3.2
Impact Factor
ISSN: 1449-1907
Int J Med Sci 2017; 14(5):412-418. doi:10.7150/ijms.18641 This issue Cite
Research Paper
hsa-miR-96 and hsa-miR-217 Expression Down-Regulates with Increasing Dysplasia in Pancreatic Intraepithelial Neoplasias and Intraductal Papillary Mucinous Neoplasms
1. Department of Pathology, Peking Union Medical College Hospital, Chinese Academy of Medical Sciences and Peking Union Medical College, Tsinghua University, Beijing 100730, China;
2. Department of Pathology, Beijing Shijitan Hospital, Capital Medical University, Beijing 100038, China.
XiaoYan Chang and ChunKai Yu contributed equally to this paper.
Received 2016-12-6; Accepted 2017-1-31; Published 2017-4-8
Abstract
AIM: To compare the clinicopathological features of pancreatic intraepithelial neoplasias (PanINs) and intraductal papillary mucinous neoplasms (IPMNs), and to investigate the role of hsa-miR-96 and hsa-miR-217 in these two lesions.
Methods: Formalin-fixed paraffin-embedded pancreatic specimens were selected in this study, including 58 cases of pancreatic intraepithelial neoplasias (PanINs), 45 cases of pancreatic ductal adenocarcinomas (PDAs), and 57 cases of intraductal papillary mucinous neoplasms (IPMNs). MiRNAs hsa-miR-96 and hsa-miR-217 were detected using locked nucleic acid in situ hybridization (LNA-ISH) with the NBT/BCIP staining system. The differences in miRNA expression among sample sets were analyzed with the Chi-squared test.
Results: PanIN-PDAs were inclined to present with higher rate of invasion (p=0.033), lymph node metastasis (p=0.0004) and poorer differentiation (p<0.001). Of the 45 PDAs, only 2 cases were within AJCC Ⅰstage, while there were 11 cases of IPMN associated carcinomas (p=0.0018). In PanIN-1, PanIN-2 and PanIN-3, the expression of hsa-miR-96 was 91.3% (22/23), 78.6%(12/17) and 22.2%(4/18) respectively, while the expression of hsa-miR-217 was 95.7%(22/23) , 70.6% (12/17) and 27.8% (5/18). In IPMN with low-grade, intermediate-grade, high-grade dysplasia, associated carcinoma, the expression of hsa-miR-96 was 67%(9/13), 64%(7/11), 43%(3/7) and 27%(7/26) respectively, while the expression of hsa-miR-217 was 77%(10/13), 64%(7/11), 29%(2/7) and 38%(10/26). The expression of hsa-miR-96 and hsa-miR-217 in PanIN-1 lesions was not significantly different from that in the normal pancreatic ductal epithelium. However, their expression in PanIN-2/3 lesions was significantly different from that in normal pancreatic ductal epithelium (P<0.01). No difference was observed between PanIN derived adenocarcinomas and IPMN-associated carcinomas.
Conclusion: IPMN associated carcinomas were in a statistically earlier stage than PanIN- PDAs at the time of operation. Abnormal expression of hsa-miR-96 and of hsa-miR-217 was observed in premalignant lesions (PanINs and IPMNs) of pancreatic carcinoma and down-regulated with increasing grades of PanINs and IPMNs. These microRNAs may serve as potentially early biomarker and act as tumor suppressor genes.
Keywords: microRNAs, hsa-miR-96, hsa-miR-217, Pancreatic ductal adenocarcinoma, Pancreatic Intraepithelial Neoplasias, Intraductal papillary mucinous neoplasms, In Situ Hybridization.
Introduction
Pancreatic ductal adenocarcinoma (PDA) is one of the most fatal of all human cancers, characterized by rapid progression and early metastasis. Pancreatic intraepithelial neoplasias (PanINs) and intraductal papillary mucinous neoplasms (IPMNs) are the most common premalignant lesions of PDAs [1]. IPMNs are grossly and radiographically visible tumors with the diameter of more than 1 cm, whereas PanINs are usually less than 0.5cm and must be diagnosed microscopically [2].
PanIN, as spectrum of ductal proliferative lesions, is graded on a three-tiered scale: PanIN-1, PanIN-2, and PanIN-3. Generally, PanIN-1 is considered to be a low-grade lesion, whereas PanIN-2 and PanIN-3 are considered to be high-grade lesions. High-grade PanINs and PDAs have been reported to display similar genetic alterations, including KRAS mutation and the silencing or inactivation of tumor suppressor genes such as p16, p53, SMAD4, and cyclin D1 [3-5] . Low-grade PanINs may present telomere shortening and some gene mutations, such as KRAS, prostate stem cell antigen (PSCA), mucin 5, and fascin.
IPMNs are slow-growing cystic tumors with a favorable prognosis, characterized by intraductal papillary proliferation of mucin-producing cells. IPMNs tend to progress from adenomatous lesions (IPMN with low- and intermediate- grade dysplasia) to high-grade dysplasia, and finally to invasive carcinomas (IPMN-associated carcinomas), which exhibit the highest degree of architectural and cytological atypia. Genetic studies have established that mutations of KRAS, PI3KCA, STK11/LKB1 were the most common molecular alterations in IPMNs [6, 7].
MicroRNA (miRNA) is an endogenous non-coding single-stranded small RNA of 19 to 24 nucleotides (nt) in length that regulate gene expression at the post-transcriptional level. By complementary base pairing with the 2-8 nt seed sequence at 5' end, mature miRNA binds to the 3'-untranslated region (3'-UTR) of the target mRNA sequence, suppressing its translation and/or leading to its degration. The resulting inhibition of gene expression plays a role in the regulation of oncogenes or tumor suppressor genes [8, 9]. In human body, 20% to 30% of protein-encoding genes are regulated by miRNAs [10]. Together with the corresponding human genes and proteins, miRNA forms a complex and precise regulatory network. In recent years, many studies have confirmed that various tumor tissues show changes in their miRNA expression profiles [11]. A growing body of literature has confirmed that miRNA have tumorigenic or tumor suppressor functions. Recent studies also identified a large number of abnormally expressed miRNAs in PDAs [12-14], for example, has-miR-96 and has-miR-217, targeting KRAS, have been proved to be down-regulated by microarray technology and quantitative PCR [12, 15, 16]. Considering KRAS is the only mutated gene found in both panINs and IPMNs, we choose these two microRNAs as our study objects.
To address the potential role of hsa-miR-96 and hsa-miR-217 in the carcinogenesis of PDA, we analyzed their expression in premalignant lesions including IPMNs and PanINs, as well as associated PDAs using the method of locked nucleic acid in situ hybridization (LNA-ISH).
Materials and Methods
Pathological assessment
Patients who underwent partial or total pancreatectomy at Peking Union Medical College Hospital were enrolled. 45 PDAs, 58 PanINs (23 PanIN1, 17 PanIN2, 18 PanIN3) and 57 IPMNs were selected. Of the 58 PanINs, 11 PanIN1, 16 PanIN2 and 18 PanIN3 were in the adjacent normal tissue of PDAs. The other PanIN2 was selected from non-tumor tissue of acinar cell carcinoma. The other PanIN1 were selected from 6 cases of chronic pancreatitis, 5 cases of mucinous cystadenomas, and 1 case of solid pseudopapillary neoplasm. All tissues were treated by 10% neutral formalin fixation for 8-24 hours, paraffin embedding, sectioning, and hematoxylin & eosin (H&E) staining. All sections were reviewed by two pathologists according to the diagnostic criteria proposed by the 2010 edition of the WHO classification system for digestive neoplasms. The cases with disagreement were consulted to a third expert pathologist.
Reagents for locked nucleic acid in situ hybridization
Hybrid hsa-miR-96, hsa-miR-217, U6 and scramble-miRNA probes were purchased from Exiqon (Vedbaek, Denmark). The hsa-miR-217, hsa-miR-96, U6, and scramble-miRNA probes were labeled with 5' digoxigenin (5'DIG). Yeast RNA was purchased from Ambion (United States). DEPC was purchased from Invitrogen (California, United States). Anti-digoxigenin alkaline phosphatase-labeled secondary anti-DIG-AP Fab and NBT/BCIP staining kit were purchased from Roche (Mannheim, Germany). The sequences of the probes are shown in Table 1.
Sequence of probes
| MiRNA | Localization of genes | Sequence of probes |
|---|---|---|
| hsa-miR-217 | 2p16 | TCCAATCAGTTCCTGATGCAGTA |
| hsa-miR-96 | 7q32.2 | AGCAAAAATGTGCTAGTGCCAAA |
| U6 | Positive Control | CACGAATTTGCGTGTCATCCTT |
| Scramble-miR | Negative Control | GTGTAACACGTCTATACGCCCA |
Method of locked nucleic acid in situ hybridization (LNA-ISH)
After the 4 μm paraffin sections were incubated at 60 ℃ for 1 hour, they were deparaffinized, and washed with DEPC-treated PBS. The slides were then incubated with 10 μg/ml of proteinase K at 37 ℃ for 10-20 minutes to remove the proteins and expose the RNA. They were next washed with PBS containing 0.2% glycine for 30 seconds to terminate the digestion and fixed with 4% paraform aldehyde for 10 minutes. Subsequently, the slides were pre-hybridized in hybridization buffer without probe for 2 hours at the probe Tm of 21 ℃. The probe was diluted to 20-60 nM in a hybridization buffer, and 20 μl of hybridization solution containing probe was added onto each slide. The slide was covered with a special cover-slip, and the edges were sealed to prevent drying. The hybridization time was 20.5 hours. After stringent washing with 2×SSC and 2×SSC containing 50% acrylamide at the hybridization temperature, the slides were blocked with a blocking buffer (PBST containing 2% goat serum and 2 mg/mL bovine albumin) for 1 hour. After the blocking solution was removed, one drop of anti-DIG-AP Fab antibody was added, and the slides were incubated overnight, followed by staining with NBT/BCIP at room temperature in a moist chamber for approximately 24 hours and routine dehydration and mounting. The slides were observed under a microscope. The U6 probe was applied to calibrate the system. The scrambled probe served as negative control, and the expression of normal pancreatic tissue served as positive control.
Measurement of results
Positive miRNA in situ hybridization was evidenced by the appearance of blue or purple granules in the cytoplasm and the nucleus of pancreatic acinar and ductal cells, serving as positive internal control. U6-positive staining was observed in the nucleus. Scramble-miRNA, serving as negative control, did not show positive staining. A semi-quantitative integration method was used to determine the results. The LNA-ISH results were scored based on the intensity of hybridization as 0 (no expression or weak expression), 1 (moderate expression), and 2 (strong expression). The scoring of the number of positive cells was designated as 0 (positive cells <1%), 1 (1%-50% positive cells), and 2 (positive cells ≥ 50%) [17]. The above two scores were multiplied, and a product of 0 indicated negative expression, 1 indicated weak positive expression, and 2-4 indicated strong expression.
Statistical analysis
All results were analyzed using SPSS statistical software. The differences between groups were compared using the χ2 test. P value < 0.05 was considered as statistically significant.
Results
PanINs and IPMNs
High grade PanINs (PanIN2 and PanIN3) were found in the surrounding area of pancreatic adenocarcinomas, while PanIN1 was located around malignant neoplasms, benigh neoplasms, and chronic pancreatitis. Of the 45 PDAs, 34 high grade PanINs and 11 low grade PanINs were found around the adenocarcinomas.
Of the 57 IPMNs, there were 13 IPMNs with low-grade dysplasia, 11 IPMNs with intermediate-grade dysplasia, 7 IPMNs with high-grade dysplasia, and 26 IPMNs with an associated invasive carcinoma (four of which were diagnosed as colloid carcinoma).
We compared the clinical and pathological features of PanIN-PDAs and IPMN with an associated carcinoma in Table 2. PanIN-PDAs demonstrated more aggressive behavior, as measured by higher percentage of outside-pancreas invasion (p=0.033), increased rate of lymph node metastases (p=0.0004) and a higher percentage of poorly differentiated carcinoma (p<0.001). There were no statistical differences in patients' sex or age between two groups. In addition, PDAs tend to be present at an advanced stage, with only 2/45 cases in AJCC I, comparing to 11/26 of IPMN associated carcinomas (p=0.0018).
The clinical and pathological features of PanIN-PDAs and IPMN associated carcinomas.
| PanIN-PDA(45) | IPMN-carcinoma(26) | |
|---|---|---|
| Sex (M/F) | 23/22 | 12/14 |
| Age (range/medium age) | 33-80 (61) | 42-79 (62) |
| Carcinoma type (ductal/ colloid/ other) | 45/0/0 | 21/4/1 |
| Differentiation (well/moderate/poorly) | 1/26/18 | 14/7/5 |
| Infiltrative scope (within pancreas/ outside) | 2/43 | 16/10 |
| Lymph node metastasis (yes/ no) | 29/16 | 6/20 |
| AJCC (Ⅰ/Ⅱ/Ⅲ/Ⅳ) | 2/28/7/8 | 11/14/1/0 |
Expression of hsa-miR-96 and hsa-miR-217 in PanINs
Table 3 indicated that with the increasing grade of PanINs, the expression of hsa-miR-96 and hsa-miR-217 tend to reduce. The expression of hsa-miR-217 showed no statistical difference between PanIN1 and normal pancreatic ductal epithelium (NPDE) (P=0.159), but a significant declination between PanIN1 and PanIN2 (P=0.028), PanIN2 and PanIN3 (P=0.011). Similar pattern was observed in has-miR-96. There were significant differences between PanIN1 and NPDE (P=0.045), PanIN2 and PanIN3 (P=0.004), though not between PanIN1 and PanIN2 (P=0.088). Both hsa-miR-96 and hsa-miR-217 expression decreased significantly in PDAs, comparing with NPDE (P<0.001). It is noteworthy that the expression of hsa-miR-96 and hsa-miR-217 in PanIN3 was close to that in PDAs (P=0.480 and P=0.286), indicating a similar pattern of miRNAs expression in these two lesions.
The expression of hsa-miR-96 and hsa-miR-217 in PanINs
| n | hsa-mir-96 | hsa-mir-217 | ||||||
|---|---|---|---|---|---|---|---|---|
| + | - | Positive (%) | + | - | Positive (%) | |||
| NPDE | 45 | 45 | 0 | 100 | 45 | 0 | 100 | |
| panIN1 | 23 | 21 | 2 | 91.3 | 22 | 1 | 95.7 | |
| panIN2 | 17 | 12 | 5 | 78.6 | 12 | 5 | 70.6 | |
| panIN3 | 18 | 4 | 14 | 22.2 | 5 | 13 | 27.8 | |
| PDA | 45 | 14 | 31 | 31.1 | 19 | 26 | 42.2 | |
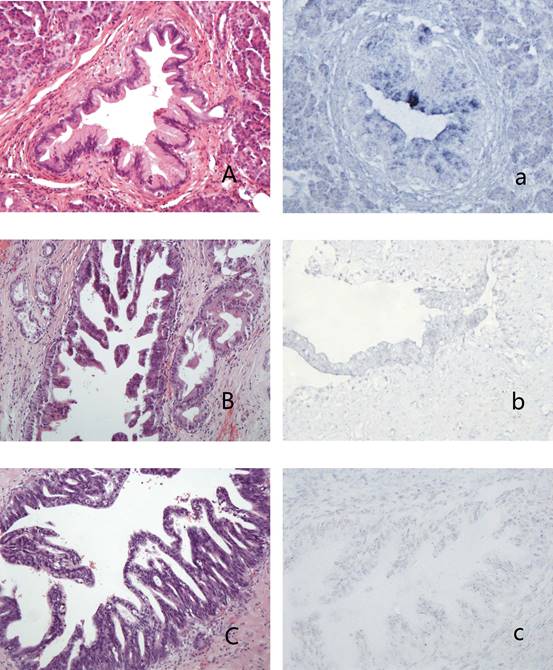
PanINs and the expression of hsa-miR-96 A-a: PanIN1b (A,HE stain) showed blue granules in the cytoplasm of lining epithelium (a); B-b: PanIN2 (B, HE stain) showed weak-stained blue granules in the nucleus and no granules in the cytoplasm of lining epithelium (b); C-c: PanIN3 (C, HE stain) showed blue granules in the nucleus and no granules in the cytoplasm of lining epithelium (c).
The expression of hsa-miR-96 and hsa-miR-217 in IPMNs
| IPMN | n | hsa-mir-96 | hsa-mir-217 | |||||
|---|---|---|---|---|---|---|---|---|
| + | - | Positive (%) | + | - | Positive (%) | |||
| low | 13 | 9 | 4 | 69 | 10 | 3 | 77 | |
| intermediate | 11 | 7 | 4 | 66 | 7 | 4 | 64 | |
| high | 7 | 3 | 4 | 43 | 2 | 5 | 29 | |
| carcinoma | 26 | 7 | 19 | 27 | 10 | 16 | 38 | |
The expression of hsa-miR-96 and hsa-miR-217 in IPMNs
The data in Table 4 showed that the expression of hsa-miR-96 and hsa-miR-217 reduced with the increasing grade of IPMNs. There was no significant difference among IPMN with low-, intermediate-, high grade dysplasia and associated carcinoma in hsa-miR-96 expression (low and intermediate grade: P=0.772; intermediate and high grade: P=0.387; high grade and carcinoma: P=0.416). Similarly, there was no significant difference among IPMN with low-, intermediate-, high grade dysplasia and associated carcinoma in the hsa-miR-217 expression (low and intermediate grade: P=0.476; intermediate and high grade: P=0.147; high grade and carcinoma: P=0.629). IPMN and associated carcinoma was significantly different in the expression of hsa-miR-96 (P=0.009), but not in the expression of has-miR-217 (P=0.086).
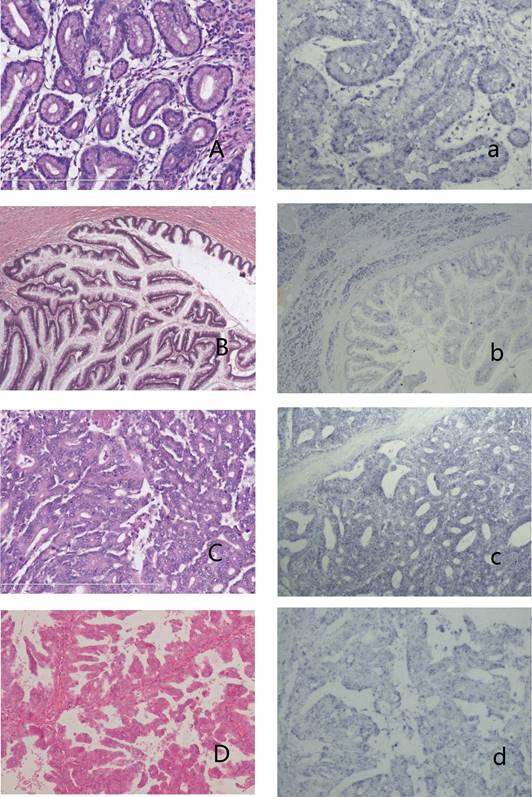
IPMNs and the expression of has-miR 217 A-a: IPMN with low-grade dysplasia (A,HE stain) showed no blue granules in the cytoplasm (a). B-b: IPMN with intermediate-grade dysplasia (B, HE stain) showed weak-stained blue granules in the cytoplasm of neoplasm cells and moderate expression in surrounding acinal cells (b). C-c: IPMN with high-grade dysplasia (C, HE stain) showed moderate expression in the cytoplasm (c). D-d: Oncocytic-type IPMN with high-grade dysplasia (D, HE stain) showed weak expression in the cytoplasm (d).
Comparison of the expression in PanIN-PDA sequence and IPMN-carcinoma sequence
Comparing the expression of hsa-miR-96 and hsa-miR-217 between PanINs and IPMNs with the same degree of epithelial proliferation, we found that there was no significantly different among PanIN1 and IPMN with low grade dysplasia (P=0.088, P=0.086), PanIN2 and IPMN with intermediate grade dysplasia (P=0.700, P=0.700), PanIN3 and IPMN with high grade dysplasia (P=0.302, P=0.968), PanIN-PDA and IPMN associated carcinoma (P=0.710, P=0.756).
Discussion
PanINs and IPMNs are the most common premalignant lesions of PDAs, playing an important role in early diagnosis and treatment. PanINs are relatively common incidental finding. More than 30% elderly people, especially those over 60s [18], have foci of PanINs in their pancreas. In addition, foci of PanINs can be found in more than 80% individuals with PDAs and 60% individuals with chronic pancreatitis [19]. PanIN3 are accompanied with pancreatic carcinoma, and do not present in pancreas without invasive carcinoma. PanIN2 can occasionally be found in pancreas without cancer. Intraductal lesions of the pancreas are being detected much more frequently. IPMNs currently account for approximately 8-20% of all resected pancreatectomy specimens [20-23], and the incidence is rising. Traditionally, PanINs are supposed to be originated from the intralobular and interlobular ductus, while IPMNs come from main duct and branch ducts. The epithelium lining small ducts and large ducts are similar in morphological features. However, the small duct-derived PanINs and the large duct-derived IPMNs show different molecular alterations. Whether there are differences in miRNAs expression between these two lesions is worth investigating.
Many studies focusing on microRNAs in pancreatic tumors have confirmed that pancreatic cancers exhibit abnormal expression of miRNAs [24], including up-regulated miR-21, miR-155, miR-221, and miR-210 [25-27], as well as down-regulated miR-148a/-148b, hsa-miR-96, and hsa-miR-217 [15,16,28]. There are more and more microRNAs with established functions and known related genes and proteins. MiR-21 and miR-155, showing aberrant expression in PanINs and IPMNs, have been widely studied. They contribute to the aggressive behavior and poor prognosis of PDAs, and may act as a marker for early detection [25, 29].
It is difficult to detect miRNA through in situ hybridization methods, particularly in paraffin-embedded tissues. Locked nucleic acid oligomers may provide high-sensitivity probes to detect miRNA [30]. LNA-ISH combined with the NBT/BCIP staining system is proved to be a useful technique for identifying of miRNA expression in formalin-fixed, paraffin-embedded (EFPE) tissues [31]. LNA-ISH becomes a useful tool to study miRNA expression in pancreatic carcinomas.
To date, a few studies of hsa-miR-96 and hsa-miR-217 in PDAs have been conducted. By miRNA microarray hybridization and quantitative PCR, our previous studies confirmed that the expression of hsa-miR-96 and hsa-miR-217 in pancreatic cancer tissue and cell lines was significantly reduced comparing with normal pancreatic tissue [15, 16]. Szafransk et al. also demonstrated that hsa-miR-217 was down-regulated in cell lines and pancreatic cancer tissues using Quantitative RT-PCR and TaqMan real-time PCR [12, 32].
In our study, the aberrant expression of hsa-miR-217 and hsa-miR-96 was concentrated in PanIN1 and IPMN with low-grade dysplasia, which are the very early phase in the carcinogenesis of PDAs. This finding indicates that hsa-miR-217 and hsa-miR-96 are early events in the development of PanINs and IPMNs, suggesting they may be markers for early detection of PDA. Morever, down-regulation of hsa-miR-217 and hsa-miR-96 with increasing stage of PanINs and IPMNs, as well as their lowest expression in PDAs, provide evidence that these microRNAs may serve as tumor suppressor.
The expression of hsa-miR-217 and hsa-miR-96 in PanIN3 showed no significant difference from PDAs, but decreased significantly in PanIN2 and PanIN1, indicating high-grade dysplasia share more similarities with carcinoma. This finding is in agreement with gene expression profile. Significant decline of miRNA-217 expression was observed with increasing degree of cell atypia in PanINs, but not in IPMNs with different degree of dysplasia.
The expression of miRNA-96 was significantly different between IPMN and IPMN associated carcinoma (p=0.009), suggesting that it may be a potential marker to distinguish benign and malignant intraductal lesions before operation, though further studies are expected. No difference was observed of miRNAs in the same grade cell atypia between PanINs and IPMNs, raising the possibility that its expression may not be associated with different histological type, but with different degree of cell atypia instead.
Previous studies revealed that hsa-miR-96 and hsa-miR-217 may regulate the expression of K-RAS [15, 16]. K-RAS mutations are detected in more than 90% of human pancreatic cancers [17], and at the early stage of PanINs and IPMNs. Therefore, it is considered to be one of the earliest genetic changes and a key factor in the progression of PDAs [33-36]. We had expected to establish a connection between miRNAs and k-ras protein level, by detecting the changes of k-ras protein expression between different grades of PanINs using immunohistochemical staining. However, no good k-ras antibody was available for a credible result.
In conclusion, with the LNA-ISH system, we found that hsa-miR-96 and hsa-miR-217 were down-regulated in PanIN1 and IPMAs, with a decline during the transition from premalignant lesions to PDAs. Therefore, hsa-miR-96 and hsa-miR-217 may play a potential tumor suppressor role in the development of PDA and may serve as biomarkers for the detection of early lesions of PDA.
Acknowledgements
Supported by the Foundation of Pathologic Research Centre of the China Academy of Medical Sciences (Project No. 2015PT320002, and Project No. 2016ZX310176-3).
Competing Interests
The authors have declared that no competing interest exists.
References
1. Scarlett CJ, Salisbury EL, Biankin AV. et al. Precursor lesions in pancreatic cancer: morphological and molecular pathology. Pathology. 2011;43(3):183-200
2. Hruban RH, Pitman MB, Klimsra DS. Intraductal neoplasms. In: (ed.) Hruban RH, Pitman MB, Klimstra DS. AFIP Atlas of Tumor Pathology, Series 4: tumors of the Pancreas. Washington, DC: American Registry of Pathology. 2007:75-110
3. Maitra A, Fukushima N, Takaori K. et al. Precursors to invasive pancreatic cancer. Adv Anat Pathol. 2005;12(2):81-91
4. Hruban RH, Maitra A, Kern SE. et al. Precursors to pancreatic cancer. Gastroenterol Clin North Am. 2007;36(4):831-49 vi
5. Fernandez-del Castillo C, Adsay NV. Intraductal papillary mucinous neoplasms of the pancreas. Gastroenterology. 2010;139(3):708-13
6. Sahin F, Maitra A, Arqani P. et al. Loss of STK11/LKB1 expression in pancreatic and biliary neoplasms. Mod Pathol. 2003;16(7):686-91
7. Schonleben F, Qiu W, Remotti HE. et al. PIK3CA, KRAS, and BRAF mutations in intraductal papillary mucinous neoplasm/carcinoma (IPMN/C) of the pancreas. Langerbecks Arch Surg. 2008;393(3):289-96
8. Du T, Zamore PD. Beginning to understand microRNA function. Cell Res. 2007;17(8):661-3
9. Standart N, Jackson RJ. MicroRNAs repress translation of m7Gppp-capped target mRNAs in vitro by inhibiting initiation and promoting deadenylation. Genes Dev. 2007;21(16):1975-82
10. Lewis BP, Burge CB, Bartel DP. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell. 2005;120(1):15-20
11. Volinia S, Calin GA, Liu CG. et al. A microRNA expression signature of human solid tumors defines cancer gene targets. Proc Natl Acad Sci U S A. 2006;103(7):2257-61
12. Szafranska AE, Davison TS, John J. et al. MicroRNA expression alterations are linked to tumorigenesis and non-neoplastic processes in pancreatic ductal adenocarcinoma. Oncogene. 2007;26(30):4442-52
13. Bloomston M, Frankel WL, Petrocca F. et al. MicroRNA expression patterns to differentiate pancreatic adenocarcinoma from normal pancreas and chronic pancreatitis. JAMA. 2007;297(17):1901-8
14. Lee EJ, Gusev Y, Jiang J. et al. Expression profiling identifies microRNA signature in pancreatic cancer. Int J Cancer. 2007;120(5):1046-54
15. Yu S, Lu Z, Liu C. et al. miRNA-96 suppresses KRAS and functions as a tumor suppressor gene in pancreatic cancer. Cancer Res. 2010;70(14):6015-25
16. Zhao WG, Yu SN, Lu ZH. et al. The miR-217 microRNA functions as a potential tumor suppressor in pancreatic ductal adenocarcinoma by targeting KRAS. Carcinogenesis. 2010;31(10):1726-33
17. Habbe N, Koorstra JB, Mendell JT. et al. MicroRNA miR-155 is a biomarker of early pancreatic neoplasia. Cancer Biol Ther. 2009;8(4):340-6
18. Kern SE, Hruban RH, Hidalqo M. et al. An introduction to pancreatic adenocarcinoma genetics, pathology and therapy. Cancer Biol Ther. 2002;1(6):607-13
19. Yonezawa S, Hiqashi M, Yamada N. et al. Precursor lesions of pancreatic cancer. Gut Liver. 2008;2(3):137-54
20. Adsay NY, Conlon KC, Zee SY. et al. Intraductal papillary-mucinous neoplasms of the pancreas: an analysis of in situ and invasive carcinomas in 28 patients. Cancer. 2002;94(1):62-77
21. Fernandez-del Castillo C, Tarqarona J, Thayer SP. et al. Incidental pancreatic cysts: clinicopathologic characteristics and comparison with symptomatic patients. Arch Surg. 2003;138(4):427-3 discussion 433-4
22. Andrejevic-Blant S, Kosmahl M, Sipos B. et al. Pancreatic intraductal papillary-mucinous neoplasms: a new and evolving entity. Virchows Arch. 2007;451(5):863-9
23. Kosmahl M, Pauser U, Peters K. et al. Cystic neoplasms of the pancreas and tumor-like lesions with cystic features: a review of 418 cases and a classification proposal. Virchows Arch. 2004;445(2):168-78
24. Schultz NA, Werner J, Willenbrock H. et al. MicroRNA expression profiles associated with pancreatic adenocarcinoma and ampullary adenocarcinoma. Mod Pathol. 2012Dec;25(12):1609-22
25. du Rieu MC, Torrisani J, Selves J. et al. MicroRNA-21 is induced early in pancreatic ductal adenocarcinoma precursor lesions. Clin Chem. 2010;56(4):603-12
26. Ryu JK, Hong SM, Karikari CA. et al. Aberrant MicroRNA-155 expression is an early event in the multistep progression of pancreatic adenocarcinoma. Pancreatology. 2010;10(1):66-73
27. Greither T, Grochola LF, Udelnow A. et al. Elevated expression of microRNAs 155,203,210 and 222 in pancreatic tumors is associated with poorer survival. Int J Cancer. 2010;126(1):73-80
28. Hanoun N1, Delpu Y, Suriawinata AA. et al. The silencing of microRNA 148a production by DNA hypermethylation is an early event in pancreatic carcinogenesis. Clin Chem. 2010;56(7):1107-18
29. Caponi S, Funel N, Frampton AE. et al. The good, the bad and the ugly: a tale of miR-101, miR-21 and miR-155 in pancreatic intraductal papillary mucinous neoplasms. Ann Oncol. 2012;24(3):734-41
30. Kloosterman WP, Wienholds E, de Bruijn E. et al. In situ detection of miRNAs in animal embryos using LNA-modified oligonucleotide probes. Nat Methods. 2006;3(1):27-9
31. Yamamichi N, Shimomura R, Inada K. et al. Locked nucleic acid in situ hybridization analysis of miR-21 expression during colorectal cancer development. Clin Cancer Res. 2009;15(12):4009-16
32. Szafranska AE, Doleshal M, Edmunds HS. et al. Analysis of microRNAs in pancreatic fine-needle aspirates can classify benign and malignant tissues. Clin Chem. 2008;54(10):1716-24
33. Tada M, Ohashi M, Shiratori Y. et al. Analysis of K-ras gene mutation in hyperplastic duct cells of the pancreas without pancreatic disease. Gastroenterology. 1996;110(1):227-31
34. Jones S, Zhang X, Parsons DW. et al. Core signaling pathways in human pancreatic cancers revealed by global genomic analyses. Science. 2008;321(5897):1801-6
35. Moskaluk CA, Hruban RH, Kern SE. p16 and K-ras gene mutations in the intraductal precursors of human pancreatic adenocarcinoma. Cancer Res. 1997;57(11):2140-3
36. Yoshizawa K, Nagai H, Sakurai S. et al. Clonality and K-ras mutation analyses of epithelia in intraductal papillary mucinous tumor and mucinous cystic tumor of the pancreas. Virchows Arch. 2002;441(5):437-43
Author contact
![]() Corresponding author: Dr. Jie Chen, Department of Pathology, Peking Union Medical College Hospital, Chinese Academy of Medical Sciences and Peking Union Medical College, Tsinghua University, Beijing 100730, China. xhblkcom Telephone: +86-10-69159362 Fax: +86-10-69159362
Corresponding author: Dr. Jie Chen, Department of Pathology, Peking Union Medical College Hospital, Chinese Academy of Medical Sciences and Peking Union Medical College, Tsinghua University, Beijing 100730, China. xhblkcom Telephone: +86-10-69159362 Fax: +86-10-69159362

 Global reach, higher impact
Global reach, higher impact