis patients and control) in each target. The statistical analysis was performed with the SPSS software version 17.0. Reverse transcription followed by polymerase chain reaction (RT-PCR) is the most suitable method for the detection and quantification of miRNA. It provides high sensitivity, good reproducibility, and a wide range quantification. Several mathematical algorithms have been developed to calculate a ratio of expression based on real-time PCR efficiency and the crossing point deviation of an unknown sample against a control. Then a software tool named REST© (relative expression software) [27] was used, which compares two o more groups and different reference and target genes. The mathematical model used is based on the correction for exact PCR efficiencies and the mean crossing point deviation between sample group(s) and control group(s). Subsequently the expression ratio results of the investigated transcripts are tested for significance by a Pair Wise Fixed Reallocation Randomisation Test © and plotted using standard error (SE) estimation via a complex Taylor algorithm. Expression variation for each gene is visualized in a whisker-box plot. In this study, the relative expression of the mature miRNAs was analyzed using the software REST. The non-parametric bootstrapping test was used to evaluate expression differences of miRNAs between cases and controls.
Conclusions
Our results show that miR-425-5p, miR-21-5p and miR-212-3p significantly decreased in centenarians and in patients with rheumatoid arthritis, compared with controls. The similarity between centenarians and patients with autoimmune diseases treated with glucocorticoids is very interesting and deserve to be investigated thoroughly. Furthermore in this work we highlight a connection between the corticosteroid treatment and miRNA expression.
The miR- 425-5p and miR-21-5p, were proven to be overexpressed in cancer progression and in chronic-degenerative inflammations like those associated with autoimmune and age-related diseases. We can speculate that centenarians have survived diseases and cancer due to their lifestyle but also to a low inflammation level. Recent studies linked altered miRNAs function to a number of diseases and age-related processes. The identification of miRNAs that modulate longevity will provide important insights into the molecular basis of aging. Considering the results obtained, the low expression of miR-425-5p, miR-21-5p and miR-212-3p seems to be of great interest in age-related processes and metabolic diseases. These results suggest that circulating miRNAs may be a biological marker of aging and could also be important for regulating longevity. Identification of stable miRNA biomarkers in serum could have great potential as a non-invasive diagnostic tool as well as enhance our understanding of physiological changes that occur with age. The low expression of miRNAs we analyzed in this work seems to be different from that reported by other authors. This finding could suggest that doesn't exist a single miRNA pathway of centenarians, but that could exist several pathways linked to different countries and lifestyles of centenarians. So it would be interesting to perform a comparison within centenarians and not only between centenarians and healthy controls.
Acknowledgements
We gratefully acknowledge the AKeA project equipe (Luca Deiana) and the Associazione “Isola dei Centenari”. Funding: L.D. was funded by Regione Autonoma della Sardegna, grant number L.R.7/2007; C.C. was funded by MIUR Progetti di Ricerca di Rilevante Interesse Nazionale - PRIN 2015 (Prot. 20157ATSLF_002). Funding provided to the institution Associazione “Isola dei Centenari” by Fondazione Banco di Sardegna, grant numbers Prot. 998/2007.0729, Prot. 906/2012-1046. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Author Contributions
Conceived and designed the experiments: Luca Deiana, Francesca Balzano. Performed the experiments: Luca Deiana, Francesca Balzano, Marta Deiana, Ciriaco Carru. Analyzed data: Luca Deiana, Francesca Balzano, Silvia Dei Giudici, Annalisa Oggiano. Contributed reagents/materials/analysis tools: Luca Deiana, Marta Deiana. Wrote the paper: Francesca Balzano, Luca Deiana, Silvia Dei Giudici, Annalisa Oggiano. Technical support: Ciriaco Carru, Angelo Zinellu, Baingio Porcu, Andrea Mannu, Sara Pinna, Nicola Deiana.
Conflicts of Interest
The authors declare no conflict of interest.
References
1. Bartel DP. MicroRNAs: Genomics, biogenesis, mechanism and function. Cell. 2004;116:281-297
2. Boehm M, Slack FA. Developmental timing microRNA and its target regulate life span in C. elegans. Science. 2005;310(5756):1954-1957
3. de Lencastre A, Pincus Z, Zhou K, Kato M, Lee SS, Slack FJ. MicroRNAs both promote and antagonize longevity in C. elegans. Curr Biol. 2010;20(24):2159-2168
4. Ibanez-Ventoso C, Yang M, Guo S, Robins H, Padgett RW, Driscoll M. Modulated microRNA expression during adult lifespan in Caenorhabditis elegans. Aging Cell. 2006;5(3):235-246
5. Ibanez-Ventoso C, Vora M, Driscoll M. Sequence Relationships among C-elegans, D-melanogaster and Human microRNAs Highlight the Extensive Conservation of microRNAs in Biology. PLoS One. 2008;3:7
6. Wang E. MicroRNA Regulation and its Biological Significance in Personalized Medicine and Aging. Current Genomics. 2009;10(3):143-143
7. Cortez MA, Bueso-Ramos C, Ferdin J, Lopez-Berestein G, Sood AK, Calin GA. MicroRNAs in body fluids-the mix of hormones and biomarkers. Nat Rev Clin Oncol. 2012;8:467-477
8. Balzano F, Deiana M, Dei Giudici S, Oggiano A, Baralla A, Pasella S, Mannu A, Pescatori M, Porcu B, Fanciulli G, Zinellu A, Carru C, Deiana L. miRNA Stability in Frozen Plasma Sample. Molecules. 2015;20(10):19030-19040 doi: 10.3390/molecules201019030
9. Olivieri F, Spazzafumo L, Santini G, Lazzarini R, Albertini MC, Rippo MR, Galeazzi R, Abbatecola AM, Marcheselli F, Monti D, Ostan R, Cevenini E, Antonicelli R, Franceschi C, Procopio AD. Age-related differences in the expression of circulating microRNAs: miR-21 as a new circulating marker of inflammaging. Mech Ageing Dev. 2012;133:675-685
10. Noren Hooten N, Fitzpatrick M, Wood WH, De S, Ejiogu N, Zhang Y, Mattison JA, Becker KG, Zonderman AB, Michele MK. Age-related changes in microRNA levels in serum. Aging. 2013;5(10):725-740
11. Jun Ma, Jun Liu, Zhiming Wang, Xixi Gu, Yue Fan, Wen Zhang, Lili Xu, Jianjun Zhang, Dingfang Cai. NF-kappaB-dependent MicroRNA-425 upregulation promotes gastric cancer cell growth by targeting PTEN upon IL-1β induction. Mol Cancer. 2014;13:40
12. Xuan Zhou, Yu Ren, Lynette Moore, Mei Me, Yongping You, Peng Xu, Baoli Wang, Guangxiu Wang, Zhifan Jia, Peiyu Pu, Wei Zhang, Chunsheng Kang. Downregulation of miR-21 inhibits EGFR pathway and suppresses the growth of human glioblastoma cells independent of PTEN status. Laboratory Investigation. 2010;90(2):144-55
13. Liu PT, Wheelwright M, Teles R, Komisopoulou E, Edfeldt K, Ferguson B, Mehta MD, Vazirnia A, Rea TH, Sarno EN, Graeber TG, Modlin RL. MicroRNA-21 targets the vitamin D-dependent antimicrobial pathway in leprosy. Nat Med. 2012;18(2):267-73 doi: 10.1038/nm.2584
14. Schetter AJ, Nguyen GH, Bowman ED, Mathe EA, Yuen ST, Hawkes JE, Croce CM, Leung SY, Harris CC. Association of inflammation-related and microRNA gene expression with cancer-specific mortality of colon adenocarcinoma. Clin Cancer Res. 2009;15:5878-588
15. Frederick J, Sheedy. Turning 21: Induction of miR-21 as a Key Switch in the Inflammatory Response. Front Immunol. 2015;6:19. doi: 10.3389/fimmu.2015.00019
16. Ruan Q, Wang P, Wang T, Qi J, Wei M, Wang S, Fan T, Johnson D, Wan X, Shi W, Sun H, Chen YH. MicroRNA-21 regulates T-cell apoptosis by directly targeting the tumor suppressor gene Tipe2. Cell Death Dis. 2014;2:e1095
17. Barnes PJ. Anti-inflammatory actions of glucocorticoids: molecular mechanisms. Clin Sci Lond. 1998;94(6):557-72
18. Smith-Vikos T, Slack FJ. MicroRNAs and their roles in aging. J Cell Sci. 2012;125:7-17 doi: 10.1242/jcs
19. Olson EN. MicroRNAs as Therapeutic Targets and Biomarkers of Cardiovascular Disease. Science Translational Medicine. 2014;6:239-239 DOI: 10.1126/scitranslmed.3009008
20. Jakob P, Landmesser U. Role of microRNAs in stem/progenitor cells and cardiovascular repair. Cardiovasc Res. 2012;93(4):614-22
21. Deiana L, Ferrucci L, Pes GM. et al. AKEntAnnos. The Sardinia Study of Extreme Longevi-ty. Aging. 1999;11(3):142-9
22. Cortez MA, Bueso-Ramos C, Ferdin J, Lopez-Berestein G, Sood AK, Calin GA. MicroRNAs in body fluids-the mix of hormones and biomarkers. Nat Rev Clin Oncol. 2012;8:467-477
23. Andersen CL, Ledet-Jensen J, Ørntoft T. Normalization of real-time quantitative RT-PCR data: a model based variance estimation approach to identify genes suited for normalization - applied to bladder- and colon-cancer data-sets. Cancer Research. 2004;64:5245-5250
24. Benz F, Roderburg C, Vargas Cardenas D, Vucur M, Gautheron J, Koch A, Zimmermann H, Janssen J, Nieuwenhuijsen L, Luedde M. et al. U6 is unsuitable for normalization of serum miRNA levels in patients with sepsis or liver fibrosis. Exp Mol Med. 2013:45 (42) doi: 10.1038/emm.2013.81
25. Rice J, Roberts H, Rai SN, Galandiuk S. Housekeeping genes for studies of plasma microRNA: A need for more precise standardization. Surgery. 2015;158:1345-1351
26. miRbase. http://www.mirbase.org/
27. Pfaffl MW, Horgan GW, Dempfle L. Relative expression software tool (REST©) forgroup-wise comparison and statistical analysis of relative expression results in real-time PCR. Nucleic Acids Res. 2002;30:9 doi 10.1093/nar/30.9.e36
Author contact
![]() Corresponding author: Luca Deiana, Viale San Pietro 43, 07100 Sassari, Sardinia (Italy). TEL: +39-079-228275, FAX: +39-079-228275, e-mail: proflucadeianacom.
Corresponding author: Luca Deiana, Viale San Pietro 43, 07100 Sassari, Sardinia (Italy). TEL: +39-079-228275, FAX: +39-079-228275, e-mail: proflucadeianacom.

 Global reach, higher impact
Global reach, higher impact