Impact Factor
ISSN: 1449-1907
Int J Med Sci 2016; 13(12):914-922. doi:10.7150/ijms.16571 This issue Cite
Research Paper
Overexpression of lncRNA H19/miR-675 promotes tumorigenesis in head and neck squamous cell carcinoma
1. Department of Otolaryngology, Head and Neck Surgery, The Second Hospital of Jilin University, Changchun 130041, P. R. China
2. Department of Respiratory Medicine, The Second Hospital of Jilin University, Changchun 130041, P. R. China.
Received 2016-6-22; Accepted 2016-10-7; Published 2016-11-9
Abstract
There is accumulating evidence indicating that long non-coding RNA H19 and its mature product miR-675 play essential roles for tumor growth and progression. However, their prognostic value in human head and neck squamous cell carcinoma (HNSCC), particular in laryngeal carcinoma, remains to be elucidated. In this study, we observed that both H19 and miR-675 were significantly overexpressed in a cohort of 65 primary tumor samples and two HNSCC cell lines. Importantly, when paired with patient follow-up data, higher expression of either H19 or miR-675 was significantly correlated with higher risk of patient relapse, and associated with worse overall survival and poor disease-free survival. Knockdown miR-675 caused significant reduction of cell viability, migratory and invasive capabilities. Taken together, these results suggest that the strong correlation of H19 overexpression together with higher miR-675 and lymph node metastases could be useful predictive markers, indicating a potentially therapeutic strategy for HNSCC patients.
Keywords: head and neck squamous cell carcinoma, H19, miR-675, prognostic predictor, proliferation.
1. Introduction
As genome-wide expression analysis has been continue to improve, led to increase insights into the molecular events underlying human biology and disease. The recent Encyclopedia of DNA Elements (ENCODE) project has identified and characterized annotated and novel RNAs transcripts that are enriched in either of nucleus and cytosol by RNA-seq [1]. Their results reveal that more than 70% of the human genome to be covered by either processed or primary transcripts, including thousands of non-coding RNAs [1-3].
Long non-coding RNAs (lncRNAs) are a novel class of non-protein coding molecules with more than 200 bases in length. Although lncRNAs share many features of miRNAs, increasing evidence suggests that lncRNAs have essential roles in diverse physiological cellular processes, such as decoys, scaffolds for interacting proteins, chromatin remodeling, post-transcriptional modifications [4]. lncRNAs deregulated expression has recently been implicated in the pathogenesis of many types of cancer. MALAT1 has been reported to be up-regulated in lung [5] and colorectal cancer [6]. Overexpression of MALAT1 increases cell proliferation, induces cell migration and correlates with tumor metastasis [6]. Other most well established lncRNA, HOTAIR has been observed overexpress in different type of cancers and is positively associated with patients' survival in hepatocellular carcinoma [7] and colorectal cancers [8]. H19 was first reported up-regulated in bladder carcinoma and has been recognized as a predict marker for early recurrence [9]. Subsequently, in a cohort of 80 archival paraffin embedded hepatic metastases samples, H19 RNA was found overexpression in 64 of 80 (80%) patients [10]. Furthermore, emerging evidence from recent studies has shown that H19 promotes human pancreatic tumors progression [11] and affects cell proliferation through c-Myc. Overexpression of H19 predicts poor prognosis in gastric cancer and non-small cell lung cancer patients [12, 13]. In contrast to miRNAs, lncRNAs also function as precursors or spongers for miRNAs. As a precursor of miR-675, the tumoriogenesis role of H19 may go through miR-675[14]. In human colon cancer cell lines and primary human colorectal cancer, both H19 and miR-675 have been found to up-regulate and affect colorectal cancer development through suppressing RB [15].
Head and neck squamous cell carcinoma (HNSCC) is the sixth most common malignant tumor worldwide [16], and laryngeal carcinoma accounts for 30% to 40% of all malignant head and neck tumors. Despite recent advances in combination with chemotherapy for more advanced diseases, the 5-year overall survival remains poor. As far as several predisposing factors are concerned, including predominantly smoking and alcohol consumption and other possible risk factors such as HPV and EBV infections, work environment and nutrition, the mechanism of carcinogenesis in the HNSCC remains elusive, underlining the importance to better understand the molecular basis and developing novel therapeutic strategies to target this disease. The oncogenic role of H19 in different cancers has been investigated. However, there is no evidence whether H19 and miR-675 are involved in human HNSCC. We therefore evaluate the expression of H19 and miR-675 and its correlation with clinicopathologic features in HNSCC patients.
2. Results
2.1. H19 and miR-675 were significantly up-regulated in HNSCC patients and associated with tumor recurrence
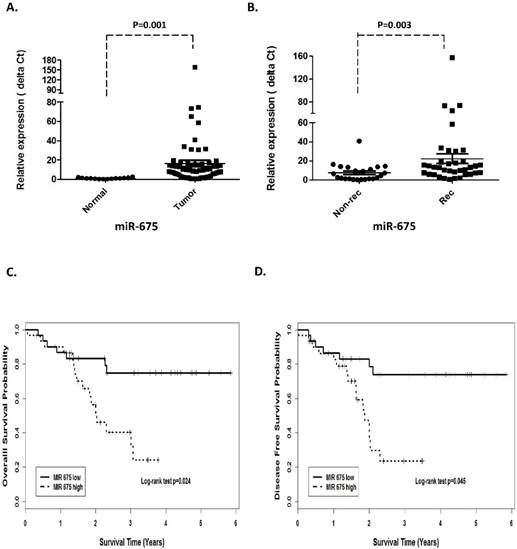
Quantitative real-time PCR was performed on 62 HNSCC (46 larynx, 14 hypopharynx and 2 oropharynx) and 19 adjacent normal tissues to assess H19 and miR-675 expression. The expression of H19 was significantly higher in the tumor cohorts compared with the adjacent normal tissues (p =0.0063, Fig 1A). Furthermore, when comparing H19 expression in recurrence with non-recurrence patient's group, H19 was significantly higher in the patients that eventually relapsed (p=0.052; Fig 1B).
Prognostic significance of H19 in patients with head neck squamous cell carcinoma. (A). Relative expression of H19 in HNSCC compared with normal tissues. (B). non-recurrence compared with recurrence samples. Kaplan-Meier analysis was performed for overall survival and disease free survival. Patients with high expression of H19 exhibited significantly worse overall survival (C) and shortened disease-free survival (D) than that with low expression of H19 as defined by log-rank test.
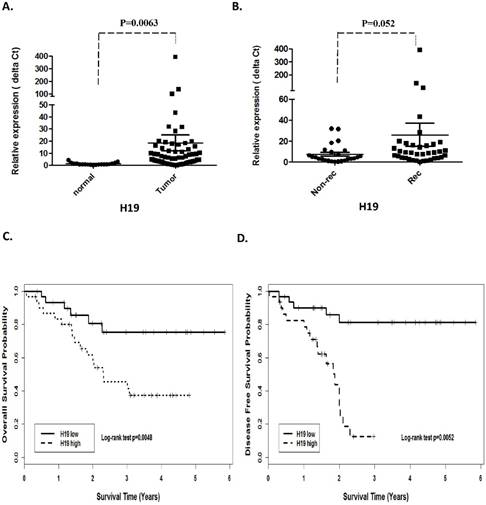
Prognostic values of miR-675 in patients with head neck squamous cell carcinoma. (A). Relative expression of miR-675 in HNSCC compared with normal tissues. (B). non-recurrence compared with recurrence samples. Kaplan-Meier analysis was performed for overall survival and disease free survival. Patients with high expression of miR-675 exhibited significantly worse overall survival (C) and shortened disease-free survival (D) than that with low expression of miR-675 as defined by log-rank test.
H19 is a precursor of miR-675 and its function could be at least through derived miR-675. Unsurprisingly, qRT-PCR revealed that miR-675 was positively concordant with H19 expression and significantly overexpressed in the tumors compared with the adjacent normal tissues (p=0.001, Fig 2A).
2.2. High expression of H19 and miR-675 were correlated with poor prognosis in HNSCC patients
In order to investigate the association between H19 expression and patient survival, the patients were dichotomized into H19 high (n=37) and low (n=25) groups by using the median expression value of H19. The Kaplan-Meier survival curves showed that patients with higher H19 expression had worse overall survival (OS) and disease-free survival (DFS) compared with those with lower H19 expression by log-rank test (p=0.0048 and p=0.0052, respectively. Fig 1 C and D). Similarly, miR-675 expression level was observed to be correlated significant for overall survival (OS) and disease-free survival (DFS). Higher expression of miR-675 was associated with patient recurrences (p =0.003, Fig 2B) and had worse overall survival (p=0.024) and DFS (p=0.045) than those with low miR-675 expression group (Fig 2C and 2D).
When comparing H19 and miR-675 expression with gender, age, tumor stage, primary tumor location and lymph node metastases, univariate analysis showed that overexpression of miR-675 significantly correlated with patients have higher expression of H19 and lymph node metastases, but not with sex, age and primary tumor location (Table 2 (A)) for both OS and DFS. By using Cox proportional hazard regression model, multivariate analysis revealed that H19 overexpression together with higher miR-675 and lymph node metastases were independent prognostic factors for poor disease-free survival (Table 2 (B); H19; p=0.017; Hazard ratio: 4.11; 95% CI between 1.27 to 13.21). Likewise, H19 was also significantly correlated with overall survival when controlled for the clinical prognostic factors (Table 2 (B); p= 0.0025). The hazard ratio for the effect of H19 on survival when controlled for clinical factors was 14.29, 95% CI was between 3.43 and 59.41.
Analysis of prognostic parameters.
| Variable | OS | DFS | ||
|---|---|---|---|---|
| (p value) | (p value) | |||
| (A). Univariate analysis | ||||
| Age at surgery (>64 v.s. <=64) | 0.78 | 0.79 | ||
| Gender (Male/Female) | 0.94 | 0.76 | ||
| Primary location (Larynx/others) | 0.59 | 0.37 | ||
| Tumor stage (T1-2/T3-4) | 0.057 | 0.059 | ||
| Relapse v.s. Non-relapse | 0.0012 | 0.0055 | ||
| High H19 (Y/N) | 0.0017 | 0.021 | ||
| High miR-675 (Y/N) | 0.021 | 0.0067 | ||
| (B). Multivariate analysis | OS (HR, 95%CI) | p value | DFS (HR, 95%CI) | p value |
| Tstage | 1.086(1.018-1.15) | 0.012 | 1.098(1.026-1.17) | 0.0064 |
| Lymph node metastases | 0.049(0.0060-0.41) | 0.0053 | 0.029(0.0033-0.25) | 0.0014 |
| High H19 | 14.29(3.43-59.41) | 0.0025 | 4.11(1.27- 13.21) | 0.017 |
| High miR-675 | 2.52(1.75-8.45) | 0.013 | 3.26(0.94-10.71) | 0.051 |
OS-overall survival; DFS-disease-free survival; HR-Hazard ratio; CI-confidence interval.
2.3. miR-675 promoted cell proliferation in HNSCC cells
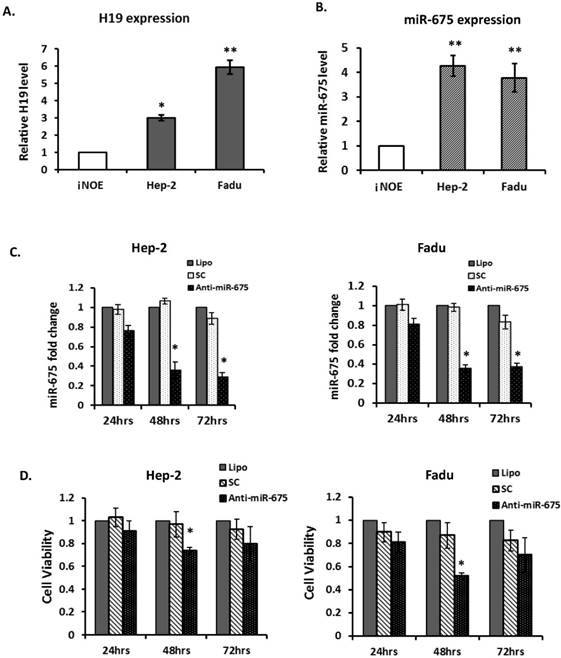
Given the evidence that overexpression of H19 and miR-675 were associated with poor clinical outcomes in HNSCC, particularly in laryngeal carcinoma, we further assessed the biological significance of depletion of miR-675 in 2 HNSCC cell lines; Hep-2, a laryngeal carcinoma line and Fadu, a hypopharyngeal carcinoma line. H19 was significantly overexpressed in two examined cancer lines, when compared with immortalized normal human oral epithelial cells (iNOE; Fig 3A). As expected, both cancer cell lines demonstrated higher level of miR-675 expression compared with that of normal (Fig 3B).
To evaluate the effects of miR-675 depletion, cells were transfected with either scramble controls (SC) or anti-miR-675. Expression of miR-675 was significantly reduced by 65% and 70% respectively, at 48 and 72 hours post-transfection (Fig 3C left) in Hep-2 cells. Similarly, in Fadu cells, miR-675 was significantly suppressed by 65% at 48 hours post-transfection and continuously declined at 72 hours (Fig 3C right). Furthermore, transfection with anti-miR-675 led to significantly decreased cell viability in both Hep-2 and Fadu cells, by 30% and 55% respectively, at 48 hours post-transfection compared with controls (Fig 3D). Taken together, these data indicate that overexpressions of H19 and miR-675 are common events in HNSCC, effective suppression of miR-675 warrant further investigation for potential therapeutic applications.
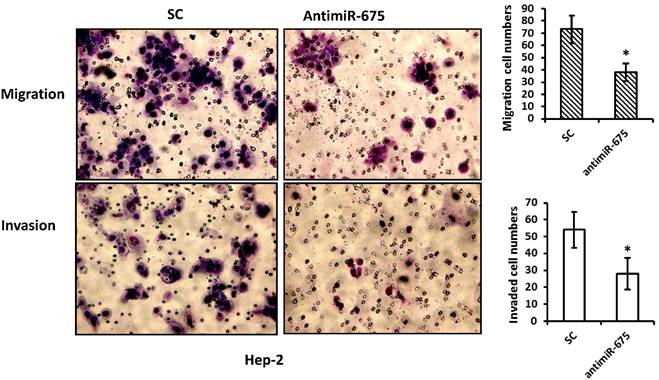
2.4. miR-675 enhanced cell migration and invasion
As miR-675 up-regulation was observed to be associated with recurrence in our patients, we performed migration and invasion assays on Hep-2 cells. Compared with their corresponding negative controls, transfection with antimiR-675 significantly reduced migrate capacities of Hep-2 cells by 40% and resulted in reduction of invasion of Hep-2 cells by 30%, as shown in Figure 4.
3. Discussion
It has become increasing evidence that lncRNAs play essential roles for human tumour growth and have been found to be key regulators of the complex protein signaling pathways underling carcinogenesis. In the present study, we found that clinically, high expression levels of H19 correlated with patient clinical outcome in our cohort of HNSCC patients. miR-675 expression was positively correlated with H19. Moreover, both high expression of H19 and miR-675 significantly correlated with poor overall survival and disease-free survival. Furthermore, H19 overexpression together with higher miR-675 and lymph node metastases were identified as independent prognostic factors for HNSCC patients. These finding further supported by the evidence of that depletion of miR-675 inhibiting laryngeal and hypopharyngeal cancer cells proliferation, migration and invasion through involvement in cellular processes.
H19 is an imprinted long non-coding RNA gene. Emerging evidence indicates that H19 is frequently overexpressed in the majority of human malignancies. Although it acts as an oncogene in breast [17], bladder [18], glioma [19] and prostate [20] cancers, some studies also show its controversial role as a tumor suppressor in different tumor types [21] [22]. In HNSCC, a study by Esteves LI [23] has shown a significant correlation between H19 expression and tumor recurrence. Consistence with their observation, H19 expression in our cohort demonstrated significant up-regulation in cancer than that in normal. Importantly, the patients with higher H19 expression had worse overall survival and disease-free survival compared with those with lower H19 expression. These results suggest that H19 contributes to oncogenesis of HNSCC.
Down-regulation of miR-675 reduced cell proliferation in squamous cell carcinoma cells. (A & B). Quantitative real-time PCR (qRT-PCR) for H19 and miR-675 expressions in head neck squamous cell carcinoma cells (Hep2 and Fadu) compared with normal epithelial cells (iNOE). (C). Equal amount of Anti-miR-675 (40 nM) or scrambled control (SC) was transfected into either Hep-2 cells or Fadu cells and significantly reduced miR-675 expression in both cells, compared with scrambled control (SC) or transfection reagent alone (Lipo). (D). Cell viability was assessed in both Hep2 and Fadu cells by the MTT assay at 24-72 hours post-transfection. Each datum represents the mean fold change ± SE in triplicates. *p<0.05. **p<0.01.
Depletion of miR-675 reduced cell migration and invasion. Top panel--- Representative images of migration assay depict migratory ability after transfection with antimiR-675 compared to scramble control. Bottom panel--- Representative images of invasive assay depict invasiveness after transfection with antimiR-675 compared to scramble control.
miR-675 is excised from first exon of H19. Both H19 and miR-675 are believed to be involved in tumoriogenesis. It was first described by Tsang WP [15], in which H19-derived miR-675 regulated primary human colorectal cancer development through down-regulation of tumor suppressor RB. Subsequently, a strong relationship between H19 and miR-675 was confirmed in various studies. H19/miR-675 targeted tumor suppressor Runt Domain Transcription Factor1 and regulated gastric cancer proliferation and development [24]. H19 derived miR-675 promoted glioma cell invasion [19] and enhanced tumorigenesis and metastasis through downregulating c-CbI/CbI-b in breast cancer cells [25].
The precise underlying mechanism of H19/miR-675 in tumorigenesis remains unclear. H19 involves in the complex biological process of oncogenesis [2, 17, 26]. Genomic imprinting is an inherited epigenetic phenomenon, loss of imprinting at the IGF2 and H19 loci play a role in the oncogenesis of head and neck carcinoma [27]. H19 expression in many cancers is usually associated with the function of methylation. H19 CBS6 methylation potentially represents a novel clinically relevant epigenetic marker for the recurrence and progression of esophageal squamous cell carcinoma [28]. There are clear evidences show that TGF-β can induce the expression of H19 along with EMT markers, such as Slug, which also contribute to tumor metastasis and dependent on H19[17]. Recent study has demonstrated that H19 functions as a miRNA sponges to promoting EMT in colorectal cancer through activating Wnt signaling pathway [29]. On one hand, H19 overexpression increases the cellular abundance of miR-675 [30] and regulates intestinal epithelial barrier function via miR-675 by interacting with RNA-binding protein HuR [31]. On the other hand, miR-675 up-regulates long noncoding RNA H19 through activating EGR1 in human liver cancer [32]. In our study, miR-675 expression was positively correlated with H19 over-expression in HNSCC. Overexpression of miR-675 together with higher H19 and lymph node metastases were independent prognostic factors for poor disease-free survival. Furthermore, our results were in line with their observation, in that knock- down miR-675 caused significant reduction of cell viability, migratory and invasive capabilities in carcinoma lines, indicating miR-675 involving in HNSCC tumor progression. Nonetheless, further studies in the function of H19/miR-675 axis in the molecular mechanism of HNSCC tumor progression in a large cohort of samples may validate the exploratory findings.
4. Material and Methods
4.1. Patient Characteristics
Sixty-five head neck squamous cell carcinoma formalin-fixed and paraffin-embedded samples were collected during the period of 2001-2011, with approval from the Second Hospital of Jilin University. Among of them, sixty-two tissue samples had sufficient tumor cell contains and have been involved in the study. The clinical, demographic and tumor-related characteristics of patients have been descripted in Table 1. The predominant primary sub-site was larynx (46 of 62, or 74.2%). The majority of patients were male (48 or 77.4 %) and median age was 63.8 years. More than half of patients (59.6%) had lymph node metastasis at the diagnosis and 71% patients had locally advanced disease with stage III and IV. The average follow-up time was 3.09 years. During the follow-up period, 25 patients were dead (40.3%) and all of the patients were treated neither with radiotherapy nor with chemotherapy prior to the surgery. Histologically confirmed, adjacent normal tissues from 19 samples were able to obtain either from separate block or from macro-dissected blocks of patients.
4.2. Sample preparation
All studied tumour (n=65) and adjacent normal (n=19) specimens were assessed on hematoxylin-and-eosin-stained (H&E) slides using standard diagnostic criteria. 62 tumor specimens were confirmed with more than 60% of malignant cells and were selected for the analysis. Based on the H&E review, the samples were macro-dissected. This study was approval from the Second Hospital of Jilin University and all patients have consented for the study.
4.3. Cell lines and transfection
The human laryngeal cancer cell line, Hep-2, was purchased from the China Center for Type Culture Collection and maintained at 37°C, 5% CO2 in RPMI 1640 complete medium (Gibco, USA) with 10% fetal bovine serum (FBS, Gibco, USA). The human hypopharyngeal squamous carcinoma cell line, Fadu, was purchased from American Type Culture Collection and were maintained in Minimum Essential Medium, supplemented with 10% Fetal Bovine Serum (FBS), 1.5 g/L bicarbonate, and 1 mM pyruvate. The immortalized normal human oral epithelial cells (iNOE) were purchased commercially and cultured in the recommended medium. All cells were maintained in a 37°C incubator with humidified 5% CO2. To knockdown endogenous miR-675 expression, Hep-2 and Fadu cells were transfected using the LipofectAMINE 2000 (Invitrogen) reverse transfection protocol, according to the manufacturer's instructions. AntagomiR-675 and anti-miR scramble negative control 1 (SC; Ambion) were transfected at a final concentration of 40 nmol/L to a 96-well plate.
Clinical Characteristics of Patients.
| Parameter | N=62 | (%) | H19 expression* | p value | mir-675 expression* | p value |
|---|---|---|---|---|---|---|
| Gender | ||||||
| Male | 48 | 77 | 20.11 | 0.36 | 18.88 | 0.36 |
| Female | 14 | 23 | 13.08 | 8.15 | ||
| Age (years) | ||||||
| >64 | 33 | 53 | 25.01 | 0.27 | 20.79 | 0.3 |
| <=64 | 29 | 47 | 11.15 | 11.51 | ||
| Primary location | ||||||
| Larynx | 46 | 74 | 22.04 | 0.77 | 17.66 | 0.92 |
| Hypo-&others | 16 | 26 | 8.41 | 12.98 | ||
| Tumor stage | ||||||
| T1-2 | 18 | 29 | 15.22 | 0.16 | 15.59 | 0.77 |
| T3-4 | 44 | 71 | 19.87 | 16.81 | ||
| Relapse | ||||||
| No | 25 | 40 | 7.48 | 0.038 | 7.61 | 0.017 |
| Yes | 37 | 60 | 25.98 | 22.44 |
* represent the median of H19 or miR-675 expression.
4.4. RNA and miRNAs extraction
Total RNA containing miRNAs were extracted from either primary tumor tissues or cell lines using RecoverAll Total Nucleic Acid Isolation kit (Ambion) according to the manufacturer's instructions. Briefly, the macrodissected FFPE samples were de-paraffinized with xylene, washed with 100% ethanol. The pellet then was digested by proteinase K. After incubation the pellet in different heating blocks, total RNA and miRNAs were isolated through a filter cartridge and treated with DNase. The samples were then stored in -80ºC until use.
4.5. Real-Time Quantification of H19 and microRNA-675 expression
For H19 mRNA expression, the reverse transcription was performed using SuperScript III Reverse Transcriptase (Invitrogen Corp.) according to the manufacturer's recommendations. qRT-PCR analyses were performed using SYBR Green Master Mix (Applied Biosystems) and the ABI PRISM 7900 Sequence Detection System (Applied Biosystems Inc., Foster City, CA, USA). As described previously [15], the primers used for H19 were: forward: 5′-TACAACCACTGCACTACCTG-3′; reverse: 5′-TGGAATGCTTGAAGGCTGCT-3′. Compared with control specimens, the relative fold change in mRNA expression was calculated using the 2-ΔΔCt method and was normalized to that of an endogenous gene (GAPDH), which was amplified in parallel as the internal control. For miR-675 expression, the RNA was reverse-transcribed with a MultiScribe reverse transcriptase (Applied Biosystems) by using a stem-loop RT primer specifically hybridises with a miR molecule. The RT products were subsequently amplified with sequence-specific primers using Taqman MicroRNA Assay kit (Applied Biosystems) by the Applied Biosystems 7900 HT Real-Time PCR system. RNU48 was amplified in parallel and used as an endogenous control.
4.6. MTT cell proliferation assay
The cell proliferation and cytotoxicity of introducing anti-miR-675 were assessed by using a 3-[4,5-dimethylthiazol-2-yl]-2,5-diphenyltetrazolium bromide (MTT) staining kit (Sigma, USA). Cells were reverse transfected with either SC control, anti-miR-675 or Lipofectamine 2000 and seeded onto 96-well plates (5 x 103 cells/per well). Cell viability was measured at 24, 48 and 72 hours' post-transfection by a microplate reader.
4.7. Cell migration and invasion assays
Cell migration and invasion were assessed by using BD BioCoat Matrigel Invasion Chambers and Control Inserts (BD Biosciences). The cells were transfected with either anti-miR-675 or SC (40nM), then seeded on either control inserts (polyethylene terephthalate membrane) or trans-well chambers with Matrigel. Two ml RPMI supplemented with 15% FBS was added to the lower chamber, served as the chemo-attractant. 0.5x104 transfected Fadu cells were re-suspended in RPMI plus 1% FBS, added to the upper chamber (0.5 mls). Twenty hours later, migrating or invading cells attached to the lower surface of the membrane insert were fixed and stained, then counted under a microscope. Relative migration was calculated by comparison with cells transfected with the negative control. The percentage invasion was calculated based on the number of cells which have invaded through the Matrigel insert, divided by the number of cells which have migrated through the control insert.
4.8. Statistical Analysis
The differences expression of H19 and miR-675 between each group were evaluated by unpaired, two-tailed Student's t-test. Overall survival (OS) was defined from the time of diagnosis to date of death. Disease-free survival (DFS) was defined from the time of diagnosis to the date of first failure. Survival curves were generated using the Kaplan-Meier method and were compared by means of the log-rank test. In the univariate model, if factors with prognostic significance, they were then further analysed in a multivariate Cox's proportional hazards regression model. A p-value of less than 0.05 was considered to be statistically significant.
5. Conclusions
In summary, our results indicate that H19 and miR-675 are significantly overexpressed in HNSCC cancer cells and patients. Higher expression of H19 and miR-675 are correlated with poor prognosis. The strong correlation of H19 overexpression together with higher miR-675 and lymph node metastases could be useful predictive markers, indicating a potentially therapeutic strategy for HNSCC patients.
Acknowledgements
The authors acknowledge the financial support of the Science and Technology R&D program of Jilin Province, China (20150101140JC).
Conflict of interest
The authors report no conflict of interest in this work.
References
1. Djebali S, Davis CA, Merkel A, Dobin A, Lassmann T, Mortazavi A. et al. Landscape of transcription in human cells. Nature. 2012;489:101-8
2. Mattick JS. Long noncoding RNAs in cell and developmental biology. Seminars in cell & developmental biology. 2011;22:327
3. Mele M, Ferreira PG, Reverter F, DeLuca DS, Monlong J, Sammeth M. et al. Human genomics. The human transcriptome across tissues and individuals. Science. 2015;348:660-5
4. Cheetham SW, Gruhl F, Mattick JS, Dinger ME. Long noncoding RNAs and the genetics of cancer. British journal of cancer. 2013;108:2419-25
5. Schmidt LH, Spieker T, Koschmieder S, Schaffers S, Humberg J, Jungen D. et al. The long noncoding MALAT-1 RNA indicates a poor prognosis in non-small cell lung cancer and induces migration and tumor growth. Journal of thoracic oncology: official publication of the International Association for the Study of Lung Cancer. 2011;6:1984-92
6. Xu C, Yang M, Tian J, Wang X, Li Z. MALAT-1: a long non-coding RNA and its important 3' end functional motif in colorectal cancer metastasis. International journal of oncology. 2011;39:169-75
7. Yang Z, Zhou L, Wu LM, Lai MC, Xie HY, Zhang F. et al. Overexpression of long non-coding RNA HOTAIR predicts tumor recurrence in hepatocellular carcinoma patients following liver transplantation. Annals of surgical oncology. 2011;18:1243-50
8. Kogo R, Shimamura T, Mimori K, Kawahara K, Imoto S, Sudo T. et al. Long noncoding RNA HOTAIR regulates polycomb-dependent chromatin modification and is associated with poor prognosis in colorectal cancers. Cancer research. 2011;71:6320-6
9. Ariel I, de Groot N, Hochberg A. Imprinted H19 gene expression in embryogenesis and human cancer: the oncofetal connection. American journal of medical genetics. 2000;91:46-50
10. Fellig Y, Ariel I, Ohana P, Schachter P, Sinelnikov I, Birman T. et al. H19 expression in hepatic metastases from a range of human carcinomas. Journal of clinical pathology. 2005;58:1064-8
11. Sorin V, Ohana P, Gallula J, Birman T, Matouk I, Hubert A. et al. H19-promoter-targeted therapy combined with gemcitabine in the treatment of pancreatic cancer. ISRN oncology. 2012;2012:351750
12. Zhang EB, Han L, Yin DD, Kong R, De W, Chen J. c-Myc-induced, long, noncoding H19 affects cell proliferation and predicts a poor prognosis in patients with gastric cancer. Medical oncology. 2014;31:914
13. Zhang E, Li W, Yin D, De W, Zhu L, Sun S. et al. c-Myc-regulated long non-coding RNA H19 indicates a poor prognosis and affects cell proliferation in non-small-cell lung cancer. Tumour biology: the journal of the International Society for Oncodevelopmental Biology and Medicine. 2016;37:4007-15
14. Matouk IJ, Halle D, Gilon M, Hochberg A. The non-coding RNAs of the H19-IGF2 imprinted loci: a focus on biological roles and therapeutic potential in Lung Cancer. Journal of translational medicine. 2015;13:113
15. Tsang WP, Ng EK, Ng SS, Jin H, Yu J, Sung JJ. et al. Oncofetal H19-derived miR-675 regulates tumor suppressor RB in human colorectal cancer. Carcinogenesis. 2010;31:350-8
16. Jemal A, Bray F, Center MM, Ferlay J, Ward E, Forman D. Global cancer statistics. CA: a cancer journal for clinicians. 2011;61:69-90
17. Matouk IJ, Raveh E, Abu-lail R, Mezan S, Gilon M, Gershtain E. et al. Oncofetal H19 RNA promotes tumor metastasis. Biochimica et biophysica acta. 2014;1843:1414-26
18. Byun HM, Wong HL, Birnstein EA, Wolff EM, Liang G, Yang AS. Examination of IGF2 and H19 loss of imprinting in bladder cancer. Cancer research. 2007;67:10753-8
19. Shi Y, Wang Y, Luan W, Wang P, Tao T, Zhang J. et al. Long non-coding RNA H19 promotes glioma cell invasion by deriving miR-675. PloS one. 2014;9:e86295
20. Ribarska T, Goering W, Droop J, Bastian KM, Ingenwerth M, Schulz WA. Deregulation of an imprinted gene network in prostate cancer. Epigenetics. 2014;9:704-17
21. Cui H, Hedborg F, He L, Nordenskjold A, Sandstedt B, Pfeifer-Ohlsson S. et al. Inactivation of H19, an imprinted and putative tumor repressor gene, is a preneoplastic event during Wilms' tumorigenesis. Cancer research. 1997;57:4469-73
22. Fukuzawa R, Umezawa A, Ochi K, Urano F, Ikeda H, Hata J. High frequency of inactivation of the imprinted H19 gene in "sporadic" hepatoblastoma. International journal of cancer. 1999;82:490-7
23. Esteves LI, Javaroni AC, Nishimoto IN, Magrin J, Squire JA, Kowalski LP. et al. DNA methylation in the CTCF-binding site I and the expression pattern of the H19 gene: does positive expression predict poor prognosis in early stage head and neck carcinomas? Molecular carcinogenesis. 2005;44:102-10
24. Zhuang M, Gao W, Xu J, Wang P, Shu Y. The long non-coding RNA H19-derived miR-675 modulates human gastric cancer cell proliferation by targeting tumor suppressor RUNX1. Biochemical and biophysical research communications. 2014;448:315-22
25. Vennin C, Spruyt N, Dahmani F, Julien S, Bertucci F, Finetti P. et al. H19 non coding RNA-derived miR-675 enhances tumorigenesis and metastasis of breast cancer cells by downregulating c-Cbl and Cbl-b. Oncotarget. 2015;6:29209-23
26. Nakao M, Sasaki H. Genomic imprinting: significance in development and diseases and the molecular mechanisms. Journal of biochemistry. 1996;120:467-73
27. Rainho CA, Kowalski LP, Rogatto SR. Loss of imprinting and loss of heterozygosity on 11p15.5 in head and neck squamous cell carcinomas. Head & neck. 2001;23:851-9
28. Gao T, He B, Pan Y, Gu L, Chen L, Nie Z. et al. H19 DMR methylation correlates to the progression of esophageal squamous cell carcinoma through IGF2 imprinting pathway. Clinical & translational oncology: official publication of the Federation of Spanish Oncology Societies and of the National Cancer Institute of Mexico. 2014;16:410-7
29. Liang WC, Fu WM, Wong CW, Wang Y, Wang WM, Hu GX. et al. The lncRNA H19 promotes epithelial to mesenchymal transition by functioning as miRNA sponges in colorectal cancer. Oncotarget. 2015;6:22513-25
30. Keniry A, Oxley D, Monnier P, Kyba M, Dandolo L, Smits G. et al. The H19 lincRNA is a developmental reservoir of miR-675 that suppresses growth and Igf1r. Nature cell biology. 2012;14:659-65
31. Zou T, Jaladanki SK, Liu L, Xiao L, Chung HK, Wang JY. et al. H19 Long Noncoding RNA Regulates Intestinal Epithelial Barrier Function via MicroRNA 675 by Interacting with RNA-Binding Protein HuR. Molecular and cellular biology. 2016;36:1332-41
32. Li H, Li J, Jia S, Wu M, An J, Zheng Q. et al. miR675 upregulates long noncoding RNA H19 through activating EGR1 in human liver cancer. Oncotarget. 2015;6:31958-84
Author contact
![]() Corresponding author: Ke Wang. Department of Respiratory Medicine, The Second Hospital of Jilin University, Changchun 130041, P. R. China. Tel: 13844097779. E-mail: kewang2016com.
Corresponding author: Ke Wang. Department of Respiratory Medicine, The Second Hospital of Jilin University, Changchun 130041, P. R. China. Tel: 13844097779. E-mail: kewang2016com.

 Global reach, higher impact
Global reach, higher impact