Impact Factor
ISSN: 1449-1907
Int J Med Sci 2017; 14(6):595-601. doi:10.7150/ijms.18996 This issue Cite
Research Paper
Gastric Juice-Based Real-Time PCR for Tailored Helicobacter Pylori Treatment: A Practical Approach
1. State Key Laboratory of Infectious Disease Prevention and Control, Collaborative Innovation Center for Diagnosis and Treatment of Infectious Diseases, Chinese Center for Disease Control and Prevention, Beijing, China;
2. Department of Gastroenterology, Peking University Third Hospital, Beijing, China.
Received 2016-12-31; Accepted 2017-3-15; Published 2017-5-15
Abstract
A gastric juice-based real-time polymerase chain reaction (PCR) assay was established to identify Helicobacter pylori infection, clarithromycin susceptibility and human CYP2C19 genotypes and to guide the choice of proton pump inhibitor (PPI), clarithromycin and amoxicillin treatment for tailored H. pylori eradication therapy. From January 2013 to November 2014, 178 consecutive dyspeptic patients were enrolled for collection of gastric biopsy samples and gastric juice by endoscopy at the Peking University Third Hospital; 105 and 73 H. pylori-positive and -negative patients, respectively, were included in this study. H. pylori infection was defined as samples with both a strongly positive rapid urease test (RUT) and positive H. pylori histology. A series of primers and probes were distributed into four reactions for identifying the H. pylori cagH gene coupled with an internal control (Rnase P gene), A2142G and A2143G mutants of the H. pylori 23S rRNA gene, and single-nucleotide polymorphisms (SNPs) G681A of CYP2C19*2 and G636A of CYP2C19*3. The E-test and DNA sequencing were used to evaluate the H. pylori clarithromycin susceptibility phenotype and genotype. The SNPs CYP2C19*2 and CYP2C19*3 were also evaluated by nucleotide sequencing. The sensitivity, specificity, positive predictive value (PPV), and negative predictive value (NPV) of this gastric juice-based real-time PCR assay were evaluated by comparing with the same measures obtained through gastric biopsy-based PCR and culture. The H. pylori diagnostic sensitivities of the culture, PCR, and gastric biopsy- and gastric juice-based real-time PCR assays were 90.48% (95/105), 92.38% (97/105), 97.14% (102/105) and 100% (105/105), respectively; the specificities of the above methods were all 100%. Higher false-negative rates were found among the gastric biopsy samples assessed by culture (10.48%, 11/105), PCR (7.62%, 8/105) and real-time PCR (2.86%, 3/105) than in gastric juice by real-time PCR. Regarding clarithromycin susceptibility, a concordance of 82.98% (78/94) and discordance of 17.02% (16/94) were observed among the different methods, discrepancies that mainly represent differences between the H. pylori clarithromycin susceptibility phenotype and genotype. Three coinfections of susceptible and resistant strains were detected, with resistant-to-susceptible ratios of 1.16, 3.44, and 8.26. The CYP2C19 genotyping results from gastric juice by real-time PCR were completely in accordance with those obtained from biopsy samples by conventional PCR. This gastric juice-based real-time PCR assay is a more accurate method for detecting H. pylori infection, clarithromycin susceptibility and CYP2C19 polymorphisms. The method may be employed to inform the choice of proton pump inhibitor (PPI), clarithromycin and amoxicillin treatment for tailored H. pylori eradication therapy.
Keywords: gastric juice, real-time PCR, tailored H. pylori eradication.
Introduction
The global consensus statement on the management of Helicobacter pylori eradication recommends standard triple therapy based on a proton pump inhibitor (PPI), clarithromycin and amoxicillin (metronidazole) as the first-line treatment [1]. However, several studies have reported that the first eradication rate was much lower than 80% [2-4]. In the Maastricht IV consensus report, clarithromycin resistance was the most important reason for eradication failure based on the standard triple therapy. Indeed, PPI-clarithromycin-containing triple therapy without prior susceptibility testing should be abandoned if the clarithromycin resistance rate in the region is greater than 15-20% [5]. A multi-centre randomized trial reported a primary resistance rate of H. pylori to clarithromycin ranging from 0 to 40% (average 23.9%), with the resistance rate varying among people from different regions; in addition, a higher secondary drug resistance rate was noted in the same population [6]. A study evaluating H. pylori antibiotic resistance in Beijing from 2000 to 2009 revealed an increase in the clarithromycin resistance of H. pylori, with an average rate of 37.2% [7]. Thus, more attention should be paid to tailored H. pylori eradication therapy, which may help to improve eradication rates and reduce H. pylori resistance.
Culture and standard susceptibility testing to antimicrobial agents should be performed in populations with a high clarithromycin resistance rate if standard clarithromycin-containing therapy is being considered [5]. However, culture and antibiotic resistance testing methods have many disadvantages, including requirements of time and laboratory equipment as well as strong technology and heavy workloads, limiting their widespread application in clinical practice. When standard susceptibility testing based on H. pylori isolation is not possible, molecular tests, such as polymerase chain reaction (PCR) or real-time PCR, can be used to detect H. pylori infection and clarithromycin resistance using gastric biopsy samples [8,9]. As H. pylori has a focal distribution in different parts of the gastric mucosa, false-negative results often occur with single-site gastric biopsy-based detection. In contrast, gastric juice, which contains constantly shed gastric epithelial cells and bacteria from the entire stomach, should be more suitable for detecting actual H. pylori infection [10].
Point mutations in the peptidyl transferase region of the 23S rRNA gene frequently confer macrolide resistance. The most common point mutations are A2143G (69.8%) and A2142G (11.7%), which account for more than 80% of clarithromycin resistance [11]. Other mutations, such as A2142C, A2115G, G2141A, T2717C, A2115G, G2141A and A2142T, are rarely observed [12, 13]. Another important effect on eradication may be polymorphisms of the cytochrome P450 (CYP2C19) gene, the genotype of which determines the metabolic rate of PPI in the human liver. For example, the CYP2C19 wild-type allele (CYP2C19*1) has high enzymatic activity compared to the mutant-type CYP2C19*2 and CYP2C19*3 alleles. CYP2C19*2 and CYP2C19*3 are located in the fifth (G681A) and fourth (G636A) exons, respectively. Accordingly, the CYP2C19 phenotype has been classified into three groups: homozygous extensive metabolizers (Hom-EMs), heterozygous extensive metabolizers (Het-EMs) and poor metabolizers (PMs) [14,15].
In this study, we established an easy and accurate diagnostic technology based on gastric juice to identify H. pylori infection, H. pylori clarithromycin susceptibility and CYP2C19 gene polymorphisms in patients.
Materials and Methods
Bacterial strains
A total of 44 DNA samples, including 28 samples from common non-H. pylori bacteria isolated from the gastric mucosa, 15 enterobacterial samples and one human tissue sample, were provided by the Department of Communicable Disease Diagnostics, National Institute for Communicable Disease Control and Prevention, Chinese Centre for Disease Control and Prevention (see the supplement, Table S1).
Patients and specimens
Patients at Peking University Third Hospital with dyspeptic symptoms were enrolled from January 2013 to November 2014. In total, 178 patients were randomly selected in this trial, including 105 cases that were both rapid urease test (RUT) strongly positive (becoming red within 2 min) and histology test (Warthin-Starry silver staining) positive and 73 cases that were negative for both RUT (no colour change within 2 hours) and histology. We considered the 105 cases as the HP-positive group and the remaining 73 cases as the HP-negative group. Of the subjects, 90 were women and 88 men, with ages ranging from 19 to 68 years (mean±SD, 41.6±12.8). Four gastric mucosa biopsies and 5-10 mL of fasting gastric juice specimens were collected by gastrointestinal endoscopy examination. None of the patients received any H. pylori eradication therapy, including antibiotics and acid-suppressive drugs (PPIs, H2-receptor antagonists, bismuth agent, or antacids). For details, refer to the supplementary information (Table S2).
Ethical considerations
The study was approved by the independent Ethics Committee of Peking University Health Science Centre (IRB00001052-0709) and by the Research Ethics Committee of the National Institute for Communicable Disease Control and Prevention (No: ICDC-2013001) and was performed in accordance with the ethical guidelines of the Declaration of Helsinki, Good Laboratory Practices and Good Clinical Practices. Written informed consent was obtained from each patient prior to study enrolment.
Sample DNA extraction, H. pylori isolation and E-test
One piece of a gastric biopsy sample was homogenized using a sterile glass homogenizer. Half of the gastric biopsy tissue homogenate was directly used for DNA extraction. Approximately 1 mL of gastric fluid was neutralized with an equivalent amount of Tris-HCl (0.67 mol/L, pH 7.4). The mixture was mixed well and centrifuged at 13,000 rpm for 10 minutes. The supernatants were removed, and the pellets were reserved. Genomic DNA was extracted using the QIAamp DNA Mini Kit (QIAGEN, Germany).
The other half of the homogeneous solution was uniformly coated onto the surface of a Karmali agar plate supplemented with 7% defibrinated sheep blood (Biotek Medical Device Co., Ltd., Beijing, China) and an appropriate H. pylori selective supplement (OXOID, England). The plates were incubated for 2-7 days in a microaerophilic environment (5% O2, 10% CO2 and 85% N2) at 37°C. The isolates were identified by Gram staining, and positivity was confirmed by urease, oxidase and catalase traits. Clarithromycin susceptibility was assessed by the E-test, with the addition of 200 μL of inoculum onto plates, which was equivalent to the McFarland 2 opacity standard (8.8 ×107), and incubation for 2 days in a microaerophilic environment at 37°C. Isolates were considered resistant when the minimal inhibitory concentration (MIC) value was more than 2 μg/mL.
Conventional PCR
Four pairs of specific primers focused on target genes were used to confirm the presence of H. pylori, H. pylori 23S rRNA gene mutations and CYP2C19*2 and CYP2C19*3 genotypes in the gastric biopsy specimens. The primer sequences are shown in table 1. All of the PCR reactions were performed in a 25-µL volume containing 12.5 μL 2× Easy Taq® PCR SuperMix (Transgene, Beijing, China), 0.5 μL forward and reverse primers (2 μL each), 2 μL template DNA and 9.5 μL nuclease-free water. The PCR amplifications were performed under the following conditions: denaturation at 94°C for 5 min, 40 cycles of denaturation at 94°C for 30 seconds, annealing at 55°C for 30 seconds, and extension at 72°C for 30 seconds, and a final extension at 72°C for 7 min. The amplification products were analysed by 1.5% agarose gel electrophoresis. Positive products were sequenced using both forward and reverse primers.
Primer sequences used for conventional PCR and sequencing
| Target gene | PCR primer (5'-3') | Product (bp) | Reference |
|---|---|---|---|
| ureB | F: AAAGAGCGTGGTTTTCATGGCG | 217 bp | [16] |
| R: GGGTTTTACCGCCACCGAATTTAA | |||
| H. pylori 23S rRNA | F: AGCGATGTGGTCTCAGCA | 444 bp | This study |
| R: CAAGGGTGGTATCTCAAGG | |||
| CYP2C19*2 | F: AATTACAACCAGAGCTTGGC | 168 bp | [17] |
| R: TATCACTTTCCATAAAAGCAAG | |||
| CYP2C19*3 | F: AAATTGTTTCCAATCATTTAGCT | 271 bp | [18] |
| R: ACTTCAGGGCTTGGTCAATA |
Primer and probe sequences and their distribution for multiple real-time PCR
| Distribution | Target gene | Primer probe name | Sequence (5'-3') | Product (bp) | GenBank No. accession No. |
|---|---|---|---|---|---|
| Reaction 1 | RnaseP | RnaseP-F | 5'-AGATTTGGACCTGCGAGCG-3' | 71 | U77665.1 |
| RnaseP-R | GAGCGGCTGTCTCCACAAGT | ||||
| RnaseP-P | VIC-TTCTGACCTGAAGGCTCTGCGCG-MGB | ||||
| cagH | cagH-F | TTATGTTAGAAATCGCTTGAGTGTCA | 98 | FR666857.1 | |
| cagH-R | CGCTTCTCAAATGATACTTAATCAATC | ||||
| cagH-P | FAM-AGGTGCTAGTAGCTAATC-MGB | ||||
| Reaction 2 | HP23SrRNA | HP23S-F | TTCAGTGAAATTGTAGTGGAGGTG | 98 | NR_076155.1 |
| HP23S-R | TCCCATTAGCAGTGCTAAGTTGTA | ||||
| HP23S-AA | FAM-AGACGGAAAGACC-MGB | ||||
| HP23S-GA | VIC-AGACGGGAAGACC-MGB | ||||
| HP23S-AG | VIC-AGACGGAGAGACC-MGB | ||||
| Reaction 3 | CYP2C19*2 | CY2-F | GCTTGGCATATTGTATCTATACCTT | 85 | NG_008384.2 |
| CY2-R | GATTCTTGGTGTTCTTTTACTTTCT | ||||
| CY2-G | FAM-ATTTCCCGGGAACC-MGB | ||||
| CY2-A | VIC-ATTTCCCAGGAACC-MGB | ||||
| Reaction 4 | CYP2C19*3 | CY3-F | AATTGAATGAAAACATCAGGATTG | 88 | NG_008384.2 |
| CY3-R | ACTGTAAGTGGTTTCTCAGGAAGC | ||||
| CY3-G | FAM-CTGGATCCAGGTAAG-MGB | ||||
| CY3-A | VIC-CCTGAATCCAGGTAAG-MGB |
Real-time PCR
Five primers and nine matching Taqman probes targeting the H. pylori cagH and 23S rRNA genes and the human RnaseP, CYP2C19*2 and CYP2C19*3 genes were designed for this assay. All sequences obtained from NCBI Entrez Nucleotide Database (http://www.ncbi.nlm.nih.gov/nuccore) were aligned using Vector NTI alignment software (http://www.lifetechnologies.com/cn/zh/home/life-science/cloning/vector-nti-software.html). The primers and probes were designed using Primer Express 3.0 software (Applied Biosystems). The sequences of the primers and probes used in this study are summarized in table 2.
The reaction mixture (20 μL) was prepared as follows: 2 μL 10× PCR buffer, 2 mM MgCl2 (Platinum® Taq DNA Polymerase, Invitrogen, Thermo Fisher, USA), 0.4 μL dNTPs (Promega, USA), 0.5 μM forward and reverse primers (Sangon Biotech, Shanghai, China), 0.2 μM probe (ABI, USA), 0.2 μL Taq DNA polymerase (Platinum® Taq DNA Polymerase, Invitrogen, Thermo Fisher, USA), 2 μL DNA template, and up to 20 μL nuclease-free water.
Evaluation of multiple real-time PCR performance
The specificity of the cagH probe in the real-time PCR was assessed using bacterial DNA from 28 common bacteria in addition to H. pylori from gastric mucosa and 15 enterobacteria.
To assess the sensitivity of this assay, we constructed recombination plasmids containing a target gene or point mutation from the reference strains. To evaluate the detection limit of each probe in this assay, a series of 10-fold dilutions of the recombination plasmids ranging from 1×109 copies/μL to 1×100 copies/μL were used as the template. Simultaneously, the correlation coefficient R and amplification efficiency of each primer/probe were determined using standard curves based on 10-fold serial dilutions of the recombinant plasmids.
To evaluate assay precision, the intra- and inter-assay variability were evaluated to reveal the corresponding repeatability and reproducibility, respectively. High, medium and low plasmid concentrations (1×107 copies/μL, 1×105 copies/μL, and 1×102 copies/μL, respectively) were used as the template. To estimate intra-experimental variation, nine positive standard plasmids with different copy numbers were detected three times in the same experiment. To determine inter-experimental variation, the same plasmids were tested on different days in three different experiments. All 178 gastric juice samples were detected by this multiple real-time PCR approach, and the results were compared with those obtained by culture and gastric biopsy-based PCR.
Statistical analysis
A P value <0.05 was considered significant. All statistical tests and figures in our study were prepared using R statistical software version 3.3.1 (http:// www.r-project.org/).
Results
Real-time PCR assay development
Bacterial DNA from 28 common gastric bacteria and 17 enterobacteria were detected with our four assays, and no positive amplification was observed. The cagH and 23S rRNA assays were only positive for H. pylori and were negative for the other bacteria. The CYP2C19*2 and CYP2C19*3 assays were only positive for the human genome. A series of 10-fold dilutions of plasmid DNA (ranging from 1×109 to 1×100 copies/μL) was used as the template and tested in four multiple real-time PCR reactions, with each plasmid concentration repeated three times. The limit of detection (LOD) of all probes was 102 copies/μL of plasmid DNA. The LODs of the cagH-prob and RnaseP-prob in the first-group PCR were 101 copies/μL, with average Ct values of 36.66 and 36.57, respectively. The LODs of the HP23S-AA, HP23S-AG, and HP23S-GA probes in the second-group PCR were 100, 100, and 101 copies/μL, with average Ct values of 37.30, 37.91, and 37.33, respectively. The LODs of the CY2-G and CY2-A probes in the third-group PCR were 101 and 102 copies/μL, with average Ct values of 35.03 and 35.20, respectively. The LODs of the CY3-G and CY3-A probes in the fourth-group PCR were 101 and 102 copies/μL, with average Ct values of 37.27 and 38.00, respectively. No significant differences were found in repeatability and reproducibility evaluations (P>0.05).
Real-time PCR performance for H. pylori infection diagnosis
In the H. pylori-positive group, the positive rates obtained using culture, PCR, and gastric biopsy- and gastric juice-based real-time PCR assays were 90.48% (95/105), 92.38% (97/105), 97.14% (102/105) and 100% (105/105), respectively. The consistency rate for all four H. pylori infection diagnostic methods was 89.52% (94/105). Four discrepancies occurred between the culture and PCR methods, including three cases with positive PCR results but negative culture results and one case with a positive culture result but a negative PCR result. Ten false-negative results were found by cultures, eight by PCR and three by gastric biopsy-based real-time PCR.
Comparison of the performances of culture, PCR and real-time PCR for the diagnosis of H. pylori infection
| Parameter | Result (%) | |||
|---|---|---|---|---|
| Culture (gastric biopsy-based) | PCR (gastric biopsy-based) | Real-time PCR (gastric biopsy-based) | Real-time PCR (gastric juice-based) | |
| Sensitivity | 90.48% (95/105)* | 92.38% (97/105)* | 97.14% (102/105)** | 100% (105/105) |
| Specificity | 100% (73/73) | 100% (73/73) | 100% (73/73) | 100% (73/73) |
| Positive predictive value | 90.48% (95/105)* | 92.38% (97/105)* | 97.14% (102/105)** | 100% (105/105) |
| Negative predictive value | 87.95% (73/83)* | 90.12% (73/81)* | 96.05% (73/76)*** | 100% (73/73) |
*: P<0.05, **: P=0.081, ***: P=0.086.
For these false-negative cases, both the H. pylori-specific ureB and the 23S rRNA gene fragments could be amplified from the corresponding gastric juice specimen, and these PCR products were confirmed by nucleotide sequencing. In the H. pylori-negative group, no false-positive results were found in the gastric biopsy or gastric juice samples by PCR or real-time PCR. The significance levels for sensitivity, specificity, positive predictive value (PPV) and negative predictive value (NPV) among the different methods are displayed in table 3.
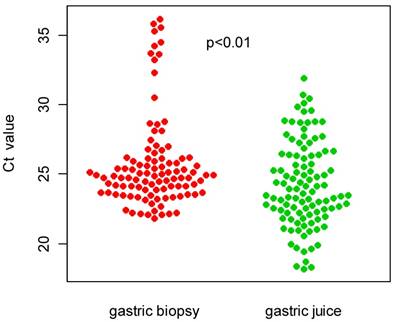
Additionally, we compared the distribution of Ct values for real-time PCR between the gastric biopsy and gastric juice specimens. As shown in figure 1, the Ct values obtained from the gastric juice samples (24.27±3.05) were significantly higher than those obtained from the gastric biopsy samples (25.75±3.32).
Comparison of Ct value distributions between gastric biopsy and gastric juice specimens.
Clarithromycin susceptibility testing
Regarding clarithromycin susceptibility, we found 82.98% (78/94) concordance among the different methods for the 94 H. pylori-positive cases, which consisted of 40 clarithromycin-susceptible cases and 38 clarithromycin-resistant cases. The discrepancies accounted for 15.24% (16/94) of the discordance among the methods. These discrepancies suggest inconsistency between the genotype and phenotype. In three cases, both resistant and susceptible genotypes were detected simultaneously by PCR and real-time PCR using both gastric biopsy and gastric juice specimens, whereas E-test results showed only phenotypic resistance or susceptibility. The resistant-to-susceptible ratios detected by real-time PCR for gastric juice were 1.16, 3.44, and 8.26.
Additionally, we found 11 cases that were culture-negative but PCR- or real-time PCR-positive. In three cases, the clarithromycin genotype obtained by real-time PCR using gastric juice was in complete agreement with the genotype based on gastric biopsies from the same patients determined by either PCR or real-time PCR. The clarithromycin genotype in another five cases identified by gastric juice-based real-time PCR was confirmed by gastric biopsy-based real-time PCR, though the PCR results were negative. Moreover, three cases were only detected by gastric juice-based real-time PCR. The details are provided in table 4.
Comparison of clarithromycin susceptibility testing by E-test, PCR and real-time PCR
| Gastric biopsy | Gastric juice | No. of gastric specimens (%) | ||
|---|---|---|---|---|
| E -test | PCR | Real-time PCR | Real-time PCR | |
| S | S | S | S | 40 (38.10%) |
| R | R | R | R | 38 (36.19%) |
| S | H | H | H | 1 (0.01%) |
| R | H | H | H | 2 (0.02%) |
| R | S | S | S | 12 (11.43%) |
| S | R | R | R | 1 (0.01%) |
| — | R | R | R | 1 (0.01%) |
| — | — | R | R | 3 (0.03%) |
| — | — | — | R | 2 (0.03%) |
| — | S | S | S | 2 (0.02%) |
| — | — | S | S | 2 (0.01%) |
| — | — | — | S | 1 (0.01%) |
| Total | 105 | |||
S, susceptible; R, resistant; H, heterogeneous.
Human CYP2C19 genotyping
The CYP2C19 genotype in all 178 patients was determined by gastric biopsy PCR coupled with nucleotide sequencing. Gastric biopsy PCR results and gastric juice-based real-time PCR provided identical results regarding CYP2C19*2 and CYP2C19*3 mutations.
Discussion
A real-time PCR method was developed to guide tailored H. pylori therapy through easy and accurate detection of H. pylori 23S rRNA gene mutations and the human CYP2C19 genotype from gastric juice samples. The gastric juice-based real-time PCR results were compared with results acquired using conventional diagnostic methods with strongly positive RUT and histology-positive biopsy specimens. Gastric juice-based real-time PCR demonstrated a higher sensitivity and NPV compared to culture and PCR using gastric biopsy samples for the diagnosis of H. pylori. Although no remarkable significance was determined regarding specificity, based on PPV and NPV evaluations for gastric biopsy-based real-time PCR (P>0.05), we can speculate that significance may be apparent when a larger sample size is assessed. Taking health economics into account, RUT and histology were performed for the diagnosis of H. pylori infection. For RUT-positive cases, gastric juice-based real-time PCR showed 100% concordance, whereas false-negative results were obtained by culture, PCR and gastric biopsy-based real-time PCR. Such false negatives may have occurred for the following reasons: ① the 'focal distribution' of H. pylori in the gastric mucosa may lead to low-level colonization or absence in some gastric niches; ② contamination by other bacteria that suppress H. pylori overgrowth; ③ the presence of non-culturable coccoid forms; ④ loss of viability during transport; and ⑤ reduced sensitivity in patients with bleeding peptic ulcers detected using classical diagnostic methods with gastric biopsies [19, 20]. Compared with these conventional diagnostic methods, our gastric juice-based real-time PCR exhibits the following prominent advantages: gastric juice reflects the real H. pylori infection status in the entire gastric environment; there is no need for viable bacteria and critical transport conditions for culture; Taqman-MGB probe real-time PCR has higher sensitivity than traditional methods [21]; and gastric fluid specimens appear to be more suitable for patients with bleeding tendencies.
Because tailored treatment based on 23S rRNA mutations and CYP2C19 polymorphisms yield a higher H. pylori eradication rate than the empirical standard triple therapy, H. pylori susceptibility to clarithromycin and the human CYP2C19 genotype should be evaluated. Culture and the E-test are often employed to determine the clarithromycin susceptibility phenotype, whereas PCR with nucleotide sequencing is used to identify 23S rRNA and CYP2C19 genotypes. However, these classic genotype methods are usually time-consuming and have notable laboratory equipment requirements, which are not applicable in daily clinical practice. In our study, the gastric juice-based real-time PCR was completed within 1 hour and 40 minutes (not including DNA extraction). We found 82.97% (78/94) concordance and 17.02% (16/94) discordance among the three methods using gastric biopsy or gastric juice samples. These discrepancies were most likely because the A2142G to A2143G ratio accounts for approximately 80% of all mutations causing clarithromycin resistance. The results are in agreement with the results reported in the literature [11]. Additionally, three mixed infections of susceptible and resistant strains were simultaneously detected by real-time PCR and PCR, whereas one infection was classified as susceptible and another two as resistant by culture. The resistant-to-susceptible ratios tested by real-time PCR were 1.16, 3.44, and 8.26. Thus, resistant strains play a major role in the entire gastric microenvironment, and we should avoid using clarithromycin when devising an administration scheme.
Despite the superior performance of our gastric juice-based real-time PCR, inevitable shortcomings exist. Some hot-spot mutations associated with clarithromycin resistance should be added to improve the detection accuracy. To enhance patient compliance and reduce discomfort, the string test can be adopted, instead of endoscopy, for collecting gastric juice. Indeed, obtaining gastric fluid specimens using the string test is suitable for large-scale population screening.
Conclusions
In summary, we established a gastric juice Taqman-MGB-based real-time PCR method that could conveniently and accurately determine the A2142G or A2143G mutation associated with clarithromycin resistance and the human CYP2C19 genotype. In this manuscript, we show that our method can overcome many flaws and deficiencies compared to the use of gastric biopsy specimens tested using various traditional detection methods. Four obvious advantages were observed: ① higher sensitivity of H. pylori diagnosis; ② low false-negative results caused by focal distribution; ③ precise instructions to assess H. pylori clarithromycin susceptibility, especially for coinfections with clarithromycin-resistant and susceptible strains; and ④ easier operation and a shorter time requirement. This gastric juice-based real-time PCR method demonstrated better performance than culture and gastric biopsy-based PCR. Thus, gastric juice-based real-time PCR is a more accurate method that can be used to guide individualized H. pylori eradication.
Supplementary Material
Supplemental table s1, table s2.
Acknowledgements
This work was supported by funding from the China Mega-Project for Infectious Disease (2011ZX10004-001), a grant from the National Technology R&D Program in the 12th Five-Year Plan of China (2012BAI06B02) and a grant from the State Key Laboratory of Infectious Disease Prevention and Control (SKLID) (2014SKLID102).
Competing Interests
The authors have declared that no competing interest exists.
References
1. Group EHpS. Current European concepts in the management of Helicobacter pylori infection. The Maastricht Consensus Report. Gut. 1997;41:8-13
2. Graham DY, Fischbach L. Helicobacter pylori treatment in the era of increasing antibiotic resistance. Gut. 2010;59:1143-53
3. Malfertheiner P, Bazzoli F, Delchier J. Helicobacter pylori eradication with a capsule containing bismuth subcitrate potassium, metronidazole, and tetracycline given with omeprazole versus clarithromycin-based triple therapy: a randomised, open-label, non-inferiority, phase 3 trial (vol 377, pg 905, 2011). Lancet. 2011;377:905-13
4. Zhiqiang Song, Liya Zhou, Jianzhong Zhang. et al. Hybrid Therapy as First-Line Regimen for Helicobacter pylori Eradication in Populations with High Antibiotic Resistance Rates. Helicobacter. 2016;21:382-8
5. Malfertheiner P, Megraud F, O'Morain CA, Atherton J, Axon AT, Bazzoli F, Gensini GF, Gisbert JP, Graham DY, Rokkas T, El-Omar EM, Kuipers EJ, European Helicobacter Study G. Management of Helicobacter pylori infection-the Maastricht IV/ Florence Consensus Report. Gut. 2012;61:646-64
6. Hong Cheng, Fulian Hu, Yong Xie. et al. Prevalence of Helicobacter pylori Resistance to Antibiotics and its Influence on the Treatment outcome in China: A Multicenter Clinical Study. Chin J Gastroenterol. 2007;12:525-30
7. Gao W, Cheng H, Hu F, Li J, Wang L, Yang G, Xu L, Zheng X. The evolution of Helicobacter pylori antibiotics resistance over 10 years in Beijing, China. Helicobacter. 2010;15:460-6
8. Chisholm SA, Owen RJ, Teare EL, Saverymuttu S. PCR-based diagnosis of Helicobacter pylori infection and real-time determination of clarithromycin resistance directly from human gastric biopsy samples. J Clin Microbiol. 2001;39:1217-20
9. Kargar M, Ghorbani-Dalini S, Doosti A, Souod N. Real-time PCR for Helicobacter pylori quantification and detection of clarithromycin resistance in gastric tissue from patients with gastrointestinal disorders. Res Microbiol. 2012;163:109-13
10. Yakoob J, Rasool S, Abbas Z, Jafri W, Abid S, Islam M, Ahmad Z. Gastric juice for the diagnosis of H pylori infection in patients on proton pump inhibitors. World J Gastroenterol. 2008;14:1539-43
11. Megraud F. H pylori antibiotic resistance: prevalence, importance, and advances in testing. Gut. 2004;53:1374-84
12. Saez J, Belda S, Santibanez M, Rodriguez JC, Sola-Vera J, Galiana A, Ruiz-Garcia M, Brotons A, Lopez-Girona E, Girona E, Sillero C, Royo G. Real-time PCR for diagnosing Helicobacter pylori infection in patients with upper gastrointestinal bleeding: comparison with other classical diagnostic methods. J Clin Microbiol. 2012;50:3233-7
13. Van der Ende A, Van Doorn LJ, Rooijakkers S, Feller M, Tytgat GN, Dankert J. Clarithromycin-susceptible and -resistant Helicobacter pylori isolates with identical randomly amplified polymorphic DNA-PCR genotypes cultured from single gastric biopsy specimens prior to antibiotic therapy. J Clin Microbiol. 2001;39:2648-51
14. Ozdil B, Akkiz H, Bayram S, Bekar A, Akgöllü E, Sandikçi M. Influence of CYP2C19 functional polymorphism on Helicobacter pylori eradication. Turk J Gastroenterol. 2010;21:23-8
15. Kuo CH, Lu CY, Shih HY, Liu CJ, Wu MC, Hu HM, Hsu WH, Yu FJ, Wu DC, Kuo FC. CYP2C19 polymorphism influences Helicobacter pylori eradication. World J Gastroenterol. 2014;20:16029-36
16. Liu J, He L, Haesebrouck F, Gong Y, Flahou B, Cao Q, Zhang J. 2015. Prevalence of Coinfection with Gastric Non-Helicobacter pylori Helicobacter (NHPH) Species in Helicobacter pylori-infected Patients Suffering from Gastric Disease in Beijing, China. Helicobacter. 2015;20:284-90
17. Griese EU, Läpple F, Eichelbaum M. 1999. Detection of CYP2C19 alleles *1, *2 and *3 by multiplex polymerase chain reaction. Pharmacogenetics. 1999;9:389-91
18. De Morais SM, Wilkinson GR, Blaisdell J, Meyer UA, Nakamura K, Goldstein JA. Identification of a new genetic defect responsible for the polymorphism of (S)-mephenytoin metabolism in Japanese. Mol Pharmacol. 1994;46:594-8
19. Colin R. et al. Low sensitivity of invasive tests for detection of Helicobacter pylori infection in patients with bleeding ulcer. Gastroenterol Clin Biol. 2000;24:31-5
20. Lo CC. et al. Polymerase chain reaction: a sensitive method for detecting Helicobacter pylori infection in bleeding peptic ulcers. World J Gastroenterol. 2005;11:3909-14
21. P. Narayanasamy. Molecular Biology in Plant Pathogenesis and Disease Management: Microbial Plant Pathogens. P. Narayanasamy. Molecular Biology in Plant Pathogenesis and Disease Management: Microbial Plant Pathogens. Germany: Springer Netherlands. 2008:7-158
Author contact
![]() Corresponding authors: Prof. Jianzhong Zhang, Tel.: 86-10-58900707, Fax: 86-10-58900700, E-mail: zhangjianzhongcn; and Prof. Liya Zhou, Tel.: 86-18910192576, Fax: 86-10-62357303. E-mail address: zhouliya123456com.
Corresponding authors: Prof. Jianzhong Zhang, Tel.: 86-10-58900707, Fax: 86-10-58900700, E-mail: zhangjianzhongcn; and Prof. Liya Zhou, Tel.: 86-18910192576, Fax: 86-10-62357303. E-mail address: zhouliya123456com.

 Global reach, higher impact
Global reach, higher impact