Impact Factor
ISSN: 1449-1907
Int J Med Sci 2017; 14(3):284-293. doi:10.7150/ijms.17861 This issue Cite
Research Paper
Upregulation of heat shock protein 70 and the differential protein expression induced by tumor necrosis factor-alpha enhances migration and inhibits apoptosis of hepatocellular carcinoma cell HepG2
1. Department of pathology, Tungs' Taichung MetroHarbor Hospital, Taichung, Taiwan.
2. Institute of Biochemistry, Microbiology, and Immunology, Chung Shan Medical University, Taichung City, Taiwan.
3. Clinical Laboratory, Chung Shan Medical University Hospital, Taichung City, Taiwan.
Received 2016-10-10; Accepted 2017-1-30; Published 2017-2-25
Abstract
Tumor necrosis factor alpha (TNFα) plays diverse roles in liver damage and hepatocarcinogenesis with its multipotent bioactivity. However, the influence of TNFα on protein expression of hepatocellular carcinoma (HCC) is incompletely understood. Therefore, we aimed to investigate the differential protein expression of HCC in response to TNFα stimulus. We observed that HepG2 cell revealed a higher resistance to TNFα-induced apoptosis as compared to the non-tumorigenic hepatocyte THLE-2. By using a label-free quantitative proteomic analysis, we found that 520 proteins were differentially expressed in the HepG2 cells exposed to TNFα, including 211 up-regulated and 309 down-regulated proteins. We further confirmed several proteins with significant expression change (TNFα/control ratio>2.0 or <0.5) by immunoblotting using specific antibodies. We also analyzed the differential expressed proteins using Gene ontology and KEGG annotations, and the results implicated that TNFα might regulate ribosome, spliceosome, antigen processing and presentation, and energy metabolism in HepG2 cells. Moreover, we demonstrated that upregulation of heat shock protein 70 (HSP70) was involved in both the promoted migration and the inhibited apoptosis of HepG2 cells in response to TNFα. Collectively, these findings indicate that TNFα alters protein expression such as HSP70, which triggering specific molecular processes and signaling cascades that promote migration and inhibit apoptosis of HepG2 cells.
Keywords: Hepatocellular carcinoma, Tumor necrosis factor-alpha, apoptosis, Label-free proteomic analysis
Introduction
Hepatocellular carcinoma (HCC) is the most common type of liver tumors that causes over 600,000 deaths per year in the world [1]. Patients with HCC usually combine with cirrhosis, thrombocytopenia, ascites and neutropenia, and the complicated combination often makes the therapies for the patients ineffectual [2]. In addition, the prognosis of HCC is poor due to the frequent resistance to current chemotherapies via the dysfunction of signaling pathways controlling cell proliferation and survival [3].
Tumor necrosis factor alpha (TNFα) possesses multipotent bioactivity that mediates various cellular responses, including proliferation, proinflammatory factor production, and cell death. Particularly, TNFα is known to involve in the pathophysiology of multiple liver disorders such viral hepatitis, alcoholic hepatitis, nonalcoholic fatty liver disease, and ischemia-reperfusion injury. The roles of TNFα in liver injury acts as not only a mediator of cell death but also an inducer of cell proliferation [4, 5]. Moreover, continuous TNFα production during chronic inflammation is reported to dysregulate diverse signaling cascades, thereby contributing to tumorigenesis [6]. For example, nuclear factor-kappa B (NF-κB) signaling that can be efficiently and specifically activated by trimeric form of TNFα has been known to be closely associated with tumorigenesis [7]. Recently, high level of TNFα has been recognized as an independent predictor of poor survival in HCC patients, and the combination of anti-TNFα treatment and 5'-fluorouracil can promote apoptosis of HCC cells [8].
In this study, we aimed to explore the differential protein expression in response to TNFα by using a label-free quantitative proteomic approach, combining liquid chromatography-tandem mass spectrometry (LC-MS/MS) and spectra counting, to identify and semi-quantitate the changes of protein expression in HCC cell HepG2. The identified proteins with significant change were subjected to gene ontology (GO), KEGG annotation analysis, and biological evaluation of the understanding of specific pathways and subcellular processes induced by TNFα.
Materials and Methods
Materials
Protease inhibitor (P2714), trypsin (T802), ammonium bicarbonate (ABC; A6141), acrylamide (A3699), Coomassie Brilliant Blue (CBB, B7920), formic acid (94318), and trifluoroacetic acid (TFA; T6508) Tris (161-0719), 4-vinylpyridine (V3204), dithiothreitol (DTT; 161-0611), sodium dodecyl sulfate (SDS), and TNFα (H8916) were purchased from Sigma-Aldrich (St. Louis, MO, USA).
Cell Culture and Treatments
Human HCC cell line HepG2 (ATCC® HB-8065™) and non-malignant liver cell line THLE-2 was obtained from the American Type Culture Collection (ATCC; Rockville, MD, USA), cultured with complete medium [MEM/EBSS (HyClone, Logan, UT, USA) containing 10% fetal bovine serum (Gibco BRL, Gaithersburg, MD, USA), 100 U/mL penicillin, 100 µg/mL streptomycin, and 1% L-glutamine, and maintained at 37°C in a humidified atmosphere of 5% CO2.
For TNFα treatment, cells were grown to 75 - 80% confluence, washed with PBS and then starved in serum-free medium for 16 hours (h). The starved cells were incubated with serum-free medium containing 10 μg/mL TNFα at 37°C for 24 h and then harvested for the subsequent experiments.
Cell Viability Assay
Cell viability was determined by MTT assay. Briefly, cells were seeded at a density of 4 x 104 cells/well in a 24-well plate and cultured for 24 h to allow the cells for attachment. After the TNFα treatment, the supernatant was aspirated and the cells were washed with phosphate-buffered saline (PBS). Then, the cells were incubated with MTT solution [5 mg/mL 3-(4,5-Dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide] for 4 h. After the incubation, the supernatant was removed and isopropanol was added. The absorbance at 563 nm solubilized was determined for the solubilized formazan using a spectrophotometer (SpectraMAX 360 pc, Molecular Devices, Sunnyvale, CA, USA). The cell viability was presented as the percentage as comparing with control. Each treatment was performed in triplicate for statistical analysis.
Flow Cytometry Analysis
Cells were synchronized at G0 phase and then incubated in complete medium to allow cell-cycle progression. After the treatments, cells were collected, fixed with 1 mL of ice-cold 70% ethanol, incubated at -20oC for 24 h, and centrifuged at 380 g for 5 min. Cell pellets were treated with l mL of cold staining solution containing 20 μg/mL propidium iodide, 20 μg/mL RNase A, and 1% Triton X-100, and incubated for 15 min in dark. After the incubation, the cells were subjected to cell cycle distribution analysis by using a FACS Calibur system (version 2.0, BD Biosciences, Franklin Lakes, NJ, USA) with CellQuest software. Each treatment was performed in triplicate for statistical analysis.
GeLC-MS/MS Analysis
A total of 50 μg of each protein fraction from each biological replicate was resolved on a 12% SDS-PAGE gel and stained with CBB. Each lane was processed into 5 gel slices and subject to pyridylethylation and in-gel trypsin digestion as described previously [9]. Peptides were extracted once in 100 μL of 1% formic acid and subsequently twice in 100 μL of 50% acetonitrile in 5% formic acid. The volume was reduced to 50 μL in a vacuum centrifuge prior to LC-MS/MS analysis.
Extracted peptides were separated using an Ultimate 3000 nanoLC system (Dionex LC-Packings, Amsterdam, The Netherlands) equipped with a 20 cm x 75 μm i.d. fused silica column custom packed with 3 μm 120 Å ReproSil Pur C18 aqua (Dr. Maisch, GMBH, Ammerbuch-Entringen, Germany). After injection, peptides were trapped at 30 μL/min on a 5 mm x 300 μm i.d. Pepmap C18 cartridge (Dionex-LC Packings, Amsterdam, The Netherlands) at 2% buffer B (buffer A, 0.05% formic acid in distilled water); buffer B, 80% ACN and 0.05% formic acid in MQ) and separated at 300 nL/min in a 10-40% buffer B gradient in 60 min. Eluting peptides were ionized at 1.7 kV in a Nano made Triversa Chip-based nanospray source using a Triversa LC coupler (Advion, Ithaca, NJ). Intact peptide mass spectra and fragmentation spectra were acquired on an LTQ FT hybrid mass spectrometer (Thermo Fisher, Bremen, Germany). Intact masses were measured at a resolution of 50,000 in the ICR cell using a target value of 1 x 106 charges. In parallel, following an FT prescan, the top 5 peptide signals (charge-states 2+ and higher) were submitted to MS/MS in the linear ion trap (3 amu isolation width, 30-millisecond activation, 35% normalized activation energy, Q-value of 0.25 and a threshold of 5000 counts). Dynamic exclusion was applied to a repeat count of 1 and an exclusion time of 30 sec.
Database Searching and System Biology Analysis
MS/MS spectra were searched against Homo sapiens (human) SwissProt 2014_07 (546,000 sequences; 194,259,968 residues) using Sequest (version 27, rev 12), which is part of the BioWorks 3.3 data analysis package (Thermo Fisher, San Jose, CA, USA). MS/MS spectra were searched with a maximum allowed deviation of 10 ppm for the precursor mass and 1 amu for fragment masses. Methionine oxidation was allowed as variable modifications and cysteine pyridylethylation was allowed as fixed modifications, two missed cleavages were allowed and the minimum number of tryptic termini was 1. After database searching, the DTA and OUT files were imported into Scaffold (versions 1.07 and 2.01) (Proteome software, Portland, OR, USA). The scaffold was used to organize the data and to validate peptide identifications using the Peptide Prophet algorithm, and only identifications with a probability >95% were retained. Subsequently, the Protein-Prophet algorithm was applied and protein identifications with a probability of >99% with 1 or 2 peptides in at least one of the samples were retained. Proteins that contained similar peptides and could not be differentiated based on MS/MS analysis alone were grouped. For each protein identified, the number of spectral counts (the number of MS/MS associated with an identified protein) was exported to Excel.
The LC-MS/MS data were uploaded from a Microsoft Excel spreadsheet onto DAVID functional annotation tools (http://david.abcc.ncifcrf.gov/tools.jsp). DAVID functional annotation tools analysis to describe common pathways or molecular connections between control and experiment. The representations of the molecular relationships between protein were generated using the Gene Ontology Analysis, based upon processes showing significant (P<0.05) association.
Immunoblotting
Cells were collected and lysed in the lysis buffer [10 mM Tris-HCl, pH7.5; containing 1% v/v Triton X-100, 150 mM NaCl, 0.5 mM EDTA, 1 mM phenylmethanesulfonylfluoride (PMSF), 1 mM NaF, 1 mM Na4P2O7, 10 μg/mL aprotinin and leupeptin (Sigma-Aldrich)]. After centrifugation to remove cell debris, the supernatants were collected for protein quantitation using BCA protein assay kit (Pierce Biotechnology). The crude proteins (30 μg/lane) were separated in 12.5% SDS-PAGE and then transferred onto nitrocellulose membrane (Millipore, Bedford, MA, USA). After blocking with 3% w/v skimmed milk, the membranes were incubated with primary antibodies for 2 h. The primary antibodies against human 14-3-3ε (No.9635), HMGB1 (D3E5, No.6893), cathepsin B (G60, No.3373), calnexin (C5C9, No.2679), HSP 70 (No.4872), histone H4 (No.2592), and α-tubulin (No.2144) were purchased from Cell Signaling Technologies (Beverly, MA, USA); α-actinin-1 (7A4, No. NBP1-48251) and sialidase-1/NEU1 (No.MAB6860) was purchased from Novus Biologicals (Littleton, CO, USA); and gelsolin (GS-2C4, No.ab11081) was purchased from Abcam (Cambridge, UK). After washing with PBS containing 0.1% v/v Tween-20, the reacted membranes were incubated with anti-IgG antibodies conjugated with peroxidase (Abcam). The detection of the antigen-antibody complex was performed by using ECL reagent (Millipore, Bedford, MA, USA) and luminescence image system (LAS-4000; Fujifilm, Tokyo, Japan).
RNA interference
Small interfering RNA (siRNA) transfection was applied to knockdown the expression of HSP70 in HepG2 cells. Specific siRNA against HSP70 and the non-specific negative control were obtained from Qiagene (Hilden, Germany) and prepared according to manufacturer's instructions, and dissolved in siRNA suspension buffer at concentration of 20 μM as previously described [10]. The HSP70 siRNA was transfected into HepG2 cells with T-Pro NTR II reagent (Ji-Feng Biotechnology CO., Ltd. Taiwan) to obtain high knockdown efficiency. Control non-silencing siRNA (sense 5'-UUC UCC GAA CGU GUC ACG UdTdT -3', antisense 5'-ACG UGA CAC GUU CGG AGA AdTdT-3'). HSP70 siRNA targeting [2 lines pooled, HSP70(a), sense 5'-CCA UUG AGG AGG UAG AUU ATT-3', antisense 5'-GTG GUA ACU CCU CCA UCU AAU-3', and HSP70(b), sense 5'-GUU ACU UCA AAG UAA AUA ATT-3', antisense 5'-ACC AAU GAA GUU UCA UUU AUU-3'].
Statistical Analysis
Data were expressed as means ± SEMs of the three independent experiments. Statistical significance analysis was determined by using 1-way ANOVA followed by Dunnett for multiple comparisons with the control or the impaired 2-tailed Student t-test. P values less than 0.05 were considered as statistically significant.
Results
HepG2 cells exhibits resistance to TNFα-induced apoptosis
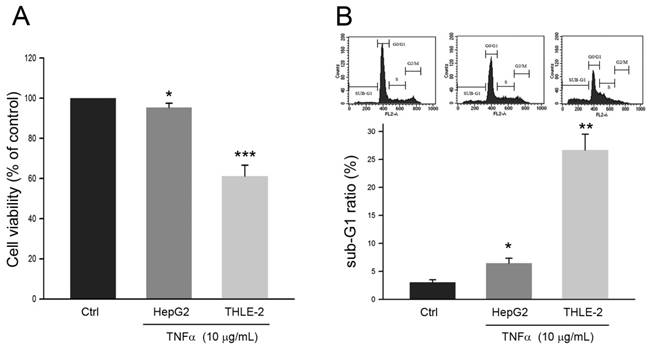
The effects of TNFα on cell viability of human HCC cell line HepG2 and non-malignant liver cell line THLE-2 were first explored. As shown in Fig. 1A, the cell viability of HepG2 was reduced to 95.3 ± 1.2% of control (P<0.05) in response to TNFα treatment at 10 μg/mL for 24 h. Comparing to the cell viability of HepG2, the cell viability of THLE-2 was reduced to 60.7 ± 3.1% of control (P<0.005) with the same TNFα treatment. Similar observations were also obtained in sub-G1 phase ratio of the two cell lines. The Sub-G1 phase ratio of HepG2 and THLE-2 cells in response to the TNFα treatment was increased to 6.2 ± 1.1% (P<0.05) and 26.1 ± 3.2% (P<0.01), respectively (Fig. 1B). Taken together, these findings showed that HepG2 cell was resistant to TNFα-induced apoptosis as comparing to the non-malignant THLE-2 cell.
Reproducible protein profiles extracted from HepG2 cell on SDS-PAGE and GeLC-MS/MS data
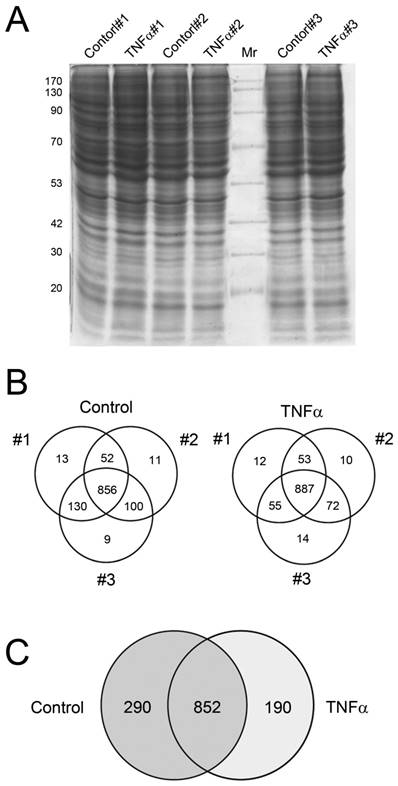
First, we assessed whether the protein extraction results in consistent protein profile. Cells were incubated with TNFα at 10 μg/mL for 24 h, and then the crude proteins were extracted, separated by 12.5% SDS-PAGE and observed by CBB staining (Fig. 2A). The resulting protein bands showed similar pattern among the three independent experimental replicates, suggesting that profiles of extracted proteins were nonsignificantly changed among the three replicates of control and TNFα treatment. The gels were then subjected to in-gel digestion for peptide extraction and following LC-MS/MS analysis for protein identification and MS/MS spectra collection. The number of proteins identified in each experiment was presented in Fig. 2B. These data suggested that each experimental replicate was performed in a highly reproducible manner. In addition, of the 1142 and 1042 proteins identified in control and TNFα-treated cells (Fig. 2B), only minor unique proteins (9 - 14) were identified in single experiment among the three replicates, implying that the protein identification for each replicate was highly reproducible and the subsequent spectra counting for semi-quantitation basing on statistical analysis could be confident. These results were consistent with the negligible change of protein patterns on SDS-PAGE (Fig. 2A), showing that the protein expression profile among the experimental replicates was highly congenial. Combining the three experimental replicates, we observed that 852 proteins were identified in both control and TNFα treatment, with 290 and 190 exclusively identified in the control and the TNFα treatment group, respectively (Fig. 2C). Collectively, our proteomic results showed that TNFα treatment contributed to a differential protein expression profile of HepG2 cell.
Effects of TNFα on HepG2 cell and THLE-2 cell. Cells were treated with PBS (Control) or TNFα at 10 μg/mL for 24 h, and then were subject to (A) MTT assay for determining cell viability and (B) cell cycle distribution analysis for determining sub-G1 phase ratio. *, ** and ***, P <0.05, 0.01 and 0.005 as compared to control, respectively.
Crude protein patterns by CBB staining (A) and the Venn diagrams showing the overlap of identified proteins in individual biological replicate in control and TNFα treatment (B). The summary of the non-redundant proteins identified between control and TNFα treatment was represented in (C).
Spectra counting quantitation shows that TNFα induces differential protein expression in HepG2 cell
Basing on the reproducible crude protein profiles, we explored whether TNFα affected protein expression in HepG2 cells. We semi-quantitated the expression level of identified proteins by using spectra counting and found that 520 of the total 1332 identified proteins basing on a minimum of 2 unique peptides were significantly altered [log2(level in TNFα treatment/level in control)>1.0 or <-1.0]. The quantitative analysis was summarized in Table 1 and the number of significantly altered proteins in each experimental replicate was also indicated. According to the analysis, 211 and 309 proteins were with up-regulated and down-regulated expression in TNFα-treated cells, respectively.
Summary of proteomic analysis for HepG2 cell by three biological replicates
| Condition | Exp | Identified proteins | Nonredundant proteins | Protein abundance with significant change |
|---|---|---|---|---|
| Control | #1 | 1051 | 1142 | 309 |
| #2 | 1019 | |||
| #3 | 1095 | |||
| TNFα | #1 | 1007 | 1042 | 211 |
| #2 | 1022 | |||
| #3 | 1028 |
Gene Ontology (GO) analysis for characterization of proteomic alteration
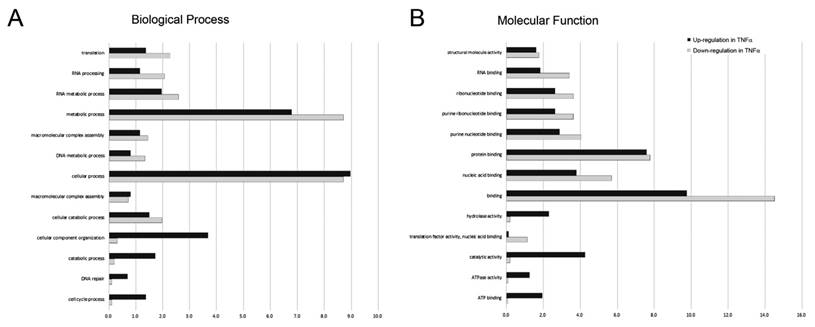
After identifying and quantitating the extracted proteins, we next investigated the biological process and molecular function linking to the proteins with significant change [log2(level in TNFα treatment/level in control)>1.0 or <-1.0] in response to TNFα treatment by using DAVID online annotation and gene ontology (GO) annotation analysis. A complete list of GO annotations for all the proteins significantly altered with the associated P values for functional enrichment is provided in Supplemental Table 1. The biological processes associating with significantly altered proteins featured by GO annotations were shown in Fig. 3A. We observed that the proteins associating with the cellular process, cellular component organization, catabolic process, DNA repair and cell cycle process were up-regulated in response to TNFα treatment. Conversely, we found that the proteins associating with translation elongation, RNA processing, and metabolic process were down-regulated in response to TNFα treatment. Similarly, we performed the molecular function analysis for the protein with significant changes (Fig. 3B) and observed that the proteins exhibiting ATP binding, ATPase activity, and hydrolase activity were up-regulated in response to TNFα treatment. On the contrary, proteins exhibiting RNA binding, nucleic acid binding, binding and translation factor activity were down-regulated in response to TNFα treatment. Taken together, we suggested that TNFα treatment suppressed specific gene transcription and the subsequent translation that promoted energy metabolism.
Gene ontology (GO) annotations for (A) biological process and (B) molecular function of proteins with up-regulated (dark bars) and down-regulated (gray bars) expression level in response to TNFα treatment. The relative number of proteins was shown as a percentage of the total number of proteins with significantly changed in each state normalized to the total number of proteins with each annotation identified in the experiment.
KEGG pathways significantly enriched in HepG2 cells in response to TNFα (Fisher Exact P-value <0.1), the number of proteins identified in each pathway, and the Fisher p-value.
| KEGGID | Term | Count | % | P-value |
|---|---|---|---|---|
| hsa03010 | Ribosome | 15 | 6.44 | 7.87E-09 |
| hsa03040 | Spliceosome | 17 | 7.30 | 2.15E-08 |
| hsa04612 | Antigen processing and presentation | 7 | 3.00 | 0.0121 |
| hsa03050 | Proteasome | 5 | 2.14 | 0.0228 |
| hsa00630 | Glyoxylate and dicarboxylate metabolism | 3 | 1.29 | 0.0460 |
| hsa00071 | Fatty acid metabolism | 4 | 1.72 | 0.0647 |
| hsa00280 | Valine, leucine and isoleucine degradation | 4 | 1.72 | 0.0812 |
| hsa00471 | D-Glutamine and D-glutamate metabolism | 2 | 0.86 | 0.0897 |
KEGG Ontology analysis for characterization of altered proteins
We also conducted KEGG (Kyoto Encyclopedia of Genes and Genomes) analysis using the KOBAS online search tool (v. 2.0) to further characterize the proteins with expression change in response to TNFα treatment. As shown in Table 2, the KEGG analysis summarized the pathways in which the identified proteins with significant change involved. We observed that TNFα treatment significantly regulated the pathways (Fisher's Exact test, P ≤0.1) involving in the ribosome, spliceosome, antigen processing and presentation, proteasome, glycoxylate and dicarboxylate metabolism, fatty acid metabolism, and metabolism of amino acids. Accordingly, we suggested that TNFα treatment may enhance RNA processing, protein degradation, and fatty acid oxidation in HepG2 cells.
Determination of protein expression level by immunoblotting
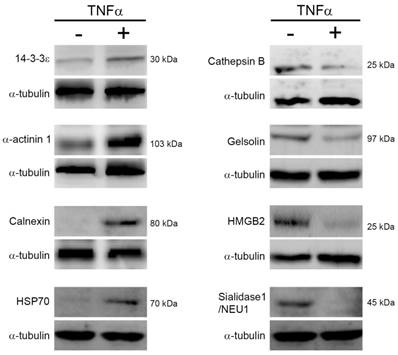
To confirm the expression changes of proteins in response to TNFα treatment, we performed immunoblotting to determine the expression level of proteins which have been reported associating with cell survival, cell cycle, protein degradation, and energy metabolism. As shown in Fig. 4, we noted that TNFα treatment altered the expression level of 14-3-3ε, α-actinin-1, calnexin, heat shock protein 70 (HSP70), cathepsin B, high mobility group protein B1 (HMGB1), gelsolin, and sialidase-1/NEU1, and the changes of these proteins were consistent with the above spectral counting quantitation. Thus, we proposed that the quantitative analysis by MS/MS spectra counting highly correlated with that by immunoblotting.
Upregulation of HSP70 involves in the promoted cell migration and apoptosis induced by TNFα
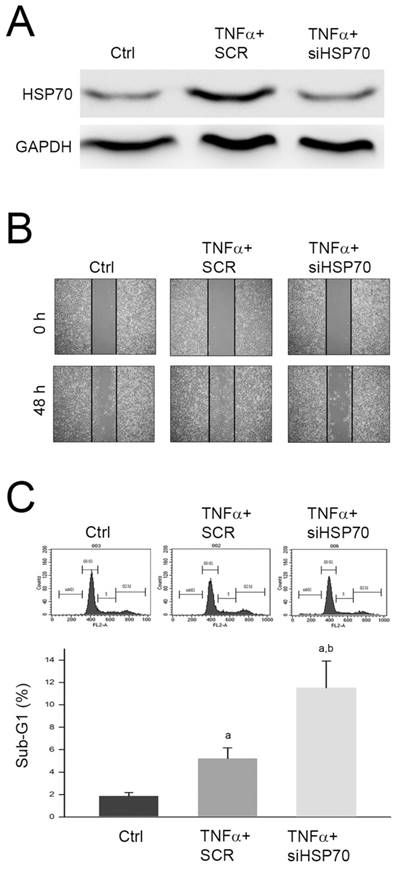
The identified proteins with significant changes in response to TNFα treatment were highly linked to the cytoskeleton and cell cycle regulation. Among the identified proteins, HSP70 expression changes have been reported to play an important role in mediating migration and apoptosis of various tumors [11-13]. Thus, we further investigated the role of upregulated HSP70 in promoted cell migration and inhibited apoptosis of HepG2 cell. We silenced the HSP70 expression by specific siRNA against HSP70 (siHSP70) and monitored the cell migration using wound healing assay. As shown in Fig. 5A, we observed that the protein level was significantly decreased by siHSP70, and the TNFα-promoted migration was inhibited by siHSP70 pretreatment (Fig. 5B). In addition, we also found that pretreatment of siHSP70 following TNFα treatment significantly increased sub-G1 phase as compared to TNFα treatment alone (Fig. 5C). Taken together, we proposed that upregulation of HSP70 involved in TNFα-promoted cell migration and TNFα-suppressed apoptosis of HepG2 cell.
Protein expression changes in response to TNFα. Cells were reacted with TNFα for 24 h, lysed for protein extraction, and the resulting crude proteins were subjected to immunoblotting. The indicated proteins were detected by using specific antibodies and chemiluminescence. Three independent immunoblotting experiments were performed for each protein and the representative images were shown. HSP70, heat shock protein 70. HMGB2, high mobility group B2.
Discussion
In the present study, we observe that TNFα stimulus certainly affects a spectrum of protein expression in human HCC cell line HepG2, which may contribute to potential cellular responses involving in the resistance of cell death and the promotion of cell migration and survival. To diminish the biological and experimental variation, we use consecutive passages of HepG2 cells and confirm the protein profiles of each extraction prior to the protein identification and quantitation using MS/MS analysis. By using GO and KEGG pathway analysis, we also explore the cellular network in which the identified proteins with significant expression change involve. We determine several protein expression levels by immunoblotting and find that the protein expression quantitation by MS/MS spectra counting and by immunoblotting is compatible. Furthermore, we demonstrate that upregulation of HSP70 expression plays an important role in the TNFα-enhanced migratory ability and TNFα-suppressed apoptosis of human HCC cell line HepG2. Collectively, we not only establish a quantitative proteomic approach that could be useful and helpful for the exploration of cellular protein expression but also provide a differential protein expression profile of and demonstrate the role of HSP70 in HepG2 cell in response to TNFα treatment.
Involvement of HSP70 in TNFα-promoted cell migration and TNFα-suppressed apoptosis of HepG2 cell. Cells were transfected with scramble RNA (SCR) or siHSP70, reacted with TNFα for 24 h, and then (A) lysed for immunodetection of HSP70; or subjected to (B) wound healing assay, or (C) cell cycle distribution analysis. a and b, P<0.05 as compared to control (Ctrl) and TNFα+SCR group, respectively.
Although we first provide the differential protein expression profile of HepG2 cell in response to TNFα, proteomic analysis of HepG2 in response to other stimuli such as mushroom polysaccharide [14] or in comparison to human hepatocyte [15] has been reported. Among these proteomic analyses for HepG2, cytoskeletal components, HSPs/chaperones, 14-3-3 family, oxidoreduction-associating proteins, and energy metabolism-associating proteins have been widely identified and investigated. Similarly, we identify 8 cytoskeletal components, 10 HSPs/chaperones, 5 14-3-3 isoforms, 7 oxidoreduction-associating proteins, and 6 energy metabolism-associating proteins (Supplementary data), and their expression levels are significantly changed in response to TNFα.
14-3-3 proteins, indicating in cell cycle process by GO analysis, modulate cellular functions via binding intracellular proteins with Ser/Thr phosphorylation-dependent domains and consequently influence subcellular localization, conformation, activity, and protein complex stability [16, 17]. Seven isoforms of 14-3-3 proteins have been identified and reported to play pivotal roles in regulating cell cycle progression, DNA repair, apoptosis, and cell adhesion and motility [16, 17]. Among the 14-3-3 isoforms, overexpression of 14-3-3ε has been reported in breast cancer [18], lung cancer [19], and HCC [20]. In addition, upregulation of 14-3-3ε has been demonstrated to protect colorectal cancer cells and endothelial cells from oxidative stress-induced apoptosis [21, 22]. Recently, 14-3-3ε has been reported to mediate cell polarization and migration via changing Zeb-1/E-cadherin expression, which contributes to HCC tumor development, progression, and metastasis [23, 24]. Our results revealed that 14-3-3ε level was increased in response to TNFα, suggesting that 14-3-3ε may play a role in the TNF-induced anti-apoptotic signals.
α-actinins are ubiquitously expressed cytoskeleton proteins that interact with various adaptor proteins such as vinculin [25] and integrins [26], and then link actin filaments to focal adhesions [27]. Among four α-actinins in mammalian cells, α-actinin-4 is primarily involved in cell motility and cancer invasion [28, 29]. Comparing to α-actinin-4, α-actinin-1 shows some different biological functions contributing to tumor development and progression [30, 31]. Our analysis revealed that TNFα treatment significantly increased α-actinin-1 and other cytoskeletal proteins such as tubulin, actin and actin-related protein 3, indicating that TNFα altered cytoskeletal protein network which may contribute to promoted cell survival.
Heat shock proteins (HSPs) and molecular chaperones play important roles in protein homeostasis, cell physiology, and protection against stressors [26, 32]. HSPs involve not only in protein folding/refolding, trafficking and degradation but also in the control of cell growth, differentiation, apoptosis and tissue repair [32]. Recently, roles of HSPs in promoting cancer cell survival and overexpression of HSPs in malignancies have been greatly noticed [33, 34]. Elevated HSP70 levels protect cancer cells from apoptosis and cellular pressures attributed to enhanced growth and accumulation of mutant proteins [35]. Abnormal levels of HSP70 have been implicated in breast cancer cell growth [36] and tumorigenesis of Rat-1 fibroblasts [37]. Similarly, we observe that TNFα upregulates HSP70 expression level, contributing to promoted cell migratory ability and suppressed apoptosis of HepG2 cell. Thus, we propose that silencing HSP70 may increase sensitivity to the antitumor drug for HCC cells as previously described for other malignancies [38, 39]. Overexpression of HSP90 has been observed in several types of tumors such as acute myeloid leukemia and is linked with poor prognosis [33, 34, 40]. HSP90 may act as an oncogenic chaperone attributing to its protective function on mutant and oncogenic proteins from degradation. Molecular chaperone calnexin is a type-I integral membrane protein in the endoplasmic reticulum (ER) which facilitates the processing of N-linked glycoprotein synthesis [41, 42]. Interestingly, HSP90 is not been identified in our proteomic analysis; therefore, the expression of HSP90 needs further investigation.
Calnexin forms heterodimeric complexes with the calnexin-associated protein disulfide isomerase ortholog ERp57 to exert its chaperone activity [43]. A Recent study is reported that calnexin and another ER-associated chaperone GRP78 are overexpressed in keratocystic odontogenic tumors [44]. Our results revealed that protein level of HSP70, HSP90 and calnexin were increased in response to TNFα treatment, suggesting that the promoted molecular chaperone expression may play an important role in the evoked prosurvival signals in HepG2 cell.
Lysosomes and cathepsins play a pivotal role in cancer cell death [45]. The lysosomal permeabilization facilitates cathepsin release, causing cell death via mitochondria-dependent apoptosis [46]. Among the lysosomal enzymes, cathepsin B is a cysteine protease mainly involved in the degradation or processing of lysosomal proteins [47], vesicle trafficking [48], cell death [49]. Our findings revealed that level of cathepsin B was significantly reduced in response to TNFα, suggesting that inhibition of lysosomal cell death may involve in the prosurvival signals evoked by TNFα in HepG2 cells.
Gelsolin is a calcium-binding protein that controls cytoskeletal actin dynamics and plays an important role in cell metabolism and survival [50]. Gelsolin is known to involve in a variety of cellular signal cascades for motility, apoptosis, proliferation, differentiation, epithelial-mesenchymal transition [51], and carcinogenesis phenotypes [52, 53]. Interestingly, gelsolin plays roles as both effector and inhibitor of apoptosis, depending on its association with different cancer types. Our findings revealed that TNFα lowered level of gelsolin in HepG2 cells; however, whether gelsolin acts as an inhibitor of apoptosis in HCC in response to TNFα needs further validation.
HMGB1 is a widely expressed and highly abundant protein and plays multiple roles in physiological and pathological processes. Thus, HMGB1 is implicated in human health and diseases [54]. Loss of HMGB1 has been reported that leads to a serial of abnormalities in the nuclear structure and function, such as genomic instability [55], abnormal gene transcription [56], and impaired DNA damage response [57]. Comparing to HMGB1, our analysis showed that level of HMGB1 was reduced in HepG2 cells with exposure to TNFα, suggesting that lowered HMGB1 may involve in carcinogenesis promoted by TNFα.
Several cancers show a tendency for decreased sialidase-1/NEU1 expression. Interestingly, sialidase-1/NEU1 expression and metastatic ability present a good inverse relationship. Cells transformed by oncogenic v-fos transfection show a severe decrease in sialidase activity and acquired higher lung metastatic potential [58]. In addition, the introduction of sialidase-1/NEU1 into Bl6 melanoma cells resulted in suppression of experimental pulmonary metastasis and tumor progression, with a decreased anchorage-independent growth and increased sensitivity to apoptosis [59]. Our findings showed that TNFα significantly reduced the level of sialidase-1/NEU1, suggesting that the decreased sialidase-1/NEU1 may induce enhanced anchorage-independent growth and anti-apoptotic signals.
Conclusion
In this study, we establish a label-free quantitative LC-MS/MS approach provide a differential protein expression profile of HepG2 cell in response to TNFα stimulus. Our results indicate that TNFα stimulus significantly modifies protein expression level of 511 cellular proteins, and the association of the 56 identified proteins with TNFα is first reported. GO and KO and KEGG pathway analysis categorize the identified proteins with significant expression changes into mainly cell cycle process, RNA process, fatty acid and amino acid metabolism, and cell death process. Furthermore, we demonstrate that inducible overexpression of HSP70 involves in the promotion of migratory ability and suppression of apoptosis in response to TNFα, suggesting that HSP70 could be a potential target to enhance sensitivity to anti-tumor drugs for HCC cells.
Supplementary Material
Supplemental table 1.
Acknowledgements
This work was supported by grant MOST105-2632-B-040-002 from the Ministry of Science and Technology, Taiwan. Flow cytometry was performed in the Instrument of Center of Chung Shan Medical University, which is supported by Ministry of Science and Technology, Ministry of Education and Chung Shan Medical University.
Abbreviations used
HCC: Hepatocellular carcinoma
HMGB1: High mobility group box 1
HSP: Heat shock protein
LC-MS: MS liquid chromatography-tandem mass spectrometry
MTT: 3-(4,5-Dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide
PBS: phosphate-buffered saline
TNFα: Tumor necrosis factor alpha
TRAIL: TNF-related apoptosis-inducing ligand
Competing Interests
The authors have declared that no competing interest exists.
References
1. Ferlay J, Parkin DM, Steliarova-Foucher E. Estimates of cancer incidence and mortality in Europe in 2008. European journal of cancer. 2010;46:765-81
2. Blum HE. Hepatocellular carcinoma: therapy and prevention. World journal of gastroenterology: WJG. 2005;11:7391-400
3. Nakanishi K, Sakamoto M, Yamasaki S. et al. Akt phosphorylation is a risk factor for early disease recurrence and poor prognosis in hepatocellular carcinoma. Cancer. 2005;103:307-12
4. Ni HM, McGill MR, Chao X. et al. Caspase Inhibition Prevents Tumor Necrosis Factor-alpha-Induced Apoptosis and Promotes Necrotic Cell Death in Mouse Hepatocytes in Vivo and in Vitro. The American journal of pathology. 2016;186:2623-36
5. Ji T, Li G, Chen J. et al. Distinct role of interleukin-6 and tumor necrosis factor receptor-1 in oval cell- mediated liver regeneration and inflammation-associated hepatocarcinogenesis. Oncotarget. 2016
6. Locksley RM, Killeen N, Lenardo MJ. The TNF and TNF receptor superfamilies: integrating mammalian biology. Cell. 2001;104:487-501
7. Wu Y, Zhou BP. TNF-alpha/NF-kappaB/Snail pathway in cancer cell migration and invasion. British journal of cancer. 2010;102:639-44
8. Keerthy HK, Mohan CD, Sivaraman Siveen K. et al. Novel synthetic biscoumarins target tumor necrosis factor-alpha in hepatocellular carcinoma in vitro and in vivo. The Journal of biological chemistry. 2014;289:31879-90
9. Kao SH, Su SN, Huang SW. et al. Sub-proteome analysis of novel IgE-binding proteins from Bermuda grass pollen. Proteomics. 2005;5:3805-13
10. Antonoff MB, Chugh R, Skube SJ. et al. Role of Hsp-70 in triptolide-mediated cell death of neuroblastoma. The Journal of surgical research. 2010;163:72-8
11. Gao Y, Han C, Huang H. et al. Heat shock protein 70 together with its co-chaperone CHIP inhibits TNF-alpha induced apoptosis by promoting proteasomal degradation of apoptosis signal-regulating kinase1. Apoptosis: an international journal on programmed cell death. 2010;15:822-33
12. Lin Y, Peng N, Zhuang H. et al. Heat shock proteins HSP70 and MRJ cooperatively regulate cell adhesion and migration through urokinase receptor. BMC cancer. 2014;14:639
13. Boroughs LK, Antonyak MA, Johnson JL, Cerione RA. A unique role for heat shock protein 70 and its binding partner tissue transglutaminase in cancer cell migration. The Journal of biological chemistry. 2011;286:37094-107
14. Chai Y, Wang G, Fan L, Zhao M. A proteomic analysis of mushroom polysaccharide-treated HepG2 cells. Scientific reports. 2016;6:23565
15. Wisniewski JR, Vildhede A, Noren A, Artursson P. In-depth quantitative analysis and comparison of the human hepatocyte and hepatoma cell line HepG2 proteomes. Journal of proteomics. 2016;136:234-47
16. Aitken A. Post-translational modification of 14-3-3 isoforms and regulation of cellular function. Seminars in cell & developmental biology. 2011;22:673-80
17. Yang X, Lee WH, Sobott F. et al. Structural basis for protein-protein interactions in the 14-3-3 protein family. Proceedings of the National Academy of Sciences of the United States of America. 2006;103:17237-42
18. Li DQ, Wang L, Fei F. et al. Identification of breast cancer metastasis-associated proteins in an isogenic tumor metastasis model using two-dimensional gel electrophoresis and liquid chromatography-ion trap-mass spectrometry. Proteomics. 2006;6:3352-68
19. Qi W, Liu X, Qiao D, Martinez JD. Isoform-specific expression of 14-3-3 proteins in human lung cancer tissues. International journal of cancer Journal international du cancer. 2005;113:359-63
20. Ko BS, Chang TC, Hsu C. et al. Overexpression of 14-3-3epsilon predicts tumour metastasis and poor survival in hepatocellular carcinoma. Histopathology. 2011;58:705-11
21. Liou JY, Lee S, Ghelani D. et al. Protection of endothelial survival by peroxisome proliferator-activated receptor-delta mediated 14-3-3 upregulation. Arteriosclerosis, thrombosis, and vascular biology. 2006;26:1481-7
22. Liou JY, Ghelani D, Yeh S, Wu KK. Nonsteroidal anti-inflammatory drugs induce colorectal cancer cell apoptosis by suppressing 14-3-3epsilon. Cancer research. 2007;67:3185-91
23. Liu TA, Jan YJ, Ko BS. et al. 14-3-3epsilon overexpression contributes to epithelial-mesenchymal transition of hepatocellular carcinoma. PloS one. 2013;8:e57968
24. Ko BS, Jan YJ, Chang TC. et al. Upregulation of focal adhesion kinase by 14-3-3epsilon via NFkappaB activation in hepatocellular carcinoma. Anti-cancer agents in medicinal chemistry. 2013;13:555-62
25. Wachsstock DH, Wilkins JA, Lin S. Specific interaction of vinculin with alpha-actinin. Biochemical and biophysical research communications. 1987;146:554-60
26. Brandvold KR, Morimoto RI. The Chemical Biology of Molecular Chaperones-Implications for Modulation of Proteostasis. Journal of molecular biology. 2015
27. Ferri KF, Kroemer G. Organelle-specific initiation of cell death pathways. Nature cell biology. 2001;3:E255-63
28. Honda K. The biological role of actinin-4 (ACTN4) in malignant phenotypes of cancer. Cell & bioscience. 2015;5:41
29. Miura N, Kamita M, Kakuya T. et al. Efficacy of adjuvant chemotherapy for non-small cell lung cancer assessed by metastatic potential associated with ACTN4. Oncotarget. 2016;7:33165-78
30. Koval OA, Tkachenko AV, Fomin AS. et al. Lactaptin induces p53-independent cell death associated with features of apoptosis and autophagy and delays growth of breast cancer cells in mouse xenografts. PloS one. 2014;9:e93921
31. Liu Z, Zhan Y, Tu Y. et al. PDZ and LIM domain protein 1(PDLIM1)/CLP36 promotes breast cancer cell migration, invasion and metastasis through interaction with alpha-actinin. Oncogene. 2015;34:1300-11
32. Niforou K, Cheimonidou C, Trougakos IP. Molecular chaperones and proteostasis regulation during redox imbalance. Redox biology. 2014;2:323-32
33. Mjahed H, Girodon F, Fontenay M, Garrido C. Heat shock proteins in hematopoietic malignancies. Experimental cell research. 2012;318:1946-58
34. Thomas X, Campos L, Le QH, Guyotat D. Heat shock proteins and acute leukemias. Hematology. 2005;10:225-35
35. Goloudina AR, Demidov ON, Garrido C. Inhibition of HSP70: a challenging anti-cancer strategy. Cancer letters. 2012;325:117-24
36. Rohde M, Daugaard M, Jensen MH. et al. Members of the heat-shock protein 70 family promote cancer cell growth by distinct mechanisms. Genes & development. 2005;19:570-82
37. Volloch VZ, Sherman MY. Oncogenic potential of Hsp72. Oncogene. 1999;18:3648-51
38. Alani B, Salehi R, Sadeghi P. et al. Silencing of Hsp70 intensifies 6-OHDA-induced apoptosis and Hsp90 upregulation in PC12 cells. Journal of molecular neuroscience: MN. 2015;55:174-83
39. Singh S, Suri A. Targeting the testis-specific heat-shock protein 70-2 (HSP70-2) reduces cellular growth, migration, and invasion in renal cell carcinoma cells. Tumour biology: the journal of the International Society for Oncodevelopmental Biology and Medicine. 2014;35:12695-706
40. Flandrin P, Guyotat D, Duval A. et al. Significance of heat-shock protein (HSP) 90 expression in acute myeloid leukemia cells. Cell stress & chaperones. 2008;13:357-64
41. Ou WJ, Bergeron JJ, Li Y. et al. Conformational changes induced in the endoplasmic reticulum luminal domain of calnexin by Mg-ATP and Ca2+. The Journal of biological chemistry. 1995;270:18051-9
42. Zapun A, Petrescu SM, Rudd PM. et al. Conformation-independent binding of monoglucosylated ribonuclease B to calnexin. Cell. 1997;88:29-38
43. Zapun A, Darby NJ, Tessier DC. et al. Enhanced catalysis of ribonuclease B folding by the interaction of calnexin or calreticulin with ERp57. The Journal of biological chemistry. 1998;273:6009-12
44. Pavli M, Farmaki E, Merkourea S. et al. Endoplasmic Reticulum Stress-Associated Chaperones, Bip/GRP78 and Calnexin are Overexpressed in Keratocystic Odontogenic Tumours. Journal of oral & maxillofacial research. 2014;5:e3
45. Aits S, Jaattela M. Lysosomal cell death at a glance. Journal of cell science. 2013;126:1905-12
46. Kroemer G, Jaattela M. Lysosomes and autophagy in cell death control. Nature reviews Cancer. 2005;5:886-97
47. McGrath ME. The lysosomal cysteine proteases. Annual review of biophysics and biomolecular structure. 1999;28:181-204
48. Ha SD, Martins A, Khazaie K. et al. Cathepsin B is involved in the trafficking of TNF-alpha-containing vesicles to the plasma membrane in macrophages. Journal of immunology. 2008;181:690-7
49. Brojatsch J, Lima H Jr, Palliser D. et al. Distinct cathepsins control necrotic cell death mediated by pyroptosis inducers and lysosome-destabilizing agents. Cell cycle. 2015;14:964-72
50. Silacci P, Mazzolai L, Gauci C. et al. Gelsolin superfamily proteins: key regulators of cellular functions. Cellular and molecular life sciences: CMLS. 2004;61:2614-23
51. Tanaka H, Shirkoohi R, Nakagawa K. et al. siRNA gelsolin knockdown induces epithelial-mesenchymal transition with a cadherin switch in human mammary epithelial cells. International journal of cancer Journal international du cancer. 2006;118:1680-91
52. Rahman M, Miyamoto H, Chang C. Androgen receptor coregulators in prostate cancer: mechanisms and clinical implications. Clinical cancer research: an official journal of the American Association for Cancer Research. 2004;10:2208-19
53. Mielnicki LM, Ying AM, Head KL. et al. Epigenetic regulation of gelsolin expression in human breast cancer cells. Experimental cell research. 1999;249:161-76
54. Kang R, Chen R, Zhang Q. et al. HMGB1 in health and disease. Molecular aspects of medicine. 2014;40:1-116
55. Giavara S, Kosmidou E, Hande MP. et al. Yeast Nhp6A/B and mammalian Hmgb1 facilitate the maintenance of genome stability. Current biology: CB. 2005;15:68-72
56. Rowell JP, Simpson KL, Stott K. et al. HMGB1-facilitated p53 DNA binding occurs via HMG-Box/p53 transactivation domain interaction, regulated by the acidic tail. Structure. 2012;20:2014-24
57. Lange SS, Mitchell DL, Vasquez KM. High mobility group protein B1 enhances DNA repair and chromatin modification after DNA damage. Proceedings of the National Academy of Sciences of the United States of America. 2008;105:10320-5
58. Miyagi T, Sato K, Hata K, Taniguchi S. Metastatic potential of transformed rat 3Y1 cell lines is inversely correlated with lysosomal-type sialidase activity. FEBS letters. 1994;349:255-9
59. Kato T, Wang Y, Yamaguchi K. et al. Overexpression of lysosomal-type sialidase leads to suppression of metastasis associated with reversion of malignant phenotype in murine B16 melanoma cells. International journal of cancer Journal international du cancer. 2001;92:797-804
Author contact
![]() Corresponding author: Shao-Hsuan Kao. Institute of Biochemistry, Microbiology, and Immunology, Chung Shan Medical University, Taichung City, Taiwan. Email: kaoshedu.tw; Tel: +886-4-24730022 ext 11681; Fax: +886-4-23248110.
Corresponding author: Shao-Hsuan Kao. Institute of Biochemistry, Microbiology, and Immunology, Chung Shan Medical University, Taichung City, Taiwan. Email: kaoshedu.tw; Tel: +886-4-24730022 ext 11681; Fax: +886-4-23248110.

 Global reach, higher impact
Global reach, higher impact