Impact Factor
ISSN: 1449-1907
Int J Med Sci 2017; 14(3):268-274. doi:10.7150/ijms.17821 This issue Cite
Research Paper
Expression of Sterol Regulatory Element-Binding Proteins in epicardial adipose tissue in patients with coronary artery disease and diabetes mellitus: preliminary study
1. Unidad de Gestión Clínica del Corazón, Hospital Universitario Virgen de la Victoria, Instituto de Investigación Biomédica de Málaga (IBIMA), Universidad de Málaga (UMA), CIBERCV Enfermedades Cardiovasculares, Málaga, Spain.
2. Unidad de Gestión Clínica de Endocrinología y Nutrición, Laboratorio del Instituto de Investigación Biomédica de Málaga (IBIMA), Hospital Universitario de Málaga (Virgen de la Victoria), Málaga, Spain. CIBER Pathophysiology of obesity and nutrition, Spain.
3. Servicio de Cardiología, Complejo Hospitalario Universitario A Coruña, Instituto de Investigación Biomédica A Coruña (INIBIC), CIBERCV Enfermedades Cardiovasculares, A Coruña. Spain.
4. Servicio de Cardiología, Hospital Universitario Puerta de Hierro-Majadahonda, Universidad Autónoma de Madrid, CIBERCV Enfermedades Cardiovasculares, Madrid, Spain.
*These authors contributed equally to this work.
Received 2016-10-5; Accepted 2016-12-20; Published 2017-2-23
Abstract
Objectives: Sterol regulatory element-binding proteins (SREBP) genes are crucial in lipid biosynthesis and cardiovascular homeostasis. Their expression in epicardial adipose tissue (EAT) and their influence in the development of coronary artery disease (CAD) and type-2 diabetes mellitus remain to be determined. The aim of our study was to evaluate the expression of SREBP genes in EAT in patients with CAD according to diabetes status and its association with clinical and biochemical data.
Methods: SREBP-1 and SREBP-2 mRNA expression levels were measured in EAT from 49 patients with CAD (26 with diabetes) and 23 controls without CAD or diabetes.
Results: Both SREBPs mRNA expression were significantly higher in patients with CAD and diabetes (p<0.001) and were identified as independent cardiovascular risk factor for coronary artery disease in patients with type-2 diabetes (SREBP-1: OR 1.7, 95%CI 1.1-2.5, p=0.02; SREBP-2: OR 1.6, 95%CI 1.2-3, p=0.02) and were independently associated with the presence of multivessel CAD, left main and anterior descending artery stenosis, and higher total and LDL cholesterol levels, and lower HDL cholesterol levels, in patients with CAD and diabetes.
Conclusions: SREBP genes are expressed in EAT and were higher in CAD patients with diabetes than those patients without CAD or diabetes. SREBP expression was associated as cardiovascular risk factor for the severity of CAD and the poor lipid control. In this preliminary study we suggest the importance of EAT in the lipid metabolism and cardiovascular homeostasis for coronary atherosclerosis of patients with diabetes and highlight a future novel therapeutic target.
Keywords: Sterol regulatory element-binding proteins (SREBP), epicardial adipose tissue, coronary artery disease, type-2 diabetes mellitus.
Introduction
The biosynthesis of cholesterol, fatty acid, and triglyceride is regulated by a family of major transcription factors, called Sterol regulatory element-binding proteins (SREBPs) [1]. They control the expression of crucial genes involved in lipogenesis and lipid uptake. Due to SREBPs play a vital role in synthesizing of lipids, its dysregulation may be intimately associated with type-2 diabetes mellitus (DM2), obesity and cardiovascular diseases [2,3].
In humans there are two SREBP genes, SREBP-1 and SREBP-2. SREBP-1 is more associated with the control of genes involved in fatty acid metabolism and SREBP-2 is closely associated with cholesterol biosynthesis and metabolism. SREBP-1 is most abundant in the liver and adrenal gland, whereas SREBP-2 is ubiquitously expressed [4].
Epicardial adipose tissue (EAT) represents a visceral fat depot located between the myocardium and the inner layer of visceral pericardium [5]. EAT participates in the energy homeostasis of the heart and the vessels. In fact, the functional EAT has been proposed to play a protector role over the myocardium or coronary arteries. However, EAT dysfunction has been implicated in the development and progression of coronary artery disease (CAD), mainly in patients with DM2, involving a more aggressive course and greater morbidity and mortality than in patients without DM2 [6].
EAT has not been fully characterized and has gained significant attention in recent years [5]. Actually, the expression of SREBP genes in EAT and the role of this tissue in the lipid biosynthesis and metabolism, and subsequently, in coronary atherosclerosis, has not been widely described. The aim of our study was to evaluate the expression of SREBP-1 and SREBP-2 in EAT in patients with CAD, stablishing the difference between patients with and without DM2. We hypothesized that SREBP genes would be expressed in EAT and would be altered according to diabetes status, playing an important role in the cardiovascular system of CAD patients. We also assessed the possible association between SREBP expression and clinical and biochemical data in patients of our cohort.
Methods
Patients
We included a total of 49 patients who underwent Coronary Artery Bypass Surgery (CAD group) and 23 patients who underwent aortic and/or mitral valve replacement (Control group). The CAD group was divided into two groups: those with DM2 (n=26) (CAD-DM2 group) and those without DM2 (n=23) (CAD-NDM2).
The CAD was defined by the presence of greater than or equal to 50% luminal diameter stenosis in at least one major epicardial artery by coronary angiogram. Multivessel disease was defined as the presence of this stenosis in two or more major epicardial arteries. Stenosis of one major epicardial artery was considered as single vessel disease. Patients of the Control group had chronic valvular heart disease, without CAD or DM2.
Exclusion criteria were acute inflammatory disease, severe infective disease and/or cancer, and women who were taking hormone replacement.
All patients gave written informed consent, and the study protocol was approved by the local Clinical Research Ethics Committee and carried out in accordance with the Declaration of Helsinki.
Biological material
Human EAT biopsy samples (average 0.2 to 0.5g) were taken near the proximal right coronary artery, approximately 1 hour after anesthesia. All the tissues were frozen immediately in liquid nitrogen and stored at -80ºC for RNA isolation.
Blood assays
On the morning of surgery, peripheral venous blood was drawn into pyrogen-free tubes with or without EDTA as an anticoagulant. For serum, the tubes were left at room temperature for 20 min and then centrifuged at 1500 g for 10 min at 4ºC. Fasting glucose, glycated hemoglobin (HbA1c), total cholesterol, low-density lipoprotein (LDL), high-density lipoprotein (HDL), triglycerides, creatinine, uric acid, glutamic-oxolacetic transaminase (GOT), glutamate-piruvate transaminase (GPT), gamma-glutamyl transferase (GGT), C-reactive protein (CRP), calcium, sodium and potassium were measured in a Dimension autoanalyzer (Dade Behring Inc., Deerfield, IL) by enzymatic methods (Randox Laboratories, Ldt., UK) in the hospital laboratory.
RNA Isolation and TaqMan Real-Time Reverse Transcription-Polymerase Chain Reaction
Adipose tissue samples were minced in TriZol reagent (Invitrogen) and homogenized completely on ice. Total RNA was extracted by chloroform and purified through RNeasy minicolumns. After on-column DNase treatment, RNA was eluted with Rnase-free water. Total RNA was quantified with a spectrophotometer (Nanodrop N-100, Thermo Scientific), and all samples had a 260/280 nm absorbance ratio ≥1.8. Reverse transcriptions were performed using 1 μg of total RNA with Transcriptor First Strand cDNA Synthesis Kit (Roche) and random hexamers in 20 μl reactions. The gene expression levels in the adipose tissue were determined by real time quantitative polymerase chain reaction (PCR) using a predesigned and validated Taqman primer/probe sets. Real-time PCR amplifications were performed on 96-well plates in reaction buffer containing Taqman Universal PCR Master Mix (No AmpErase UNG, Applied Biosystems, USA), 150 nM Taqman probe, 900 nM primers, and 22.5 ng cDNA. PCR reaction conditions were 48°C for 30 minutes, 95°C for 10 minutes, followed by 40 cycles of 95°C for 15 seconds and 60°C for 1 minute using an ABI 7500 Fast Detection System (Applied Biosystems). Data were obtained as Ct values according to the manufacturer's guidelines (the cycle number at which logarithmic PCR plots cross a calculated threshold line) and were used to determine ΔCt values (ΔCt = Ct of the target gene minus Ct of the housekeeping gene). Cyclophilin A transcripts were amplified in the same reaction to normalize for variance in input RNA. mRNA expression levels relative to cyclophilin A were calculated by the 2-ΔCt method. All tests were performed in duplicate. A negative control, RNA amplification without previous retrotranscription, was done to test for possible genomic DNA contamination.
Statistical analysis
Normality of continuous variables was checked by means of the Kolmogorov-Smirnov test. Continuous variables are summarized as mean ± SD. Discrete variables are presented as frequencies and percentages. Comparison between the results of the different groups was made with the analysis of variance (ANOVA) and chi-square test for continuous and categorical data, respectively. The post hoc analysis was done with the Bonferroni test. Logistic regression models were used in order to identify independent factors (Odds ratio [OR]; 95% Confidence Interval) for CAD in patients with DM2 associated with SREBP-1 and SREBP-2 expression, as well as to control for confounding factors. Statistical analyses were performed with SPSS for Windows version 15 (SPSS Inc. Chicago, IL, USA). Values were considered to be statistically significant when P<0.05.
Results
General characteristics of the patients
Among the 49 patients with CAD in our study cohort, 53.1% (n=26) had DM2 (CAD-DM2 group). Table 1 lists clinical and biochemical differences between patients with CAD according to diabetes status and Control group. Patients with CAD and DM2 were more likely to have hypertension and dyslipidemia, higher levels of glucose, HbA1c, total and LDL cholesterol, triglycerides and C-reactive protein, and lower HDL-cholesterol levels than those without DM2 and controls. Angiotensin converting enzyme inhibitors/Angiotensin II receptor blockers were more often used in CAD-DM2 and Control group, and Aspirin and Statins in CAD patients with and without DM2.
Clinical and biochemical characteristics of patients with coronary artery disease according to diabetes status and control group.
| Variables N (%) | CAD-DM2 (n=26) | CAD-NDM2 (n=23) | p value* | CONTROL (n=23) | p value** | p value*** | p value |
|---|---|---|---|---|---|---|---|
| Age, years | 64.4 ± 10.2 | 65.1 ± 10.8 | 0.225 | 62 ± 10 | 0.201 | 0.187 | 0.204 |
| Male gender | 20 (76.9%) | 17 (73.9%) | 0.245 | 15 (65.2%) | 0.09 | 0.100 | 0.166 |
| Smoking | 18 (69.2%) | 16 (69.6%) | 0.344 | 12 (52.2%) | 0.06 | 0.07 | 0.07 |
| Body mass index, kg/m2 | 29 ± 6 | 28.4 ± 5 | 0.102 | 27.8 ± 4.1 | 0.101 | 0.124 | 0.113 |
| Obesity | 14 (53.8%) | 11 (47.8%) | 0.127 | 10 (43.4%) | 0.08 | 0.114 | 0.144 |
| Hypertension | 23 (88.5%) | 18 (78.3%) | 0.203 | 17 (74%) | 0.333 | 0.533 | 0.293 |
| Dyslipidemia | 21 (80.8%) | 18 (78.3%) | 0.121 | 16 (69.6%) | 0.08 | 0.09 | 0.09 |
| Cerebrovascular disease | 2 (7.7%) | 1 (4.3%) | 0.141 | 2 (8.7%) | 0.119 | 0.09 | 0.186 |
| Left ventricular ejection fraction, % | 55 ± 6 | 53 ± 6 | 0.288 | 52 ± 6 | 0.288 | 0.601 | 0.257 |
| Left ventricular ejection fraction ≤40% | 5 (19.2%) | 4 (17.4%) | 0.199 | 4 (17.4%) | 0.201 | 0.249 | 0.209 |
| Medications | |||||||
| Aspirin | 23 (88.5%) | 20 (87%) | 0.185 | 8 (34.8%) | 0.01 | 0.01 | 0.02 |
| Statins | 20 (76.9%) | 17 (73.9%) | 0.108 | 15 (65.2%) | 0.06 | 0.09 | 0.09 |
| ACEI/ARB | 23 (88.5%) | 16 (69.6) | 0.04 | 19 (82.6%) | 0.107 | 0.04 | 0.03 |
| Beta-blocker | 21 (80.8%) | 19 (82.6) | 0.201 | 18 (78.3%) | 0.113 | 0.101 | 0.155 |
| Biochemical data | |||||||
| Glucose, mg/dL | 151 ± 38 | 108 ± 33 | 0.01 | 103 ± 29 | 0.01 | 0.217 | 0.02 |
| HbA1c, % | 7.9 ± 1 | 5.8 ± 0.5 | 0.01 | 5.5 ± 0.5 | 0.01 | 0.188 | 0.01 |
| Total cholesterol, mg/dL | 189 ± 31 | 159 ± 28 | 0.02 | 160 ± 30 | 0.02 | 0.201 | 0.01 |
| LDL cholesterol, mg/dL | 122 ± 26 | 100 ± 20 | 0.04 | 103 ± 21 | 0.04 | 0.199 | 0.04 |
| HDL cholesterol, mg/dL | 30 ± 6 | 49 ± 9 | 0.03 | 44 ± 9 | 0.02 | 0.214 | 0.03 |
| Triglycerides, mg/dL | 203 ± 60 | 157 ± 42 | 0.03 | 148 ± 40 | 0.03 | 0.108 | 0.03 |
| Creatinine, mg/dL | 1.14 ± 0.51 | 1.17 ± 0.76 | 0.244 | 1 ± 0.32 | 0.277 | 0.199 | 0.281 |
| Uric acid, mg/dL | 5.4 ± 2 | 6.1 ± 2.1 | 0.141 | 6.2 ± 2.2 | 0.219 | 0.109 | 0.207 |
| GOT, IU/L | 32.8 ± 21 | 29.3 ± 27 | 0.218 | 34 ± 28 | 0.111 | 0.221 | 0.281 |
| GPT, IU/L | 40.1 ± 35 | 37 ±29 | 0.109 | 42.7 ± 34 | 0.241 | 0.247 | 0.194 |
| GGT, IU/L | 60 ± 33 | 58.3 ± 31 | 0.285 | 67.7 ± 32 | 0.189 | 0.112 | 0.188 |
| CRP, mg/dL | 51.6 ± 41 | 31.3 ± 30 | 0.03 | 19.2 ± 22 | 0.01 | 0.02 | 0.02 |
| Calcium, mg/dL | 8.5 ± 0.7 | 8.6 ± 0.7 | 0.334 | 8.9 ± 0.8 | 0.147 | 0.218 | 0.222 |
| Potassium, mmol/L | 4.3 ± 0.5 | 4 ± 0.5 | 0.311 | 4.1 ± 0.5 | 0.213 | 0.201 | 0.218 |
| Sodium, mmol/L | 138 ± 3.6 | 137 ± 3 | 0.321 | 139 ± 3.7 | 0.244 | 0.299 | 0.274 |
Values are shown as mean ± SD and frequencies (percentages). Comparison between the results of the different groups was made with the analysis of variance (ANOVA) and chi-square test for continuous and categorical data, respectively. The post hoc analysis was done with the Bonferroni test. Values were considered to be statistically significant when P<0.05.
p value: overall comparison for all groups. p value*: CAD-DM2 vs CAD-NDM2 comparison. p value** CAD-DM2 vs CONTROL comparison. p value*** CAD-NDM2 vs CONTROL comparison.
ACEI: Angiotensin Converting Enzyme Inhibitor; ARB: Antiotensin II Receptro Blocker; CAD-DM2: Coronary Artery Disease-Type2-Diabetes Mellitus; CAD-NDM2: Coronary Artery Disease-Non Type2-Diabetes Mellitus; CRP: C-Reactive Protein; GGT: Gamma-Glutamyl Transferase; GOT: Glutamic-Oxolacetic Transaminase; GPT: Glutamate-Piruvate Transaminase; Hb1ac: glycated hemoglobin; HDL: High-Density Lipoprotein; IU/L: international units/liter; kg/m2: kilogram/square metre; LDL: Low-Density Lipoprotein; mg/dL: milligram/deciliter; mmol/L: milimol/liter.
Coronary artery disease characteristics grouped by diabetes status.
| Variable N (%) | CAD-DM2 (n=26) | CAD-NDM2 (n=23) | p value |
|---|---|---|---|
| Multivessel coronary disease | 20 (76.9%) | 14 (61%) | 0.02 |
| >50% stenosis left main artery | 17 (65.4%) | 11 (48%) | 0.01 |
| >50% stenosis anterior descending artery | 24 (92.3%) | 17 (74%) | 0.01 |
| >50% stenosis circumflex artery | 19 (73%) | 14 (60.9%) | 0.02 |
| >50% stenosis right coronary artery | 20 (76.9%) | 15 (65.2%) | 0.02 |
Values are shown as frequencies (percentages). Comparison between the results of the different groups was made with chi-square test. Values were considered to be statistically significant when P<0.05.
CAD-DM2: Coronary Artery Disease-Type2-Diabetes Mellitus; CAD-NDM2: Coronary Artery Disease-Non Type2-Diabetes Mellitus.
Patients of CAD-DM2 group were more likely to present multivessel coronary disease and major coronary stenosis (Table 2).
SREBP mRNA expression in EAT and comparison between CAD-DM2, CAD-NDM2 and Control group
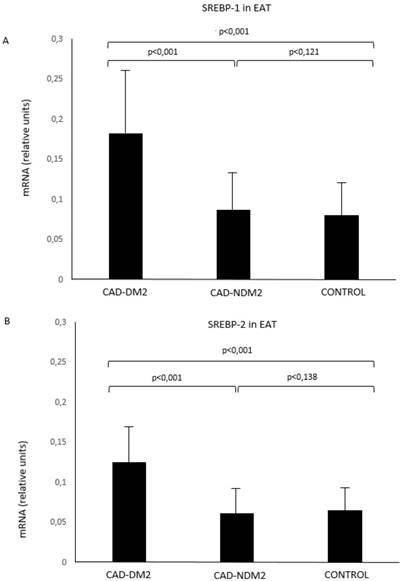
SREBP-1 and SREBP-2 mRNA in EAT were significantly higher in patients with CAD and DM2 compared with CAD-NDM2 (p<0.001) and control patients (p<0.001). No SREBP expression differences were found between CAD-NDM2 and Control group (Figure 1).
Association between SREBP mRNA expression in EAT and biochemical and clinical variables according to diabetes status
Cardiovascular risk factors, such as the presence of hypertension and dyslipidemia; biochemical parameters, such as glucose, Hb1ac, triglycerides, total, LDL and HDL cholesterol; and SREBP-1 and SREBP-2 mRNA expression, were identified as independent factors for CAD in patients with DM2. These results are presented in Table 3. The coefficient of determination for this regression model was 0.66. In addition, both SREBP-1 and SREBP-2 expression were independently associated with the presence of multivessel coronary disease, left main artery and anterior descending artery stenosis, and higher triglycerides, total and LDL cholesterol levels, and lower HDL cholesterol levels, in patients with CAD and DM2 when compared with patients without DM2 and controls (Table 4). The coefficient of determinations for this model was 0.60. Other clinical and biochemical variables were not significant.
SREBP-1 (A) and SREBP-2 (B) mRNA expression in EAT comparison between groups. CAD: coronary artery disease; DM2: type-2 diabetes mellitus; EAT: epicardial adipose tissue; SREBP: Sterol Regulatory Element-Binding Protein
Factors for coronary artery disease in patients with type-2 diabetes mellitus.
| Variable | OR (95% CI) | p value | B coefficient |
|---|---|---|---|
| Hypertension | 2.3 (1.5-4.1) | 0.02 | 0.541 |
| Dyslipidemia | 3.4 (1.7-5.4) | <0.01 | 0.688 |
| Glucose, mg/dL | 4.4 (1.8-5.8) | 0.002 | 0.718 |
| Hb1ac, % | 4.7 (1.8-6.2) | <0.001 | 0.722 |
| Total cholesterol, mg/dL | 3 (1.3-4.9) | 0.02 | 0.587 |
| LDL cholesterol, mg/dL | 2.6 (1.7-4.6) | 0.02 | 0.581 |
| HDL cholesterol, mg/dL | 2.8 (1.4-5) | 0.02 | -0.499 |
| Triglycerides, mg/dL | 1.9 (1.1-3.5) | 0.03 | 0.551 |
| SREBP-1 expression, RU | 1.7 (1.1-2.5) | 0.02 | 0.518 |
| SREBP-2 expression, RU | 1.6 (1.2-3) | 0.02 | 0.509 |
OR (95% CI) and B coefficient are shown.
Logistic regression analysis for CAD in patients with DM2. Values were considered to be statistically significant when P<0.05.
95% CI: 95% Confidence Interval; Hb1ac: glycated hemoglobin; HDL: High-Density Lipoprotein; LDL: Low-Density Lipoprotein; mg/dL: milligram/deciliter; OR: Odds Ratio; RU: Relative Units, SREBP: Sterol Regulatory Expression Binding Protein
Factors associated with SREBP-1 and SREBP-2 expression in patients with coronary artery disease and diabetes mellitus.
| OR (95% CI) | p value | B coefficient | |
|---|---|---|---|
| SREBP-1 expression, RU | |||
| Multivessel coronary disease | 1.6 (1.2-3.8) | 0.03 | 0.487 |
| Left main artery stenosis | 1.3 (1.1-3.4) | 0.04 | 0.458 |
| Anterior descending artery stenosis | 1.3 (1.1-3.8) | 0.04 | 0.438 |
| Total cholesterol, mg/dL | 1.8 (1.2-4.7) | 0.03 | 0.517 |
| LDL cholesterol, mg/dL | 1.6 (1.2-3.6) | 0.03 | 0.521 |
| HDL cholesterol, mg/dL | 1.8 (1.3-4.2) | 0.03 | -0.499 |
| Triglycerides, mg/dL | 2.8 (1.2-4.5) | 0.02 | 0.561 |
| SREBP-2 expression, RU | |||
| Multivessel coronary disease | 1.3 (1.1-3.4) | 0.04 | 0.431 |
| Left main artery stenosis | 1.2 (1.1-3.5) | 0.04 | 0.437 |
| Anterior descending artery stenosis | 1.3 (1.1-3.6) | 0.04 | 0.444 |
| Total cholesterol, mg/dL | 2.1 (1.3-4.6) | <0.01 | 0.576 |
| LDL cholesterol, mg/dL | 2.6 (1.5-4.9) | <0.01 | 0.601 |
| HDL cholesterol, mg/dL | 2.5 (1.3-4.7) | <0.01 | -0.576 |
| Triglycerides, mg/dL | 1.4 (1.1-4.6) | 0.04 | 0.461 |
OR (95% CI) and B coefficient are shown.
Logistic regression analysis for SREBP expression in patients with coronary artery disease and type2-diabetes mellitus when compared with patients without type2-diabetes mellitus and control group. Values were considered to be statistically significant when P<0.05.
95% CI: 95% Confidence Interval; HDL: High-Density Lipoprotein; LDL: Low-Density Lipoprotein; mg/dL: milligram/deciliter; OR: Odds Ratio; RU: Relative Units, SREBP: Sterol Regulatory Expression Binding Protein
Discussion
Our study found that SREBP genes are expressed in EAT and this expression was significantly higher in patients with CAD and DM2. Hypertension, dyslipidemia (high triglycerides, total and LDL-cholesterol and low HDL cholesterol levels), diabetes status (high fasting glucose and Hb1ac levels) and SREBP-1 and SREBP-2 mRNA levels were associated as cardiovascular risk factor for CAD in patients with DM2. Moreover, SREBP expression in EAT was independently associated with the severity of CAD (presence of multivessel coronary disease, and left main artery and anterior descending artery stenosis) and poor lipid control (high levels triglycerides, total and LDL cholesterol and low HDL cholesterol) in patients with DM2.
These findings are important because the expression of SREBP genes had not been previously describe in EAT. This study is also important because it adds to the relatively limited number of studies that have explored the role of this tissue in the lipid metabolism, and subsequently, in coronary atherosclerosis and cardiovascular disease. In addition, this study is unique focused on SREBPs expression, crucial genes involved in lipogenesis, adipocyte development and cholesterol homeostasis, in patients with CAD separated by DM status and associated with clinical and biochemical variables.
Several studies have shown that EAT is associated with the development and progression of coronary atherosclerosis, mainly through a dysbalance of pro/anti-inflammatory adipokines production in pathological conditions, as diabetes status, speculating about the cardiovascular implication of EAT in the DM2 [6-8]. Even, EAT has been proposed to participate in the heart energy homeostasis [9-11] and, an increase volume of this tissue has been demonstrated to be correlated with the extent and severity of CAD [8,11,12]. However, a functional EAT would play a protector role over the myocardium or coronary arteries in healthy humans [13].
Although studies have shown that the dysregulation of lipid homeostasis is closely associated with DM2 and cardiovascular disease, the molecular mechanism and regulation of lipid homeostasis is extremely complicated and poorly understood. Additionally, a lot of genes and different types of tissues involved in this process still remain to be discovered [14,15].
As our results, prior studies have investigated the association between the expression of SREBP and other genes in different tissues, mainly in liver and adipose tissue, and cardiovascular diseases [16-8]. SREBPs overexpression has been implicated with insulin resistance, carbohydrate and lipid metabolism, and has been incriminated in the development of human metabolic physiopathology such as obesity, DM2, atherosclerosis, increased fatty acid secretion, and metabolic syndrome [19-21]. Even, in an extensive study performed by Marfella et al [22], was evidenced a significant correlation between myocardium SREBP expression and myocyte lipid accumulation in patients with metabolic syndrome what might contribute to heart dysfunction. Similarly to previous reports, in the present study, lipid parameters were associated with the expression of SREBPs, contributing to the high cardiovascular risk, although this expression was explored in different tissue.
It is well known that CAD is the result of complex interactions among genetic, metabolic, and environmental risk factors. As regulators of cholesterol biosynthesis, SREBPs have been proven to be associated with CAD, helping in dissecting the molecular pathophysiology of CAD. In our study, in accordance with the results obtained in other studies [23-5], we found association between SREBPs expression and extent of coronary lesions. In this line, Karasawa et al [26] also showed that the overexpression of SREBP accelerated aortic atheroma formation and Friedlander et al [17] found an association between SREBP and the risk of myocardial infarction in among men. Another study, published by Robinet et al [27], related SREBP with early-stage carotid atherosclerosis in subjects with a risk of cardiovascular event but without detectable change in plasma lipid levels. So, these findings support a role of SREBPs in the development of cardiovascular disease.
Given the role of SREBPs as regulators of essential lipid homeostasis, their expression in different tissues but specifically in EAT and their clinical implication should be deeply characterized as an important first step for future studies. In addition, the knowledge in this field could have therapeutic implications. Regulation of SREBP overexpression could be a promising way of treating cardiovascular diseases, specialty in patients with CAD and DM2.
This preliminary study is limited by the small number of recruited patients and because our data are from a single hospital. In addition, only small EAT biopsy samples were taken, being insufficient for a proteins determination. However, our study preserves its validity because it benefits from a well-designed study protocol and has been carried out using well-stablished methods. The hypothesis that EAT SREBPs expression was involved in CAD in patients with DM2 as a cardiovascular risk factor and its association with clinical variables and lipid parameters would need to be confirmed in further research.
Conclusions
SREBP-1 and SREBP-2 genes are expressed in EAT. This expression was significantly higher in CAD patients with DM2 than in those without DM2, and it was associated as cardiovascular risk factor for the severity of CAD and the poor lipid control. These preliminary findings suggest the importance of EAT and the SREBP expression in this tissue for the cardiovascular homeostasis, and subsequently, for coronary atherosclerosis of patients with DM2 and highlight a future novel therapeutic target.
Acknowledgements
The authors thank the Cardiovascular Surgery Department of the Virgen de la Victoria Hospital of Malaga for their contribution in collecting samples. We are also grateful to Alicia Guerrero for her technical assistance.
This work was supported by grants from the Spanish Ministry of Health (FIS) (PI13/02542, PI11/01661) and Spanish Cardiovascular Research Network (RD12/0042/0030)/CIBERCV Enfermedades Cardiovasculares co-founded by Fondo Europeo de Desarrollo Regional (FEDER). Dr. Luis M. Pérez-Belmonte has the “Contrato Post-MIR Jordi Soler” from Spanish Cardiovascular Research Network (RD12/0042/0030)/CIBERCV Enfermedades Cardiovasculares.
Competing Interests
The authors have declared that no competing interest exists.
References
1. Goldstein JL, DeBose-Boyd RA, Brown MS. Protein sensors for membrane sterols. Cell. 2006;124:35-46
2. Brown MS, Goldstein JL. The SREBP pathway: regulation of cholesterol metabolism by proteolysis of a membrane-bound transcription factor. Cell. 1997;89:331-40
3. Brown MS, Goldstein JL. A proteolytic pathway that controls the cholesterol content of membranes, cells, and blood. Proc Natl Acad Sci USA. 1999;96:11041-8
4. Sato R. Sterol metabolism and SREBP activation. Arch Biochem Biophys. 2010;501:177-81
5. Iacobellis G, Corradi D, Sharma AM. Epicardial adipose tissue: anatomic, biomolecular and clinical relationships with the heart. Nat Clin Pract Cardiovasc Med. 2005;2:536-43
6. Berry C, Tardif JC, Bourassa MG. Coronary heart disease in patients with diabetes: part I: recent advances in prevention and noninvasive management. J Am Coll Cardiol. 2007;49:631-42
7. Baker AR, Silva NF, Quinn DW. et al. Human epicardial adipose tissue expresses a pathogenic profile of adipocytokines in patients with cardiovascular disease. Cardiovasc Diabetol. 2006;5:1
8. Shimabukuro M, Hirata Y, Tabata M. et al. Epicardial adipose tissue volume and adipocytokine imbalance are strongly linked to human coronary atherosclerosis. Arterioscler Thromb Vasc Biol. 2013;33:1077-84
9. Marchington JM, Pond CM. Site-specific properties of pericardial and epicardial adipose tissue: the effects of insulin and high-fat feeding on lipogenesis and the incorporation of fatty acids in vitro. Int J Obes. 1990;14:1013-22
10. Iozzo P. Myocardial, perivascular, and epicardial fat. Diabetes care. 2011;34:371-9
11. Yerramasu A, Dey D, Venuraju S. et al. Increased volume of epicardial fat is an independent risk factor for accelerated progression of sub-clinical coronary atherosclerosis. Atherosclerosis. 2012;220:223-30
12. Ahn SG, Lim HS, Joe DY. et al. Relationship of epicardial adipose tissue by echocardiography to coronary artery disease. Heart. 2008;94:e7
13. Chang L, Villacorta L, Li R. et al. Loss of perivascular adipose tissue on peroxisome proliferator-activated receptor-gamma deletion in smooth muscle cells impairs intravascular thermoregulation and enhances atherosclerosis. Circulation. 2012;126:1067-78
14. Bornfeldt KE, Tabas I. Insulin resistance, hyperglycemia, and atherosclerosis. Cell Metab. 2011;14:575-85
15. Reaven GM. Insulin resistance: the link between obesity and cardiovascular disease. Med Clin North Am. 2011;95:875-92
16. Jump DB. Fatty acid regulation of gene transcription. Crit Rev Clin Lab Sci. 2004;41:41-78
17. Friedlander Y, Schwartz SM, Durst R, Meiner V, Robertson AS. et al. SREBP-2 and SCAP isoforms and risk of early onset myocardial infarction. Atherosclerosis. 2008;196:896-904
18. Biddinger SB, Almind K, Miyazaki M. et al. Effects of diet and genetic background on sterol regulatory element-binding protein-1c, stearoyl-CoA desaturase 1, and the development of the metabolic syndrome. Diabetes. 2005;54:1314-23
19. Horton JD, Shimomura I, Ikemoto S. et al. Overexpression of sterol regulatory element binding protein-1a in mouse adipose tissue produces adipocyte hypertrophy, increased fatty acid secretion, and fatty liver. J Biol Chem. 2003;278:36652-60
20. Kolehmainen M, Vidal H, Alhava E. et al. Sterol regulatory element binding protein 1c (SREBP-1c) expression in human obesity. Obes Res. 2001;9:706-12
21. Oberkofler H, Fukushima N, Esterbauer H. et al. Sterol regulatory element binding proteins: relationship of adipose tissue gene expression with obesity in humans. Biochim Biophys Acta. 2002;1575:75-81
22. Marfella R, Di Filippo C, Portoghese M. et al. Myocardial lipid accumulation in patients with pressure-overloaded heart and metabolic syndrome. J Lipid Res. 2009;50:2314-23
23. Duan X, Zhu W, Li Y. et al. The effect of sterol regulatory element-binding protein 2 polymorphism on the serum lipid in northern Chinese subjects. J Lipid Res. 2005;46:252-7
24. Fan YM, Karhunen PJ, Levula M. et al. Expression of sterol regulatory element-binding transcription factor (SREBF) 2 and SREBF cleavage-activating protein (SCAP) in human atheroma and the association of their allelic variants with sudden cardiac death. Thromb J. 2008;6:17
25. Zhao X, Feng D, Wang Q. et al. Regulation of lipogenesis by cyclin-dependent kinase 8-mediatedcontrol of SREBP-1. J Clin Invest. 2012;122:2417-27
26. Karasawa T, Takahashi A, Saito R. et al. Sterol regulatory element-binding protein-1 determines plasma remnant lipoproteins and accelerates atherosclerosis in low-density lipoprotein receptor deficient mice. Arterioscler Thromb Vasc Biol. 2011;31:1788-95
27. Robinet P, Védie B, Chironi G. et al. Characterization of polymorphic structure of SREBP-2 gene: role in atherosclerosis. Atherosclerosis. 2003;168:381-7
Author contact
![]() Corresponding authors: Luis M. Pérez-Belmonte MD, PhD. Address: Unidad de Gestión Clínica del Corazón, Hospital Clínico Universitario Virgen de la Victoria. Campus Universitario de Teatinos, s/n. Málaga, Spain. Phone: 0034951032672. E-mail: luismiguelpb1984com. Manuel Jiménez-Navarro. Address: Unidad de Gestión Clínica del Corazón, Hospital Clínico Universitario Virgen de la Victoria. Campus Universitario de Teatinos, s/n. Málaga, Spain. Phone: 0034951032672. E-mail: jimeneznavarroes.
Corresponding authors: Luis M. Pérez-Belmonte MD, PhD. Address: Unidad de Gestión Clínica del Corazón, Hospital Clínico Universitario Virgen de la Victoria. Campus Universitario de Teatinos, s/n. Málaga, Spain. Phone: 0034951032672. E-mail: luismiguelpb1984com. Manuel Jiménez-Navarro. Address: Unidad de Gestión Clínica del Corazón, Hospital Clínico Universitario Virgen de la Victoria. Campus Universitario de Teatinos, s/n. Málaga, Spain. Phone: 0034951032672. E-mail: jimeneznavarroes.

 Global reach, higher impact
Global reach, higher impact