Impact Factor
ISSN: 1449-1907
Int J Med Sci 2017; 14(1):53-57. doi:10.7150/ijms.17027 This issue Cite
Research Paper
Polymorphisms in MicroRNA Binding Sites Predict Colorectal Cancer Survival
1. Department of Pharmacy, Zhongxiao Branch, Taipei City Hospital, Taipei, Taiwan.
2. Department of Colorectal Surgery, China Medical University Hospital, Taichung, Taiwan.
3. Division of Colorectal Surgery, Department of Surgery, Chung Shan Medical University Hospital, Taichung, Taiwan.
4. Institute of Medicine, Chung Shan Medical University, Taichung, Taiwan.
5. Department of Obstetrics and Gynecology, China Medical University Hospital, Taichung, Taiwan.
6. Department of Pharmacy, China Medical University, Taichung, Taiwan.
7. Sex Hormone Research Center, China Medical University Hospital, Taichung, Taiwan.
8. Department of Nursing, Asia University, Taichung, Taiwan.
Received 2016-7-29; Accepted 2016-11-1; Published 2017-1-1
Abstract
Background: MicroRNAs (miRNAs) mediate negative regulation of target genes through base pairing, and aberrant miRNA expression has been described in cancers. We hypothesized that single nucleotide polymorphisms (SNPs) within miRNA target sites might influence clinical outcomes in patients with colorectal cancer.
Methods: Sixteen common SNPs within miRNA target sites were identified, and the association between these SNPs and overall survival was assessed in colorectal cancer patients using Kaplan-Meier analysis, Cox regression model, and survival tree analysis.
Results: Survival tree analysis identified a higher-order genetic interaction profile consisting of the RPS6KB1 rs1051424 and ZNF839 rs11704 that was significantly associated with overall survival. The 5-year survival rates were 74.6%, 62.7%, and 57.1% for the low-, medium-, and high-risk genetic profiles, respectively (P = 0.006). The genetic interaction profile remained significant even after adjusting for potential risk factors. Additional in silico analysis provided evidence that rs1051424 and rs11704 affect RPS6KB1 and ZNF839 expressions, which in turn is significantly correlated with prognosis in colorectal cancer.
Conclusion: Our results suggest that the genetic interaction profiles among SNPs within miRNA target sites might be prognostic markers for colorectal cancer survival.
Keywords: colorectal cancer, survival, microRNAs, single nucleotide polymorphism, genetic interaction.
Introduction
Colorectal cancer is the most commonly diagnosed cancer in Taiwan and the third most common cancer worldwide [1]. Despite considerable progress in the early detection and treatment of colorectal cancer, the overall 5-year survival rate remains below 65% [2]. Therefore, identification of novel prognostic biomarkers is essential for the development of personalized therapeutic strategy that can help improve colorectal cancer survival.
MicroRNAs (miRNAs) are a class of diverse, small non-coding RNAs, approximately 25 nucleotides in length, which regulate gene expression by binding complementary sequences of target mRNAs in the 3'-untranslated region (3'-UTR), leading to mRNA cleavage or translation repression [3]. Aberrant expression of miRNAs has been described across a range of cancers including colorectal cancer [4]. Furthermore, several miRNAs have been proposed as prognostic biomarkers for colorectal cancer [5]. Single nucleotide polymorphisms (SNPs) situated in the 3'-UTR of miRNA target genes may affect miRNA-mRNA interaction and the subsequent target gene expression. While several studies have reported the association between miRNA SNPs and cancer risks [6, 7], only a few have investigated the relationship of these SNPs to the clinical outcomes of cancers. Therefore, this study sought to evaluate the prognostic significance of 16 common SNPs inside miRNA target sites on colorectal cancer survival.
Patients and Methods
Patient recruitment and data collection
A total of 188 newly diagnosed, histologically confirmed colorectal cancer patients were enrolled at China Medical University Hospital, Taiwan between 2001 and 2007. Patients' clinical and follow-up data were collected from medical records. Overall survival was defined as the interval from diagnosis to death from any cause. This study was approved by the Institutional Review Board of China Medical University Hospital. All participants provided written informed consent, and the study was carried out in accordance with approved guidelines.
SNP selection and genotyping
We identified SNPs within miRNA target sites by intersection HapMap SNPs CHB (Han Chinese in Beijing, China) table with TS (TargetScan) miRNA sites table [8] from the UCSC table browser (NCBI36/hg18) [9]. SNPs with a minor allele frequency less than 0.3 in HapMap CHB population were excluded. We initially selected 18 SNPs in miRNA target sites for analysis. Genomic DNA was extracted from peripheral blood using the QIAamp DNA Blood Mini Kit (Qiagen, Valencia, CA, USA) and stored at -80°C until the time of study. Genotyping was performed at the National Center for Genome Medicine, Taiwan, using the Agena Bioscience iPLEX matrix-assisted laser desorption/ionization time-of-flight mass-spectrometry technology, as described previously [10]. The average genotype call rate for these SNPs was 97.1%. SNPs that did not conform to Hardy-Weinberg equilibrium (P < 0.001) was removed (N = 2). Thus, a total of 16 SNPs were included for further statistical analyses.
Statistical analysis
Patient clinicopathologic characteristics were summarized as number and percentage of patients or median and interquartile range (IQR) of values. The associations of individual SNPs and clinicopathologic characteristics with overall survival were assessed using log-rank tests. Higher order SNP-SNP interactions were evaluated using survival tree analysis by STREE software (http://c2s2.yale.edu/software/stree/), which uses recursive partitioning to identify subgroups of individuals with similar risk [11]. Patients were categorized into low-, medium-, and high-risk groups based on the survival tree analysis. Multivariate Cox regression was carried out to determine the interdependency of genetic interactions and other known prognostic factors such as carcinoembryonic antigen (CEA) levels, tumor stage, lymphovascular invasion, perineural invasion, and lymph node involvement [12-14]. The Statistical Package for the Social Sciences software version 22.0.0 (IBM, Armonk, NY) was used for other statistical analyses. A two-sided P value of < 0.05 was considered statistically significant.
Bioinformatics analysis
Genotype-Tissue Expression (GTEx) data were used to identify the correlation between SNPs and gene expression levels [15]. The publicly available SurvExpress database [16] and The Cancer Genome Atlas (TCGA) dataset for colorectal adenocarcinoma [17] were utilized in order to analyze RPS6KB1 and ZNF839 gene expressions and clinical outcomes.
Results
The clinical characteristics and their associations with overall survival in 188 colorectal cancer patients are shown in Table 1. During the median follow-up of 60.1 months, 61 patients died and the 5-year overall survival rate was 69.3%. Higher CEA levels, tumor stage, positive lymphovascular invasion, perineural invasion, and lymph node involvement were significantly associated with lower survival rate (P ≤ 0.005).
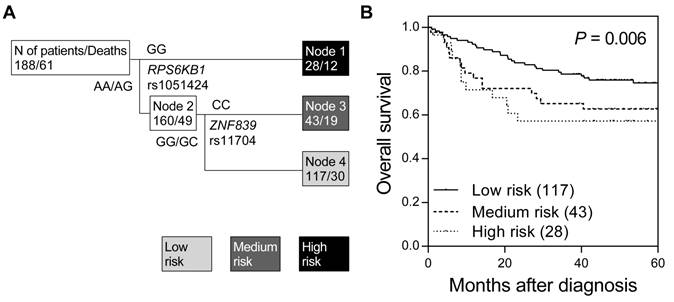
We first assessed the associations between 16 miRNA target site SNPs and overall survival using the log-rank test (Table S1). None of the tested SNPs were associated with overall survival in colorectal cancer, while only a marginal association was observed for RPS6KB1 rs1051424 (P = 0.075). Therefore, we further investigated whether higher order SNP-SNP interactions among these miRNA target site SNPs could affect colorectal cancer survival. Survival tree analysis identified a higher order genetic interaction between RPS6KB1 rs1051424 and ZNF839 rs11704, and the final tree structure identified 3 terminal nodes with low-, medium-, and high-risk of mortality (Figure 1A). The overall survival decreased in accordance with an increase in risk classification by miRNA target site SNPs (log-rank P = 0.006, Table 2 and Figure 1B). When using low-risk node 4 as the reference group (major allele carriers of RPS6KB1 rs1051424 and ZNF839 rs11704), the hazard ratio (HR) was 2.03 [95% confidence interval (CI), 1.14-3.62; Table 2] for medium-risk node 3, and 2.14 (95% CI, 1.09-4.19) for high-risk node 1 (P for trend = 0.007).
To evaluate the predictive abilities of the genetic interaction profile beyond the clinical characteristics to influence overall survival, we performed a multivariate analysis, adjusting for CEA levels, tumor stage, lymphovascular invasion, perineural invasion, and lymph node involvement. After adjusting for these predictors, the genetic interaction profile remained significant (P for trend = 0.009; Table 2). These data suggest that the genetic interaction profile among miRNA target site SNPs might be an independent predictor for colorectal cancer survival.
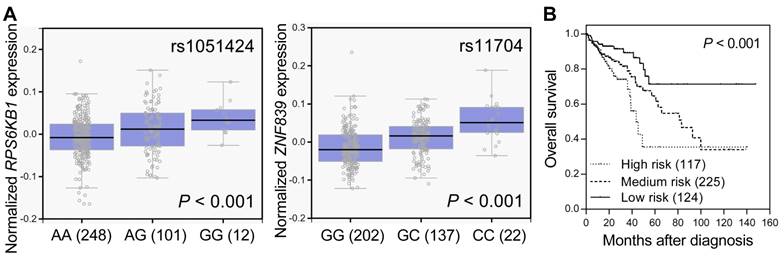
SNPs located in miRNA target sites might affect miRNA binding, the expression of their host genes, and subsequent disease progression. We used the GTEx database to investigate whether rs1051424 and rs11704 were associated with the expressions of RPS6KB1 and ZNF839, respectively. Individuals carrying a genotype with the variant G at rs1051424 showed a trend of increased expression of RPS6KB1, and individuals carrying a genotype with the variant C at rs11704 also showed a trend of increased expression of ZNF839, compared with those with the wild-type homozygous genotypes (P < 0.001, Figure 2A). We further evaluated the prognostic value of RPS6KB1 and ZNF839 expression in colorectal cancer survival using the publicly available TCGA dataset. The risk score for each patient was derived from RPS6KB1 and ZNF839 gene expression and their Cox regression coefficients, and was used to divide patients into low-, medium-, or high-risk groups using an optimization algorithm for the minimum P value. There was a significant decrease in patient survival with increasing risk score (log-rank P < 0.001, Figure 2B). These data suggest that the RPS6KB1 and ZNF839 gene expression signature might be able to predict the prognosis of patients with colorectal cancer.
Demographic and clinical characteristics of colorectal cancer patients.
| Characteristics | Overall survival | |||
|---|---|---|---|---|
| N (%)* | N of deaths* | 5-year survival rate, % | P† | |
| All patients | 188 | 61 | 69.3 | |
| Age, years | ||||
| Median (IQR) | 65 (54-73) | |||
| <65 | 90 (47.9) | 30 | 67.6 | 0.823 |
| ≥65 | 98 (52.1) | 31 | 70.9 | |
| Gender | ||||
| Male | 97 (51.6) | 37 | 63.4 | 0.092 |
| Female | 91 (48.4) | 24 | 75.6 | |
| CEA, μg/L | ||||
| Median (IQR) | 4.6 (1.8-14.4) | |||
| <5 | 89 (51.4) | 13 | 86.4 | <0.001 |
| ≥5 | 84 (48.6) | 40 | 53.9 | |
| Differentiation | ||||
| Well | 39 (22.0) | 10 | 74.0 | 0.147 |
| Moderate | 117 (66.1) | 38 | 69.6 | |
| Poor | 21 (11.9) | 10 | 52.4 | |
| Stage‡ | ||||
| I-III | 152 (81.3) | 36 | 78.2 | <0.001 |
| IV | 35 (18.7) | 25 | 29.8 | |
| Lymphovascular invasion | ||||
| Negative | 136 (72.3) | 32 | 78.3 | <0.001 |
| Positive | 52 (27.7) | 29 | 45.6 | |
| Perineural invasion | ||||
| Negative | 161 (85.6) | 44 | 74.1 | <0.001 |
| Positive | 27 (14.4) | 17 | 40.1 | |
| Lymph node involvement | ||||
| Negative | 98 (54.4) | 23 | 78.8 | 0.005 |
| Positive | 82 (45.6) | 35 | 58.5 | |
Abbreviations: CEA, carcinoembryonic antigen; IQR, interquartile range.
*Column subtotals do not sum to N of patients and N of deaths due to missing data.
†P values were calculated using the log-rank test.
‡According to the American Joint Committee on Cancer - Cancer Staging Manual (version 6.0).
P < 0.05 are in boldface.
Potential higher order SNP-SNP interactions among miRNA target site polymorphisms in colorectal cancer. (A) Survival tree analysis revealed the interactions between miRNA target site SNPs, RPS6KB1 rs1051424, and ZNF839 rs11704. (B) Kaplan-Meier curves of overall survival based on the survival tree analysis. Numbers in parentheses indicate the number of patients.
Cox proportional hazards analysis of genetic interactions among miRNA target site SNPs with overall survival.
| Groups | N (%) | N of deaths | 5-year survival rate, % | P | Univariate analysis | Multivariate analysis* | ||
|---|---|---|---|---|---|---|---|---|
| HR (95% CI) | P | HR (95% CI) | P | |||||
| Low risk | 117 (62.2) | 30 | 74.6 | 0.006 | 1.00 | 1.00 | ||
| Medium risk | 43 (22.9) | 19 | 62.7 | 2.03 (1.14-3.62) | 0.016 | 1.89 (0.99-3.59) | 0.053 | |
| High risk | 28 (14.9) | 12 | 57.1 | 2.14 (1.09-4.19) | 0.026 | 2.57 (1.15-5.73) | 0.021 | |
| Trend | 1.53 (1.12-2.08) | 0.007 | 1.65 (1.13-2.40) | 0.009 | ||||
Abbreviations: CEA, carcinoembryonic antigen; HR, hazard ratio; CI, confidence interval.
*CEA levels, tumor stage, lymphovascular invasion, perineural invasion, lymph node involvement, and genetic risk classification by miRNA target site SNPs† were included in the multivariate analysis.
†miRNA target site SNPs refer to RPS6KB1 rs1051424 and ZNF839 rs11704.
P < 0.05 are in boldface.
Functional analyses of RPS6KB1 rs1051424 and ZNF839 rs11704. (A) Correlation of rs1051424 genotypes with RPS6KB1 expression (left) and rs11704 genotypes with ZNF839 expression (right) (GTEx dataset). Numbers in parentheses indicate the number of cases. (B) The prognostic value of RPS6KB1 and ZNF839 expression in colorectal cancer. Kaplan-Meier curves of overall survival based on the risk score generating from RPS6KB1 and ZNF839 gene expression in TCGA dataset. Numbers in parentheses indicate the number of patients.
Discussion
Taking advantage of our study designs of miRNA target sites and genome-wide approaches, we have identified a higher-order genetic interaction profile consisting of RPS6KB1 rs1051424 and ZNF839 rs11704 that was associated with overall survival in colorectal cancer patients. We further confirmed functional relevance of these SNPs/genes by assessing their correlation with patients' prognosis in publicly available datasets.
The SNPs rs1051424 and rs11704 are located in putative miRNA target sites within the 3'-UTR of RPS6KB1 and ZNF839, respectively. RPS6KB1, ribosomal protein S6 kinase B1, is a member of the ribosomal S6 kinase family of serine/threonine kinases regulated by phosphatidylinositol 3-kinase (PI3K)/Akt/mammalian target of rapamycin (mTOR) pathway. Activated RPS6KB1 phosphorylates ribosomal protein S6 leading to an increase in protein synthesis, cell growth/size, and cell survival [18]. It has been shown that RPS6KB1 on chromosome 17q23 is frequently amplified and overexpressed in certain cancers, particularly breast cancer, and this phenotype correlates with poor prognosis [19]. Moreover, previous studies have demonstrated that Akt, mTOR, and RPS6KB1 are significantly activated in colorectal cancer tumors [20, 21]. The function of the ZNF839, zinc finger protein 839, is not well characterized, but it has been identified as a renal-cell carcinoma antigen involving in the humoral immune response to cancer [22]. Bioinformatic analysis using SNPinfo [23] revealed that rs1051424 A to G and rs11704 G to C transitions lost binding sites for multiple miRNAs. These transitions would be expected to result in higher expressions of RPS6KB1 and ZNF839, which is consistent with the results of our expression quantitative trait loci analysis, that individuals carrying rs1051424 G and rs11704 C alleles had increased expressions of RPS6KB1 and ZNF839 (Figure 2A). In addition, the risk score derived from RPS6KB1 and ZNF839 gene expression correlated significantly with overall survival in TCGA colorectal adenocarcinoma dataset (Figure 2B). Together, the effect of these SNPs on colorectal cancer survival might be a result of their influence on the expressions of RPS6KB1 and ZNF839, altering PI3K/Akt/mTOR signaling and host immunity.
In summary, this study systematically evaluated common SNPs within miRNA target sites in relation to the survival of colorectal cancer patients. A higher-order genetic interaction profile among SNPs in miRNA target sites was identified as a prognostic predictor for colorectal cancer, even after adjusting for other clinical factors. However, this study is limited by its relatively small sample size and the use of multiple comparisons. Moreover, our findings might be less applicable to other ethnic groups due to our homogeneous Taiwanese study population. Further investigations of the biological relevance of these SNPs/genes with larger populations are needed to validate our findings.
Supplementary Material
Table S1.
Abbreviations
miRNA, microRNA; SNP, single nucleotide polymorphism; UTR, untranslated region; IQR, interquartile range; CEA, carcinoembryonic antigen; PI3K, phosphatidylinositol 3-kinase; mTOR, mammalian target of rapamycin; HR, hazard ratio; CI, confidence interval.
Acknowledgements
This work was supported by the Ministry of Science and Technology of Taiwan (grant number: 102-2628-B-039-005-MY3) and the China Medical University (grant number: CMU105-S-42). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript. We thank the National Center for Genome Medicine, Ministry of Science and Technology of Taiwan, for technical support. The results published here are based in part on data generated by the GTEx and TCGA projects.
Competing Interests
The authors have declared that no competing interest exists.
References
1. Torre LA, Bray F, Siegel RL. et al. Global cancer statistics, 2012. CA Cancer J Clin. 2015;65:87-108
2. Siegel R, Desantis C, Jemal A. Colorectal cancer statistics, 2014. CA Cancer J Clin. 2014;64:104-17
3. Lewis BP, Burge CB, Bartel DP. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell. 2005;120:15-20
4. Cummins JM, He Y, Leary RJ. et al. The colorectal microRNAome. Proc Natl Acad Sci U S A. 2006;103:3687-92
5. Ju J. miRNAs as biomarkers in colorectal cancer diagnosis and prognosis. Bioanalysis. 2010;2:901-6
6. Landi D, Gemignani F, Naccarati A. et al. Polymorphisms within micro-RNA-binding sites and risk of sporadic colorectal cancer. Carcinogenesis. 2008;29:579-84
7. Yu Z, Li Z, Jolicoeur N. et al. Aberrant allele frequencies of the SNPs located in microRNA target sites are potentially associated with human cancers. Nucleic Acids Res. 2007;35:4535-41
8. Grimson A, Farh KK, Johnston WK. et al. MicroRNA targeting specificity in mammals: determinants beyond seed pairing. Mol Cell. 2007;27:91-105
9. Karolchik D, Hinrichs AS, Furey TS. et al. The UCSC Table Browser data retrieval tool. Nucleic Acids Res. 2004;32:D493-6
10. Huang SP, Huang LC, Ting WC. et al. Prognostic significance of prostate cancer susceptibility variants on prostate-specific antigen recurrence after radical prostatectomy. Cancer Epidemiol Biomarkers Prev. 2009;18:3068-74
11. Zhang HP, Singer B. Recursive partitioning and applications. New York: Springer. 2010
12. Ting WC, Chen LM, Huang LC. et al. Impact of interleukin-10 gene polymorphisms on survival in patients with colorectal cancer. J Korean Med Sci. 2013;28:1302-6
13. Ting WC, Chen LM, Pao JB. et al. Genetic polymorphisms of matrix metalloproteinases and clinical outcomes in colorectal cancer patients. Int J Med Sci. 2013;10:1022-7
14. Ting WC, Chen LM, Pao JB. et al. Common genetic variants in Wnt signaling pathway genes as potential prognostic biomarkers for colorectal cancer. PLoS One. 2013;8:e56196
15. Consortium GT. The Genotype-Tissue Expression (GTEx) project. Nat Genet. 2013;45:580-5
16. Aguirre-Gamboa R, Gomez-Rueda H, Martinez-Ledesma E. et al. SurvExpress: an online biomarker validation tool and database for cancer gene expression data using survival analysis. PLoS One. 2013;8:e74250
17. Cancer Genome Atlas Research N. Integrated genomic analyses of ovarian carcinoma. Nature. 2011;474:609-15
18. Bahrami BF, Ataie-Kachoie P, Pourgholami MH. et al. p70 Ribosomal protein S6 kinase (Rps6kb1): an update. J Clin Pathol. 2014;67:1019-25
19. Barlund M, Monni O, Kononen J. et al. Multiple genes at 17q23 undergo amplification and overexpression in breast cancer. Cancer Res. 2000;60:5340-4
20. Johnson SM, Gulhati P, Rampy BA. et al. Novel expression patterns of PI3K/Akt/mTOR signaling pathway components in colorectal cancer. J Am Coll Surg. 2010;210:767-76 76-8
21. Nozawa H, Watanabe T, Nagawa H. Phosphorylation of ribosomal p70 S6 kinase and rapamycin sensitivity in human colorectal cancer. Cancer Lett. 2007;251:105-13
22. Scanlan MJ, Gordan JD, Williamson B. et al. Antigens recognized by autologous antibody in patients with renal-cell carcinoma. Int J Cancer. 1999;83:456-64
23. Xu Z, Taylor JA. SNPinfo: integrating GWAS and candidate gene information into functional SNP selection for genetic association studies. Nucleic Acids Res. 2009;37:W600-5
Author contact
![]() Corresponding authors: Te-Ling Lu, Department of Pharmacy, China Medical University, 91 Hsueh-Shih Road, Taichung 404, Taiwan. Tel: +886-4-22053366 ext. 5703; Fax: +886-4-22031075; E-mail: lutlcmu.edu.tw; or Bo-Ying Bao, Department of Pharmacy, China Medical University, 91 Hsueh-Shih Road, Taichung 404, Taiwan. Tel: +886-4-22053366 ext. 5126; Fax: +886-4-22031075; E-mail: baocmu.edu.tw.
Corresponding authors: Te-Ling Lu, Department of Pharmacy, China Medical University, 91 Hsueh-Shih Road, Taichung 404, Taiwan. Tel: +886-4-22053366 ext. 5703; Fax: +886-4-22031075; E-mail: lutlcmu.edu.tw; or Bo-Ying Bao, Department of Pharmacy, China Medical University, 91 Hsueh-Shih Road, Taichung 404, Taiwan. Tel: +886-4-22053366 ext. 5126; Fax: +886-4-22031075; E-mail: baocmu.edu.tw.

 Global reach, higher impact
Global reach, higher impact